Rab14 binds in a GTP-dependent manner to RUFY1/Rabip4, which had been originally identified as a Rab4 effector. We suggest that Rab14 and Rab4 act sequentially; Rab14 is required for recruitment of RUFY1 onto endosomes and subsequent RUFY1 interaction with Rab4 may allow endosomal tethering and fusion.
Abstract
The small GTPase Rab14 localizes to early endosomes and the trans-Golgi network, but its cellular functions on endosomes and its functional relationship with other endosomal Rab proteins are poorly understood. Here, we report that Rab14 binds in a GTP-dependent manner to RUFY1/Rabip4, which had been originally identified as a Rab4 effector. Rab14 colocalizes well with Rab4 on peripheral endosomes. Depletion of Rab14, but not Rab4, causes dissociation of RUFY1 from endosomal membranes. Coexpression of RUFY1 with either Rab14 or Rab4 induces clustering and enlargement of endosomes, whereas a RUFY1 mutant lacking the Rab4-binding region does not induce a significant morphological change in the endosomal structures even when coexpressed with Rab14 or Rab4. These findings suggest that Rab14 and Rab4 act sequentially, together with RUFY1; Rab14 is required for recruitment of RUFY1 onto endosomal membranes, and subsequent RUFY1 interaction with Rab4 may allow endosomal tethering and fusion. Depletion of Rab14 or RUFY1, as well as Rab4, inhibits efficient recycling of endocytosed transferrin, suggesting that Rab14 and Rab4 regulate endosomal functions through cooperative interactions with their dual effector, RUFY1.
INTRODUCTION
Early endosomes are multifunctional organelles that regulate membrane transport between the plasma membrane and various intracellular compartments (Mukherjee et al., 1997). After arrival at early endosomes, endocytosed solutes, membrane proteins, and lipids are either returned to the plasma membrane, as in the case of recycling receptors and bulk membrane constituents, or are transported to late endosomes and lysosomes for degradation. Another transport route connects early endosomes to the trans-Golgi network (TGN). From early endosomes, recycling components can return directly to the plasma membrane (the fast recycling route) or can return through perinuclear recycling endosomes (the slow recycling route) (Hopkins, 1983; Yamashiro and Maxfield, 1984; Schmid et al., 1988; Mayor et al., 1993; Grant and Donaldson, 2009).
Rab proteins form the largest group within the Ras superfamily of small GTPases, which function as molecular switches by cycling between their inactive GDP–bound and active GTP–bound forms (Pereira-Leal and Seabra, 2001; Zerial and McBride, 2001; Pfeffer and Aivazian, 2004). Human cells express more than 60 different Rab GTPases that regulate intracellular trafficking and maintain organellar identity by controlling budding, transport, tethering, docking, and fusion of vesicular intermediates. Thus, Rab GTPases oversee the vectorial transport of proteins and membranes between organelles (Zerial and McBride, 2001).
Endosomal Rab proteins (such as Rabs 4, 5, 7, 9, and 11) are best characterized by their localization and functional interactions with effector proteins. Several Rab effectors have been identified, including Rabaptin-4 and -5, Rabenosyn, and Rabip4/RUFY1 for Rab4 (Vitale et al., 1998; Nagelkerken et al., 2000; Cormont et al., 2001; Yang et al., 2002); Rabaptin-5, Rabex, Rabenosyn, EEA1, phosphoinositide 3-kinases (PI3K), and phosphoinositide phosphatases for Rab5 (Horiuchi et al., 1997; Gournier et al., 1998; Christoforidis et al., 1999a,b; Nielsen et al., 2000; Shin et al., 2005); and FIP family members for Rab11 (Hales et al., 2001; Prekeris et al., 2001; Lindsay et al., 2002; Shiba et al., 2006). Rab5 plays important roles in clathrin-mediated endocytosis and in homotypic endosome–endosome fusion (Gorvel et al., 1991; Bucci et al., 1992). Rab4 and Rab11 regulate recycling of receptors from early endosomes to the cell surface via distinct pathways. Rab4 functions at the level of early sorting endosomes (van der Sluijs et al., 1992; Daro et al., 1996; Sheff et al., 1999; de Wit et al., 2001), and Rab11 is involved in the trafficking of cargo through perinuclear recycling endosomes (Ullrich et al., 1996; Ren et al., 1998). Rab7 and Rab9 are associated with trafficking to late endosomes and the TGN, respectively (Feng et al., 1995; Barbero et al., 2002).
Rab14 localizes to the Golgi complex and endosomes and participates in membrane trafficking between the Golgi and endosomes (Junutula et al., 2004; Proikas-Cezanne et al., 2006). Rab14 also plays a role in the fusion between early endosomes and phagosomes during phagocytosis, and in lysosomal fusion in Dictyostelium (Harris and Cardelli, 2002; Kyei et al., 2006). Despite accumulating data on Rab14 functions, the molecular mechanisms underlying cargo sorting and membrane recycling on endosomes regulated by Rab14, as well as its functional relationship with other endosomal Rab proteins, are largely unknown.
Here, we report that Rab14 engages in a GTP-dependent interaction with RUFY1 (the human counterpart of mouse Rabip4), which had been originally identified as a Rab4 effector. Furthermore, we show lines of evidence indicating that Rab14 regulates RUFY1 recruitment onto endosomal membranes, and Rab4 in turn allows endosomal fusion.
MATERIALS AND METHODS
Plasmids
The entire coding sequences of human Rab14, Rab7, Rab2, Rab4b, Rab8, Rab10, Rab11, and RUFY1 cDNAs were obtained by PCR amplification of human liver cDNA libraries (Invitrogen, Carlsbad, CA), and cloned into N-terminally hemagglutinin (HA)-tagged pcDNA3, pEGFP-C1 (Invitrogen), and N-terminally mCherry-tagged pcDNA3 vector (a fragment encoding mCherry amplified by polymerase chain reaction from pcDNA3.1-mCherry-α-tubulin construct which was kindly provided by Roger Tsien [University of California-San Diego]). Rab14(Q70L) and Rab14(S25N) mutants were generated by site-directed mutagenesis (QuickChange Site-Directed Mutagenesis, Stratagene, La Jolla, CA) and cloned into pGEX-6P-1 vector (GE Healthcare Bioscience, Piscataway, NJ), N-terminally HA tagged pcDNA3, and pEGFP-C1. Rab2(Q65L), Rab6(Q72L), Rab7(Q67L), Rab8(Q67L), Rab10(Q68L) and Rab11(Q70L) mutants were generated by site-directed mutagenesis (QuickChange Site-Directed Mutagenesis, Stratagene) and cloned into vector pGEX-6P-1 (GE Healthcare Bioscience). EGFP-Rab4 was generated by in-frame cloning of human Rab4 cDNAs (Shiba et al., 2002) into vector pEGFP-C1. Rab4(Q67L) (Shiba et al., 2002) was subcloned into vector pGEX-6P-1. A EGFP-Rab5 construct was kindly provided by Marino Zerial (Max Planck Institute, Dresden, Germany). Deletions of RUFY1 (residues 1-420, residues 1-506, or residues 507-600) were obtained by a PCR-based strategy. A deletion of 11 amino acids (residues 507-517) of RUFY1 was generated by PCR-based mutagenesis. These deletion mutants were cloned into vector pEGFP-C1. The construction of GST-Rab5(Q79L) and GST-Rab11(Q70L) was described previously (Shiba et al., 2002, 2006). For yeast two-hybrid analyses, RUFY1 was cloned into vector pGADT7 (Clontech, Mountain View, CA). Full-length Rab2(Q65L) and Rab mutants lacking the C-terminal three or four amino acids [Rab4(Q67L), Rab5(Q79L), Rab6(Q72L), Rab7(Q67L), Rab8(Q67L), Rab10(Q68L), Rab11(Q70L), Rab14(Q70L), Rab14(WT), and Rab14(S25N)] were cloned into vector pGBKT7 (Clontech).
Antibodies and Reagents
Polyclonal rabbit antibodies were raised against the human Rab14 C-terminus (residues 165-211). For the immunogen, GST-fused C-terminus of Rab14 recombinant proteins was expressed and purified from Escherichia coli BL21(DE3)-Codon Plus (Invitrogen), cleaved with PreScission (GE Healthcare), and conjugated with keyhole limpet hemocyanin (Pierce, Rockford, IL). Rabbit serum against Rab14 was affinity-purified using GST-fused Rab14 C-terminus protein immobilized on Sulfolink beads (Pierce). The mouse monoclonal antibodies used in this study are anti-EEA1 (clone 14), anti-Lamp-1 (H4B3), anti-Rab4 (clone 7), and anti-Rab5 (clone 1; BD Biosciences); anti-transferrin receptor (TfnR; H68.4; Zymed, South San Francisco, CA); anti-GFP (JL-8; Molecular Probes, Eugene, OR); anti-red fluorescent protein (RFP; 3G5; MBL International, San Diego, CA); and anti-LBPA (kind gift from Kobayashi Toshihide, RIKEN, Wako, Japan). The mouse polyclonal anti-RUFY1 antiserum, the rabbit polyclonal anti-red fluorescent protein (RFP), and the rat monoclonal anti-HA (3F10) antibody were purchased from Abnova (Taipei City, Taiwan), MBL International, and Roche Diagnostics (Alameda, CA), respectively. Secondary antibodies (AlexaFluor488-, 555-, or 647-conjugated anti-rabbit or anti-mouse) were purchased from Molecular Probes. Secondary antibodies (Cy3-conjugated anti-rat and peroxidase-conjugated anti-mouse or anti-rabbit) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Yeast Two-Hybrid Analysis
Yeast transformation, two-hybrid screening and assays were performed using the MATCHMAKER two-hybrid system (Clontech, Palo Alto, CA). The yeast AH109 cells were transformed using lithium acetate–based method with pGBKT7-Rab14(Q70L) lacking the C-terminal four amino acids and grown on synthetic medium lacking tryptophan. The transformant was then mated with Y187 cells transformed with a MATCHMAKER HeLa cDNA library (Clontech). The mated cells were plated on synthetic medium lacking tryptophan, leucine, and histidine and containing 5 mM 3-aminotriazole. After 5 d of incubation, library plasmids from colonies were rescued into E. coli DH5α. For interaction analysis, cotransformed AH109 with RUFY1 and several Rab proteins were plated on synthetic medium lacking tryptophan, leucine, histidine, and adenine.
Cell Culture, RNA Interference Suppression, Tfn Uptake, and Immunofluorescence Analysis
HeLa cells were cultured in minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Knockdown of Rab14, Rab4, or RUFY1 were performed as described previously (Ishizaki et al., 2006, 2008). Briefly, pools of small interfering RNAs (siRNAs) directed at the mRNA regions covering nucleotide residues 1-648 of Rab14, 1-657 of Rab4a, 1-658 of Rab4b, and 637-1518 of RUFY1 (when the A residue of the initiation Met codon is assigned as residue 1) were prepared using BLCK-iT RNA interference (RNAi) TOP Transcription kit and a BLOCK-iT Dicer RNAi kit (Invitrogen). siRNA oligonucleotide was synthesized by Dharmacon (Boulder, CO); CAACUACUCUUACAUCUUU for Rab14 (Kyei et al., 2006). Control siRNA was also purchased from Dharmacon. Cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen) and incubated for 24 h. The transfected cells were then transferred to a culture dish containing coverslips, further incubated for 48 h, and processed for immunofluorescence and immunoblot analyses as described previously (Shin et al., 1997; Ishizaki et al., 2008; Nishimoto-Morita et al., 2009). For cytosol extraction, cells were treated with 0.05% saponin in microtubule-stabilizing buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 5 mM EGTA) for 5 min at room temperature before fixation. For detection of endogenous RUFY1 using mouse polyclonal anti-RUFY1 antibody, cells were fixed and permeabilized with methanol for 5 min at −20°C. For detection of endogenous Rab14 using the rabbit polyclonal anti-Rab14 antibody that we raised for this study, cells were fixed with 3% paraformaldehyde and incubated with the antibody in 0.05% saponin for 1 h at room temperature. For detection of both endogenous RUFY1 and Rab14, cells were treated with 0.05% saponin in microtubule-stabilizing buffer before fixation with 3% paraformaldehyde and incubated with the antibodies in 0.05% saponin for 1 h at room temperature.
Tfn internalization and recycling assays were carried out using protocols modified from those reported by Shin et al. (2004). HeLa cells were transfected with a pool of siRNAs for LacZ as a control or treated with siRNAs for Rab14, RUFY1, and Rab4a and Rab4b. At 72 h after transfection, cells were serum-starved for 4 h in MEM medium containing 0.2% BSA and then incubated with AlexaFluor488-conjugated Tfn (Molecular Probes) at 37°C for 5 min. After cells were washed with acid (0.5% acetic acid, 0.5 M NaCl, pH 3.0) on ice, cells were incubated in medium containing unlabeled holo-Tfn for indicated times at 37°C and processed for immunofluorescence analysis. To analyze the amount of surface-bound Tfn, HeLa cells were prepared as described above and incubated with AlexaFluor488-conjugated Tfn at 4°C for 50 min. After cells washed with ice-cold phosphate-buffered saline (PBS), cells were incubated in medium without labeled Tfn for indicated times at 37°C and processed for immunofluorescence analysis. The fluorescence intensity of Tfn was quantified using the IP-Lab 4.0 software (Solution Systems, Funabashi, Japan). The total intensity and extracted bright segments number were normalized against total counted cells. Immunofluorescence analysis was performed using Axiovert 200 MAT microscope (Carl Zeiss, Thornwood, NY) for epifluorescence images and using an LSM Pascal microscope (Carl Zeiss) and FV1000 microscope (Olympus, Melville, NY) for confocal images. Deconvolution analysis was performed using an AxioVision deconvolution software (Carl Zeiss).
Pulldown Assay
The QL and SN mutants of Rab proteins fused to the C-terminus of glutathione S-transferase (GST) were expressed in E. coli BL21-Codon Plus(DE3) (Stratagene) cells by treatment of 0.1 mM IPTG for 3 h at 30°C and purified using glutathione-Sepharose4B beads (GE Healthcare). Nucleotide loading onto GST-fused Rab proteins was performed as previously described (Christoforidis et al., 1999). HeLa cells were transfected with an expression vector for N-terminally enhanced green fluorescent protein (EGFP)-tagged full-length or truncation mutant of RUFY1. The cells were then lysed in cell lysis buffer (20 mM HEPES, pH 7.4, 0.1 M NaCl, 5 mM MgCl2, 1% Triton X-100, 1 mM DTT) containing Complete, EDTA-free protease inhibitor mixture (Roche Diagnostics) and centrifuged at maximum speed in a microcentrifuge for 20 min at 4°C. The supernatant was precleared with glutathione-Sepharose 4B beads and then incubated with the GST-Rabs (50 μg) with prebound to glutathione-Sepharose4B beads in the presence of 100 μM GDP or GTPγS. Beads were washed six times with the lysis buffer containing 0.25% Triton X-100 instead of 1% Triton X-100 supplemented with 10 μM GDP or GTPγS. Bound materials were eluted by boiling in an SDS-PAGE sample buffer and resolved on a SDS-polyacrylamide gel and subjected to Western blot analysis.
Immunoprecipitation
HeLa cells were transfected with expression vectors for N-terminally EGFP-tagged full-length of RUFY1 and either N-terminally mCherry-tagged Rab14 (Q70L) or mCherry alone. The cells were then lysed in IP buffer (20 mM HEPES, pH 7.4, 0.1 M NaCl, 5 mM MgCl2, 1% NP-40, 10% glycerol) containing Complete, EDTA-free protease inhibitor mixture (Roche Diagnostics), and 200 μM GTPγS and centrifuged at maximum speed in a microcentrifuge for 20 min at 4°C. The supernatant was precleared with Dynabeads-protein G and then incubated with the mouse monoclonal anti-RFP antibody (3G5) for 4 h at 4°C. The lysate were then incubated with Dynabeads-protein G for additional 2 h at 4°C. Beads were washed eight times with IP buffer supplemented with 50 μM GTPγS. Bound materials were eluted by boiling in an SDS-PAGE sample buffer and resolved on a SDS-polyacrylamide gel and subjected to Western blot analysis using rabbit polyclonal anti-RFP and mouse monoclonal anti-GFP antibodies.
RESULTS
GTP-dependent Interaction of Rab14 and Rab4 with RUFY1/Rabip4
Rab14 localizes to the Golgi complex and endosomes (Junutula et al., 2004; Proikas-Cezanne et al., 2006; Kelly et al., 2009; also see supplementary results and Supplementary Figures S1 and S2), but its cellular functions are poorly understood. To better understand the molecular machinery involving Rab14, we performed a yeast two-hybrid screening to identify Rab14-binding proteins. Among the positive clones obtained by screening a HeLa cell library using a GTP-locked mutant of Rab14, Rab14(Q70L), as bait, we focused on RUFY1, the human counterpart of mouse Rabip4, which had been originally identified as a Rab4 effector (Cormont et al., 2001; Yang et al., 2002).
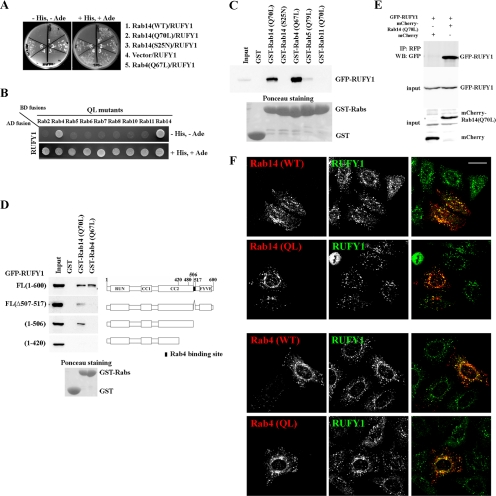
In the yeast two-hybrid assay, RUFY1 interacted with Rab14(Q70L) as well as with a GTP-locked mutant (Q67L) of Rab4, but not with wild type (WT) or a GDP-bound form (S25N) of Rab14 (Figure 1A). RUFY1 did not interact with the GTP-bound forms of other Rab proteins tested, including Rab2, Rab5, Rab6, Rab7, Rab8, Rab10, and Rab11 (Figure 1B), demonstrating that the interactions of RUFY1 with Rab14 and Rab4 are specific.
Figure 1.
Interaction of RUFY1 with Rab14 or Rab4. Yeast (AH109) transformants expressing the indicated combination of constructs were streaked (A) or spotted (B) onto plates lacking leucine and tryptophan, with or without histidine and adenine (+His, +Ade or −His, −Ade). (C and D) Pulldown assays for interaction of RUFY1 with Rab proteins. Lysates from HeLa cells transiently transfected with the EGFP-RUFY1 construct indicated were pulled down with the indicated GST-fusion protein. Bound proteins were subjected to SDS-PAGE and analyzed by immunoblotting using anti-GFP antibody. (E) HeLa cells expressing EGFP-tagged RUFY1 and either mCherry-tagged Rab14(Q70L) or mCherry were lysed and immunoprecipitated with anti-RFP antibody. Bound materials were subjected to SDS-PAGE and analyzed by immunoblotting using anti-RFP or anti-GFP antibody. (F) HeLa cells expressing HA-tagged Rab14(WT), Rab14(Q70L), Rab4(WT), or Rab4(Q67L) were fixed with methanol and doubly stained with anti-RUFY1 and anti-HA (3F10) antibodies. RUN domain (for RPIP8, UNC-14, and NESCA); cc, coiled-coil region; FYVE domain (for Fab1p, YOTB, Vac1p, and EEA1). Bar, 20 μm.
Next, we confirmed these interactions biochemically using a GST pulldown assay (Figure 1C). A GST-fused recombinant protein of each Rab mutant was immobilized on glutathione-Sepharose beads, charged with either GTPγS or GDP, and incubated with HeLa cell lysates expressing EGFP-tagged RUFY1. Consistent with the yeast two-hybrid data, RUFY1 was efficiently pulled down with Rab14(Q70L) and Rab4(Q67L), but not significantly with Rab14(S25N) or a GTP-bound form of Rab5 or Rab11. Moreover, EGFP-tagged RUFY1 was coimmunoprecipitated with mCherry-tagged Rab14(Q70L) but not with mCherry alone (Figure 1E), supporting the interaction between Rab14 and RUFY1 in vivo.
Rab14 and Rab4 Interact with Distinct Regions of RUFY1
We next asked whether the binding regions of RUFY1 for Rab14 and Rab4 are overlapping or separated. To this end, deletion mutants of RUFY1 were constructed (Figure 1D, right) and subjected to pulldown assays with GST-Rab14(Q70L) or -Rab4(Q67L) (left). Full-length RUFY1 was pulled down with both Rab14(Q70L) and Rab4(Q67L). In contrast, RUFY1(1-506) and RUFY1(Δ507-517), both of which lack a minimal Rab4-binding region determined in a previous study (Cormont et al., 2001; Figure 1D, black area in right side), were pulled down with Rab14(Q70L) but not Rab4(Q67L) (Figure 1D), indicating that the minimal Rab4-binding region is not necessary to interact with Rab14. When RUFY1 was further truncated from the C-terminus [RUFY1(1-420)], the truncation mutant could not bind to either Rab14 or Rab4, indicating that RUFY1 interaction with Rab14 requires at least the region encompassing residues 421-506. Therefore, we conclude that the regions of RUFY1 essential for binding to Rab14 and Rab4 are separated.
Colocalization of RUFY1 with Rab14 on Endosomes
We next compared the intracellular localization of Rab14 and RUFY1. HeLa cells transfected with an expression plasmid encoding HA-tagged Rab14(WT), Rab14(Q70L), Rab4(WT), or Rab4(Q67L) were immunostained for HA and endogenous RUFY1 and examined by confocal microscopy. We observed significant colocalization of endogenous RUFY1 with HA-Rab14(WT), HA-Rab14(Q70L), HA-Rab4(WT), and HA-Rab4(Q67L) on punctate endosomal structures (Figure 1E). Thus, RUFY1 interacts in a GTP-dependent manner with Rab14 as well as Rab4, and colocalizes with Rab14 and Rab4 on endosomes.
Localization of Rab4 and Rab14 Are Largely Overlapped
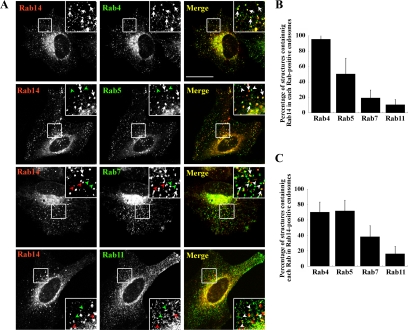
We next compared the intracellular localization of Rab14 with that of other endosomal Rab proteins, including early endosomal Rab5 and Rab4, late endosomal Rab7, and recycling endosomal Rab11. To this end, we coexpressed HA-tagged Rab14 and EGFP-tagged endosomal Rabs and quantified the overlap between Rab14 and each Rab protein (Figure 2). Rab14 was colocalized well with Rab4 on the peripheral endosomes and partially colocalized with Rab5 (Figure 2A, arrows); however, it was poorly colocalized with Rab7 and Rab11 in the cell periphery (Figure 2A, arrowheads).
Figure 2.
Overlap of endosomes positive for Rab14 and Rab4. (A) HeLa cells were transiently cotransfected with HA-Rab14 and either EGFP-Rab4, EGFP-Rab5, EGFP-Rab7, or EGFP-Rab11, and processed for immunofluorescence microscopy. Arrows indicate endosomes with overlapping staining for Rab14 and another Rab protein. Green arrowheads, Rab5-, Rab7-, or Rab11-positive endosomes; and red arrowheads, Rab14-positive endosomes. To quantify colocalization, peripheral Rab14-positive endosomes (B) or peripheral endosomes positive for the Rab protein indicated (C) were used as reference. The quantification is based on two independent experiments, using 200–400 endosomes in 6–10 transfected cells. Bar, 20 μm.
We express this colocalization in two ways: as a percentage of each type of Rab-positive endosome also positive for Rab14 (Figure 2B) or as the percentage of Rab14-positive endosomes also positive for each of the other Rab proteins (Figure 2C). More than 90% of Rab4-positive endosomes and ∼50% of Rab5-positive endosomes were positive for Rab14 (Figure 2B). In considerable contrast, only ∼20 and ∼10% of endosomes harboring Rab7 and Rab11, respectively, were positive for Rab14. Reciprocally, ∼70% of Rab14-positive endosomes were positive for Rab5 or Rab4 (Figure 2C). These observations suggest that most Rab4-positive endosomes contain Rab14 and that some population of Rab14-positive endosomes lacks Rab4. Although we cannot exclude the possibility for the overestimation of colocalization among Rabs because of their exogenous expression, we conclude that Rab14 colocalizes primarily with Rab4, but shows rather broader distribution on early endosomes.
RUFY1 Association with Endosomal Membranes Requires Rab14
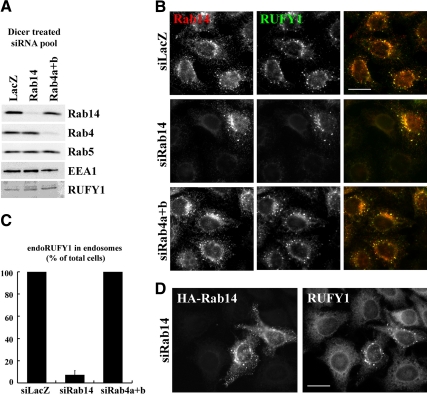
Membrane recruitment of RUFY1 does not require binding to Rab4, even though it localizes to Rab4-positive early endosomes (Cormont et al., 2001). We were therefore interested in investigating whether Rab14 is required for RUFY1 recruitment onto endosomes. For this purpose, we knocked down Rab14 or Rab4 expression in HeLa cells by RNAi, using pools of siRNAs (see Materials and Methods). Specific and efficient knockdown of Rab14 and Rab4 was confirmed by immunoblotting (Figure 3A) and immunofluorescence analyses (Figure 3B); we could not examine depletion of Rab4 by immunofluorescence analysis, because available antibodies did not work well in immunofluorescence. Endogenous RUFY1 was colocalized on endosomes with endogenous Rab14 (Figure 3B) as well as plasmid-expressed Rab14 (Figure 1F).
Figure 3.
Depletion of Rab14, but not Rab4, abolishes endosomal localization of RUFY1. HeLa cells were treated with a pool of siRNAs for LacZ, Rab14 or both Rab4a and Rab4b. After 72 h, the cells were lysed to be processed for immunoblot analysis using antibodies to indicated proteins (A) or fixed with 3% PFA, permeabilized, and immunostained for Rab14 (B). The siRNA-treated cells were permeabilized with saponin before fixation with PFA and immunostained for Rab14 and RUFY1 using rabbit polyclonal anti-Rab14 and mouse polyclonal anti-RUFY1 antibodies. (C) The number of cells in which RUFY1 localized to endosomes was counted and normalized with total counted cell numbers. The graph is a representative of four independent experiments. (D) HeLa cells treated with siRNAs for Rab14 were transfected with a plasmid encoding HA-Rab14, fixed, and immunostained with anti-RUFY1 and anti-HA antibodies. Bars, 20 μm.
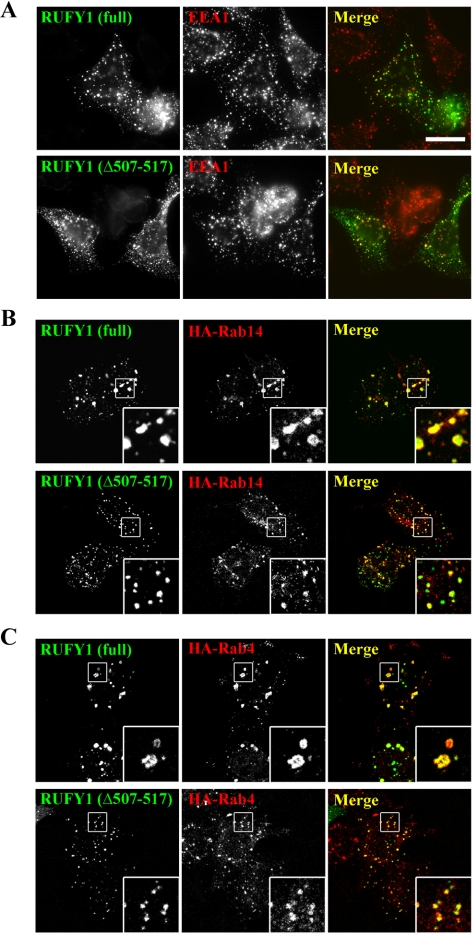
As shown in Figure 3, B and C, knockdown of Rab14 by the siRNA pool abolished RUFY1 localization on endosomes, even though the knockdown did not alter the RUFY1 protein level (Figure 3A). The endosomal localization of RUFY1 was rescued by overexpression of HA-tagged Rab14 (Figure 3D). Furthermore, essentially the same RUFY1 redistribution was observed in cells treated with a double-stranded oligonucleotide siRNA for Rab14 (Supplementary Figure S3, A and B), instead of the siRNA pool. These observations confirm that the RUFY1 redistribution resulted from specific depletion of Rab14 and not from an off-target effect. In contrast, knockdown of Rab4 did not significantly affect the endosomal localization of RUFY1; this is in line with a previous report showing that a RUFY1 construct lacking the Rab4-binding region, RUFY1(Δ507-517), retained its ability to associate with endosomes (Cormont et al., 2001; also see Figure 7A).
Figure 7.
Coexpression of RUFY1(Δ507-517) and either Rab14 or Rab4 does not induce expansion of early endosomes. (A) HeLa cells transfected with a plasmid encoding EGFP-RUFY1 or EGFP-RUFY1(Δ507-517) were stained with anti-EEA1 antibody. (B and C) HeLa cells were cotransfected with a combination of plasmids encoding either EGFP-RUFY1 or EGFP-RUFY1(Δ507-517) and either HA-Rab14 (B) or HA-Rab4 (C), and stained with anti-HA antibody. Bar, 20 μm.
Despite considerable effects on the RUFY1 localization, Rab14 knockdown did not significantly affect the localization of EEA1 (an early endosomal marker), TfnR (an early/recycling endosomal membrane protein), Lamp-1 (a transmembrane protein of late endosomes/lysosomes), or golgin-245 (a TGN marker; Supplementary Figure S3C), indicating that the endosomal and TGN structures themselves are intact under the Rab14-depleted conditions. Therefore, it is likely that RUFY1 was dissociated from endosomal membranes when Rab14 was depleted.
Coexpression of Either Rab14 or Rab4 and RUFY1 Induces Enlargement of Endosomes
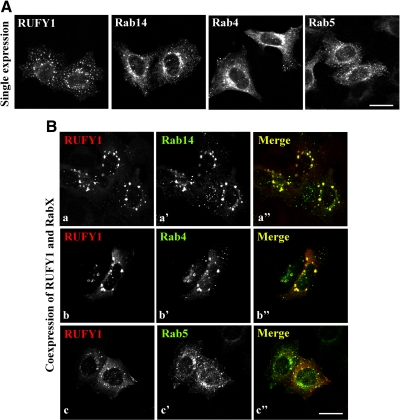
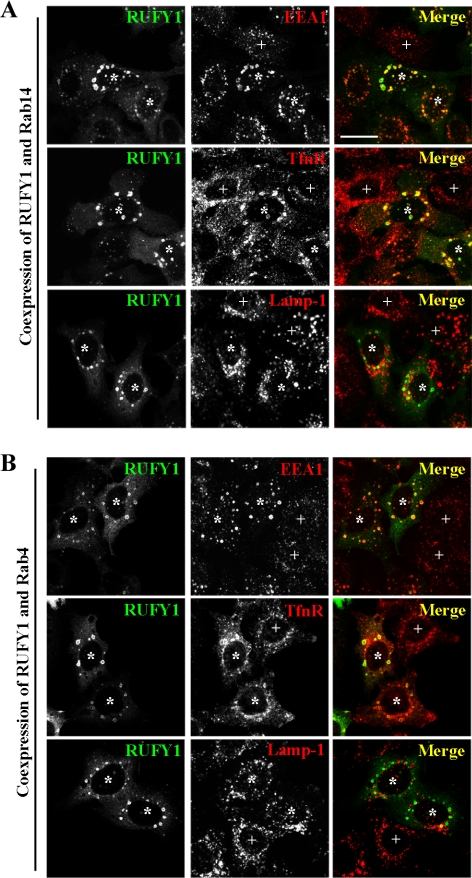
In the course of the cotransfection experiments, we noticed that coexpression of RUFY1 and Rab14 induces expansion of endosomes (Figure 4B, a–a″), compared with single expression; the expression of RUFY1, Rab14, Rab4, or Rab5 alone did not cause significant increase in the endosomal size (Figure 4A). Coexpression of RUFY1 and Rab4 also induced expansion of endosomes (Figure 4B, b–b″), but coexpression of RUFY1 and either Rab5 or Rab11 did not (Figure 4B, c–c″ and data not shown). These results are consistent with the previous report showing the enlargement of endosomes by coexpression of Rab4(Q67L) and RUFY1 in CHO cells (Cormont et al., 2001). To characterize the enlarged endosomes, we stained cells coexpressing RUFY1 and either Rab14 or Rab4 for several endosomal markers, including early endosomal EEA1, early/recycling endosomal TfnR, and late endosomal Lamp-1 (Figure 5, A and B). The enlarged endosomes colocalize almost completely with EEA1 and partially with TfnR, but not with Lamp-1, indicating that these endosomes are derived primarily from early endosomes.
Figure 4.
Coexpression of RUFY1 and either Rab14 or Rab4 induces expansion of endosomes. (A) HeLa cells were transfected with a plasmid encoding HA-RUFY1, EGFP-Rab14, EGFP-Rab4, or EGFP-Rab5. In (a), the cells were stained with antibody to HA. (B) HeLa cells were cotransfected with plasmids encoding HA-RUFY1 and either EGFP-Rab14 (a), EGFP-Rab4 (b), or EGFP-Rab5 (c), and stained with anti-HA antibody. Bars, 20 μm.
Figure 5.
The enlarged endosomes are derived from early endosomes. HeLa cells cotransfected with plasmids encoding EGFP-RUFY1 and HA-Rab14 (A) or HA-Rab4 (B) were stained with anti-EEA1, anti-TfnR, or anti-Lamp-1 antibody. *, transfected cells; +, nontransfected cells. Bars, 20 μm.
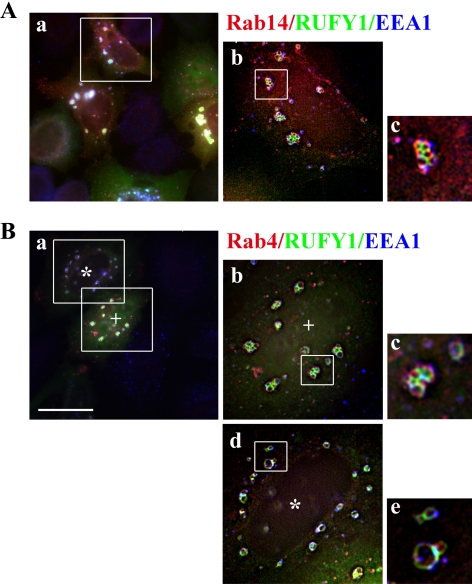
By deconvolution analysis, we often found grape-like clusters of endosomes in cells coexpressing RUFY1 and either Rab14 or Rab4 (Figure 6, A, b and c, and B, b and c). Moreover, RUFY1 and Rab4 coexpression sometimes induced vacuole-like endosomes in addition to grape-like ones (Figure 6B, d and e). Therefore, Rab14 and Rab4 may cooperatively participate in endosomal tethering and fusion through their interactions with RUFY1.
Figure 6.
Deconvolution analysis of the enlarged endosomes. HeLa cells were cotransfected with plasmids encoding EGFP-RUFY1 and HA-Rab14 (A) or HA-Rab4 (B) and doubly stained with anti-HA and anti-EEA1 antibodies. (Ab, Bb and Bd) Deconvolved images of the boxed areas in Aa and Ba (two different areas, * and +, were analyzed) using Carl Zeiss AxioVision software. (Ac, Bc and Be) Digital magnification of the boxed areas in Ab, Bb and Bd, respectively. Bar, 20 μm.
RUFY1(Δ507-517) Does Not Induce Endosomal Enlargement
EGFP-RUFY1 and EGFP-RUFY1(Δ507-517) were present on punctate structures throughout the cytoplasm that were partially colocalized with EEA1, indicating their association with early endosomes. Consistent with a previous report (Cormont et al., 2001), EGFP-RUFY1-positive endosomes were larger in size than EGFP-RUFY1(Δ507-517)-positive endosomes (Figure 7A), suggesting that, probably through interacting with endogenous Rab4, exogenously expressed RUFY1 allows enlargement of endosomes. We therefore asked whether interaction of RUFY1 with Rab4 is required for the endosomal expansion that is induced by coexpression of RUFY1 and Rab14 or Rab4 (Figure 4B). In stark contrast to RUFY1, coexpression of RUFY1(Δ507-517) with either Rab4 or Rab 14 failed to induce the expansion of endosomes (Figure 7, B and C), suggesting that the expansion of endosomes requires the RUFY1 interaction with Rab4.
Rab14 and RUFY1 Are Required for Efficient Tfn Recycling
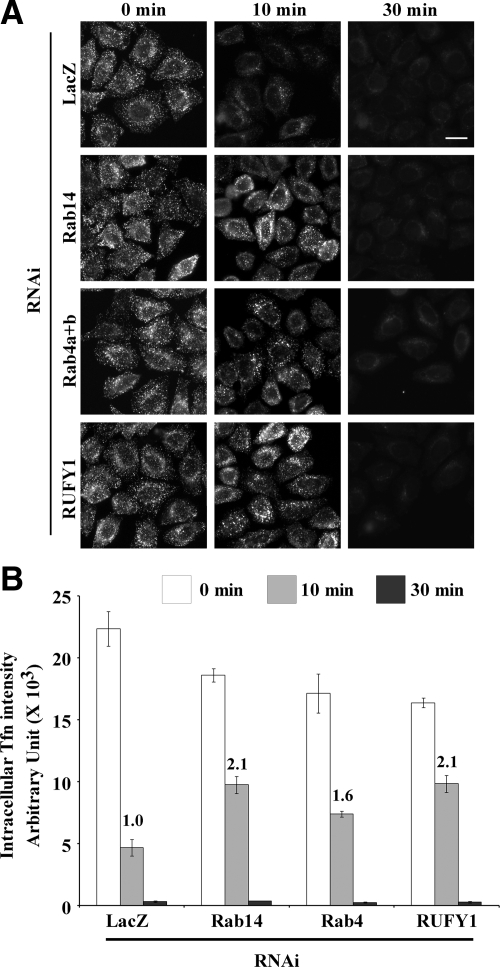
Given that Rab4 plays a role in fast recycling of Tfn from early endosomes back to the plasma membrane (van der Sluijs et al., 1992; Sheff et al., 1999), we asked whether Rab14 and RUFY1 are involved in the Tfn recycling route. First, we examined the involvement of Rab14 and RUFY1 in the Tfn endocytosis. HeLa cells treated with siRNA for Rab14, Rab4, or RUFY1 (see Supplementary Results and Supplementary Figure S4) were incubated with fluorescently labeled Tfn for 50 min at 4°C and then allowed to internalize Tfn at 37°C for indicated times (Supplementary Figure S5, A and B). Neither the amount of cell surface-bound Tfn (0 min) nor internalized Tfn at early time points (2 and 5 min) was significantly affected. Next, we examined the involvement of Rab14 and RUFY1 in the Tfn recycling pathway. HeLa cells treated with siRNA for Rab14, Rab4, or RUFY1 were allowed to internalize fluorescently labeled Tfn for 5 min at 37°C, under conditions in which internalized Tfn primarily reaches early endosomes but not perinuclear recycling endosomes. After acid washing of the cells to remove surface-bound Tfn, the labeled Tfn was chased by nonlabeled Tfn for the indicated times (Figure 8). Internalization of Tfn was not significantly affected by depletion of Rab14, Rab4, or RUFY1 (Figure 8, 0 min, and Supplementary Figure S5, A and B). After a 10-min chase, the Tfn signals were markedly decreased in control cells by virtue of active Tfn recycling, probably from early endosomes. In considerable contrast, a significant level of Tfn signals remained inside cells depleted of Rab14, Rab4, or RUFY1 (Figure 8A). After a 30-min chase, however, the Tfn signals almost completely disappeared in all of the depleted cells. Quantification of the Tfn signal intensity within the cells revealed that the level of residual Tfn signals after a 10-min chase approximately doubled in cells depleted of Rab14 or RUFY1 and increased to a lesser extent in Rab4-depleted cells (Figure 8B). These results together suggest that Rab14 and RUFY1 are not required for internalization, but are required for efficient recycling of Tfn to the cell surface, probably from early endosomes.
Figure 8.
Rab14 and RUFY1 are required for the efficient Tfn recycling. (A) HeLa cells treated with a pool of siRNAs for LacZ, Rab14, both Rab4a and Rab4b, or RUFY1 were incubated with AlexaFluor488-conjugated Tfn at 37°C for 5 min to internalize Tfn, acid-washed on ice to remove surface-bound Tfn, and further incubated at 37°C for indicated times to allow its transport and recycling to the cell surface. Bar, 20 μm. (B) Pixel intensity of AlexaFluor488-conjugated Tfn was estimated at the indicated time. The values are the mean ± SD of 100–200 cells at 0 min, 300–500 cells at 10 min, and >50 cells at 30 min. The graph is a representative of three independent experiments. Numbers indicate fold increase of the Tfn intensity at the 10-min time point.
DISCUSSION
Here, we have demonstrated that Rab14 interacts with RUFY1, previously identified as a Rab4 effector, and is required for RUFY1 recruitment onto endosomes and efficient recycling of Tfn.
The regions of RUFY1 essential for binding to Rab14 and Rab4 appear to be separated. The difference in binding sites may give rise to the functional variation between Rab14 and Rab4 on endosomes. Indeed, Rab14, but not Rab4, is necessary for the RUFY1 recruitment onto endosomes (Figure 3). Consistent with this, RUFY1(Δ507-517), which is unable to interact with active Rab4, retains its ability to localize to endosomes (Figure 7A; also see Cormont et al., 2001). RUFY1 has a FYVE domain that mediates interaction with phosphatidylinositol-3-phosphate [PtdIns(3)P]; treatment of cells with wortmannin, a PI3K inhibitor, has been shown to prevent endosomal localization of RUFY1 (Mari et al., 2001; Fouraux et al., 2004). However, the FYVE domain is dispensable for endosomal localization of RUFY1 (Mari et al., 2001; Yang et al., 2002; Fouraux et al., 2004), similar to the case of the Hrs FYVE domain (Hayakawa and Kitamura, 2000). Taken together, these data indicate that Rab14 is the primary determinant of RUFY1 recruitment onto endosomes and that the FYVE domain may help RUFY1 targeting to PtdIns(3)P-enriched early endosomes.
Most Rab4-positive endosomes include Rab14, and 70% of Rab14-positive endosomes includes Rab4 (Figure 2), indicating that Rab14 is distributed more broadly in the endosomal system than Rab4. Some Rab14-positive endosomes possessing RUFY1 are colocalized with most Rab4-positive endosomes, accordingly, Rab14 together with RUFY1 might be prerequisite for Rab4 function. Indeed, the data presented in this study suggest that Rab14 and RUFY1 are involved in Rab4-dependent Tfn recycling (Figure 8).
We also found that enlargement of early endosomes mediated by RUFY1 requires its interaction with Rab4 (Figure 7). In contrast to coexpression of RUFY1 and either Rab14 or Rab4, coexpression of the RUFY1(Δ507-517) mutant did not induce the enlargement of endosomes (Figure 7), supporting that the endosomal expansion can be mainly attributed to the interaction between RUFY1 and Rab4. Although some clustered endosomes were occasionally observed in cells coexpressing the RUFY1(Δ507-517) mutant and either Rab14 or Rab4, most of the endosomes were smaller in size than in cells coexpressing wild-type RUFY1 with Rab14 or Rab4. Thus, the interaction of RUFY1 with Rab4 participates in endosomal clustering and enlargement, possibly by stabilizing RUFY1 on endosomal membranes. On the basis of these data, we propose sequential interactions and a functional relationship among Rab14, Rab4 and RUFY1. First, activated Rab14 recruits RUFY1 onto endosomes, and Rab4 is in turn recruited onto the RUFY1-localizing endosomal microdomains. These sequential interactions may stabilize RUFY1 on endosomal membranes and promote endosomal tethering and fusion.
Rab4 participates in fast recycling of Tfn from early endosomes, and Rab11 is involved in slow recycling from recycling endosomes back to the plasma membrane, (van der Sluijs et al., 1992; Sheff et al., 1999). When Tfn was internalized for 5 min and chased for 10 min, the Tfn recycling was delayed in cells depleted of Rab14 or RUFY1 and, to a lesser extent, in Rab4-depleted cells. In contrast, when Tfn was chased for 30 min, internalized Tfn was almost completely recycled back to the cell surface in cells depleted of Rab14, Rab4, or RUFY1, comparable to the recycling observed in the control cells (Figure 8, A and B). These observations together argue that Rab14 and RUFY1 as well as Rab4 are involved in the fast recycling of Tfn from early endosomes.
Several studies indicate that Rab4 is an important player in Glut4 trafficking in adipocytes, skeletal muscle and cardiomyocytes (Kaddai et al., 2008). It has been suggested that RUFY1 and Rab14 also play a role in Glut4 trafficking in adipocytes and muscle cells, respectively (Mari et al., 2006; Ishikura et al., 2007). Therefore, it is tempting to speculate that Rab14, RUFY1, and Rab4 might cooperate in Glut4 trafficking as well as Tfn recycling. Moreover, Rab14 has been implicated in phagosome–phagosome fusion or phagosome–early endosome fusion (Kyei et al., 2006). It is thus an open question whether the ability of Rab14 to enhance fusogenicity is also cooperatively regulated by RUFY1 and/or Rab4.
Our results on the sequential and cooperative regulation of Rab14 and Rab4 through the dual effector, RUFY1, have important implications for signal transition between two different Rab proteins residing on the same compartment. Biochemical and structural dissection of the characteristics of the binding among Rab14, RUFY1, and Rab4, as well as study of the dynamics of Rab14, RUFY1, and Rab4 by live image analysis, will improve our understanding of signal transition between Rab membrane microdomains in subcellular compartments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marino Zerial and Roger Tsien for kindly providing the EGFP-Rab5 construct and the pRSETB-mCherry construct, respectively. This study was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Society for the Promotion of Science; the Special Coordination Fund for Promoting Science and Technology of MEXT; the Targeted Proteins Research Program; the Hayashi Memorial Foundation for Female Natural Scientists; the Uehara Memorial Foundation; and the NOVARTIS Foundation (Japan).
Abbreviations used:
- EGFP
enhanced green fluorescent protein
- PtdIns(3)P
phosphatidylinositol-3-phosphate
- RNAi
RNA interference
- Rabip4
Rab4 interacting protein
- siRNA
small interfering RNA
- Tfn
transferrin
- TfnR
transferrin receptor
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0074) on June 9, 2010.
REFERENCES
- Barbero P., Bittova L., Pfeffer S. R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C., Parton R. G., Mather I. H., Stunnenberg H., Simons K., Hoflack B., Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., McBride H. M., Burgoyne R., D., Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999a;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S. C., Waterfield M. D., Backer J. M., Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999b;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Cormont M., Mari M., Galmiche A., Hofman P., Le Marchand-Brustel Y. A FYVE-finger-containing protein, Rabip4, is a Rab4 effector involved in early endosomal traffic. Proc. Natl. Acad. Sci. USA. 2001;98:1637–1642. doi: 10.1073/pnas.031586998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daro E., van der Sluijs P., Galli T., Mellman I. Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc. Natl. Acad. Sci. USA. 1996;93:9559–9564. doi: 10.1073/pnas.93.18.9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit H., Lichtenstein Y., Kelly R. B., Geuze H. J., Klumperman J., van der Sluijs P. Rab4 regulates formation of synaptic-like microvesicles from early endosomes in PC12 cells. Mol. Biol. Cell. 2001;12:3703–3715. doi: 10.1091/mbc.12.11.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Press B., Wandinger-Ness A. Rab7, an important regulator of late endocytic membrane traffic. J. Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouraux M. A., Deneka M., Ivan V., van der Heijden A., Raymackers J., van Suylekom D., van Venrooij W. J., van der Sluijs P., Pruijn G.J.M. rabip4′ Is an Effector of rab5 and rab4 and regulates transport through early endosomes. Mol. Biol. Cell. 2004;15:611–624. doi: 10.1091/mbc.E03-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvel J.-P., Chavrier P., Zerial M., Gruenberg J. Rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- Gournier H., Stenmark H., Rybin V., Lippe R., Zerial M. Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J. 1998;17:1930–1940. doi: 10.1093/emboj/17.7.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B. D., Donaldson J. G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales C. M., Griner R., Hobdy-Henderson K. C., Dorn M. C., Hardy D., Kumar R., Navarre J., Chan E. K., Lapierre L. A., Goldenring J. R. Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Harris E., Cardelli J. RabD, a Dictyostelium Rab14-related GTPase, regulates phagocytosis and homotypic phagosome and lysosome fusion. J. Cell Sci. 2002;115:3703–3713. doi: 10.1242/jcs.00050. [DOI] [PubMed] [Google Scholar]
- Hayakawa A., Kitamura N. Early endosomal localization of hrs requires a sequence within the proline- and glutamine-rich region but not the FYVE finger. J. Biol. Chem. 2000;275:29636–29642. doi: 10.1074/jbc.M002696200. [DOI] [PubMed] [Google Scholar]
- Hopkins C. R. Intracellular routing of transferrin and transferrin receptors in epidermoid carcinoma A431 cells. Cell. 1983;35:321–330. doi: 10.1016/0092-8674(83)90235-0. [DOI] [PubMed] [Google Scholar]
- Horiuchi H., Lippe R., McBride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Ishikura S., Bilan P. J., Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem. Biophys. Res. Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- Ishizaki R., Shin H.-W., Mitsuhashi H., Nakayama K. Redundant Roles of BIG2 and BIG1, Guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol. Biol. Cell. 2008;19:2650–2660. doi: 10.1091/mbc.E07-10-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki R., Shin H. W., Iguchi-Ariga S. M., Ariga H., Nakayama K. AMY-1 (associate of Myc-1) localization to the trans-Golgi network through interacting with BIG2, a guanine-nucleotide exchange factor for ADP-ribosylation factors. Genes Cells. 2006;11:949–959. doi: 10.1111/j.1365-2443.2006.00991.x. [DOI] [PubMed] [Google Scholar]
- Junutula J. R., De Maziere A. M., Peden A. A., Ervin K. E., Advani R. J., van Dijk S. M., Klumperman J., Scheller R. H. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol. Biol. Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddai V., Le Marchand-Brustel Y., Cormont M. Rab proteins in endocytosis and Glut4 trafficking. Acta Physiol. 2008;192:75–88. doi: 10.1111/j.1748-1716.2007.01787.x. [DOI] [PubMed] [Google Scholar]
- Kelly E. E., Horgan C. P., Adams C., Patzer T. M., Ni Shuilleabhain D. M., Norman J. C., McCaffrey M. W. Class I Rab11-family interacting proteins are binding targets for the Rab14 GTPase. Biol. Cell. 2009;102:51–62. doi: 10.1042/BC20090068. [DOI] [PubMed] [Google Scholar]
- Kyei G. B., Vergne I., Chua J., Roberts E., Harris J., Junutula J. R., Deretic V. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. EMBO J. 2006;25:5250–5259. doi: 10.1038/sj.emboj.7601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A. J., Hendrick A. G., Cantalupo G., Senic-Matuglia F., Goud B., Bucci C., McCaffrey M. W. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 2002;277:12190–12199. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- Mari M., Macia E., Le Marchand-Brustel Y., Cormont M. Role of the FYVE Finger and the RUN domain for the subcellular localization of Rabip4. J. Biol. Chem. 2001;276:42501–42508. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- Mari M., Monzo P., Kaddai V., Keslair F., Gonzalez T., Le Marchand-Brustel Y., Cormont M. The Rab4 effector Rabip4 plays a role in the endocytotic trafficking of Glut 4 in 3T3–L1 adipocytes. J. Cell Sci. 2006;119:1297–1306. doi: 10.1242/jcs.02850. [DOI] [PubMed] [Google Scholar]
- Mayor S., Presley J. F., Maxfield F. R. Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J. Cell Biol. 1993;121:1257–1269. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Ghosh R. N., Maxfield F. R. Endocytosis. Physiol. Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- Nagelkerken B., Van Anken E., Van Raak M., Gerez L., Mohrmann K., Van Uden N., Holthuizen J., Pelkmans L., Van Der Sluijs P. Rabaptin4, a novel effector of the small GTPase rab4a, is recruited to perinuclear recycling vesicles. Biochem. J. 2000;346(Pt 3):593–601. [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto-Morita K., Shin H.-W., Mitsuhashi H., Kitamura M., Zhang Q., Johannes L., Nakayama K. Differential effects of depletion of ARL1 and ARFRP1 on membrane trafficking between the trans-Golgi network and endosomes. J. Biol. Chem. 2009;284:10583–10592. doi: 10.1074/jbc.M900847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal J. B., Seabra M. C. Evolution of the rab family of small GTP-binding proteins, J. Mol. Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Davies J. M., Scheller R. H. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001;276:38966–38970. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T., Gaugel A., Frickey T., Nordheim A. Rab14 is part of the early endosomal clathrin-coated TGN microdomain. FEBS Lett. 2006;580:5241–5246. doi: 10.1016/j.febslet.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Ren M., Xu G., Zeng J., De Lemos-Chiarandini C., Adesnik M., Sabatini D. D. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S. L., Fuchs R., Male P., Mellman I. Two distinct sub-populations of endosomes involved in membrane recycling and transport to the lysosomes. Cell. 1988;52:73–83. doi: 10.1016/0092-8674(88)90532-6. [DOI] [PubMed] [Google Scholar]
- Sheff D. R., Daro E. A., Hull M., Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Koga H., Shin H.-W., Kawasaki M., Kato R., Nakayama K., Wakatsuki S. Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc. Natl. Acad. Sci. USA. 2006;103:15416–15421. doi: 10.1073/pnas.0605357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Takatsu H., Shin H. W., Nakayama K. Gamma-adaptin interacts directly with Rabaptin-5 through its ear domain. J. Biochem. 2002;131:327–336. doi: 10.1093/oxfordjournals.jbchem.a003107. [DOI] [PubMed] [Google Scholar]
- Shin H.-W., et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.-W., Morinaga N., Noda M., Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol. Biol. Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. W., Shinotsuka C., Torii S., Murakami K., Nakayama K. Identification and subcellular localization of a novel mammalian dynamin-related protein homologous to yeast Vps1p and Dnm1p. J. Biochem. 1997;122:525–530. doi: 10.1093/oxfordjournals.jbchem.a021784. [DOI] [PubMed] [Google Scholar]
- Ullrich O., Reinsch S., Urbe S., Zerial M., Parton R. G. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P., Hull M., Huber L. A., Male P., Goud B., Mellman I. Reversible phosphorylation-dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J. 1992;11:4379–4389. doi: 10.1002/j.1460-2075.1992.tb05538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G., Rybin V., Christoforidis S., Thornqvist P., McCaffrey M., Stenmark H., Zerial M. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro D. J., Maxfield F. R. Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell. 1984;37:389–400. doi: 10.1016/0092-8674(84)90414-8. [DOI] [PubMed] [Google Scholar]
- Yang J., Kim O., Wu J., Qiu Y. Interaction between tyrosine kinase Etk and a RUN domain- and FYVE domain-containing protein RUFY1. J. Biol. Chem. 2002;277:30219–30226. doi: 10.1074/jbc.M111933200. [DOI] [PubMed] [Google Scholar]
- Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.