We present a novel strategy to enforce cytokine-independent, constitutive signaling of heterodimeric gp130 receptor complexes. Replacing the extracellular domain of gp130-type receptors by IL-15/IL-15R is sufficient to heterodimerize gp130-like receptors and as a consequence leading to sustained cytokine-independent receptor activation.
Abstract
Naturally ligand independent constitutively active gp130 variants were described to be responsible for inflammatory hepatocellular adenomas. Recently, we genetically engineered a ligand-independent constitutively active gp130 variant based on homodimerization of Jun leucine zippers. Because also heterodimeric complexes within the gp130 family may have tumorigenic potential, we seek to generate ligand-independent constitutively active heterodimers for all known gp130-receptor complexes based on IL-15/IL-15Rα-sushi fusion proteins. Ligand-independent heterodimerization of gp130 with WSX-1, LIFR, and OSMR and of OSMR with GPL led to constitutive, ligand-independent STAT1 and/or STAT3 and ERK1/2 phosphorylation. Moreover, these receptor combinations induced transcription of the STAT3 target genes c-myc and Pim-1 and factor-independent growth of stably transduced Ba/F3-gp130 cells. Here, we establish the IL-15/IL-15Rα-sushi system as a new system to mimic constitutive and ligand-independent activation of homo- and heterodimeric receptor complexes, which might be applicable to other heterodimeric receptor families. A mutated IL-15 protein, which was still able to bind the IL-15Rα-sushi domain, but not to β- and γ-receptor chains, in combination with the 2A peptide technology may be used to translate our in vitro data into the in vivo situation to assess the tumorigenic potential of gp130-heterodimeric receptor complexes.
INTRODUCTION
The IL-6 family consists of the ten cytokines interleukin (IL)-6, IL-11, IL-27, IL-31, CNTF, CT-1, neuropoctin (NP), leukemia inhibitory factor (LIF), NNT-1, and OSM. Only IL-6 and IL-11 induce the formation of a gp130 homodimer, whereas signaling by CNTF, CT-1, NP, LIF, and NNT-1 results in the formation of a glycoprotein 130 kDa (gp130)/leukemia inhibitory factor receptor (LIFR) heterodimer. OSM can induce the formation of a dimer of gp130 with LIFR or the related protein, oncostatin M receptor (OSMR; Taga and Kishimoto, 1997). IL-27 exclusively signals via a heterodimer comprising gp130 and the WSX-1 receptor (Pflanz et al., 2004). The cytokine IL-31 induces the formation of a heterodimer of GPL (gp130-like receptor) and OSMR (Dillon et al., 2004).
The formation of heterodimeric receptor complexes containing the gp130/WSX-1 (interleukin 27 receptor with a WSX signature motif: W-S-X-W-S), gp130/LIFR, gp130/OSMR, or GPL/OSMR receptors leads to intracellular activation of Janus kinase (JAK) /Tyk tyrosine kinases as well as the signal transducer and activator of transcription (STAT) family of transcription factors such as STAT1 and STAT3. Furthermore, the activation leads to stimulation of the RAS/RAF/mitogen-activated protein (MAP) kinase pathways (Hibi et al., 1996; Heinrich et al., 2003).
The extracellular parts of gp130, WSX-1, LIFR, OSMR, and GPL receptors consist of combinations of cytokine binding and/or immunglobulin-like domains and three fibronectin-like domains (Scheller et al., 2006). Although the binding of the cytokine is achieved by the cytokine-binding and the immunglobulin-like domains (Boulanger et al., 2003), the role of the three fibronectin-like domains is less clear. Truncation of these domains results in gp130 molecules devoid of signaling capacity of homodimeric gp130 (Kurth et al., 2000). For gp130 it has been speculated that after ligand binding these domains are needed to position the transmembrane domains of the receptor complexes in close proximity and to induce signaling (Skiniotis et al., 2005). Importantly, no crystal structural information is available for signal induction of the full-length homodimeric and heterodimeric receptor complexes of gp130/gp130, gp130/WSX-1, gp130/LIFR, gp130/OSMR, and GPL/OSMR. 3D reconstruction of the full-length gp130/LIF-R/CNTF-Rα/CNTF-complex and the gp130/IL-6R/IL-6-complex using crystal structural and electron microscopy data showed a close juxtaposition of the intracellular domains of the LIFR/gp130-heterodimer and the gp130/gp130-homodimer, respectively (Skiniotis et al., 2008).
Recently, we demonstrated leucine zipper–mediated ligand-independent dimerization of gp130 (Stuhlmann-Laeisz et al., 2006). Interestingly, a marked activation of the IL-6 signaling pathway in inflammatory hepatocellular adenomas was directly caused by gain-of-function mutations within the gp130 receptor chain, which led to ligand-independent constitutively active gp130 molecules (Rebouissou et al., 2008). The leucine zippers used to ligand-independently activate gp130 are naturally found in transcription factors, such as Fos and Jun, which form the AP1 transcription complex. Naturally, Fos and Jun leucine zippers form heterodimers, but the Jun leucine zipper can also form stable homodimers, albeit with significantly lower stability. The Fos leucine zipper forms an unstable homodimer, which provides a thermodynamic driving force for preferential heterodimer formation with Jun leucine zipper (O'Shea et al., 1989; Patel et al., 1996). However, we found that Fos leucine zippers formed homodimers, which are sufficient to induce ligand-independent signaling in murine cytokine-dependent cells stably transduced with a cDNA coding for an Fos-gp130 construct.
IL-15 is a member of the small four α-helix bundle family of cytokines. IL-15 was discovered by its ability to mimic IL-2–mediated T-cell proliferation. IL-15 targets the specific α-chain receptor IL-15Rα (Bulfone-Paus et al., 2006). The exceptionally high-affinity binding of IL-15 to IL-15Rα is mediated by the sushi domain of IL-15Rα (IL-15Rα-sushi), in which a large interface of ionic interactions facilitates the interaction of the IL-15 and IL-15Rα, whereas interactions within the gp130 cytokine/cytokine receptor family are facilitated by hydrophobic interaction (Wang et al., 2005; Lorenzen et al., 2006).
We reasoned that this type of interaction, which explains the exceptionally high affinity of the IL-15/IL-15Rα complex, is well suited for forced heterodimerization of the gp130 receptor with LIFR, OSMR, and WSX-1 as well as GPL with OSMR. We show ligand-independent receptor complex activation of gp130/WSX-1, gp130/LIFR, gp130/OSMR, and GPL/OSMR heterodimers by replacing the entire extracellular portion of gp130, WSX-1, LIFR, OSMR, and GPL with IL-15 and/or the IL-15Rα-sushi domain. Our data indicate that all heterodimeric gp130-type receptor complexes are activated by a similar mechanism in which close juxtaposition of the intracellular receptor domains is sufficient for signal induction.
MATERIALS AND METHODS
Cells and Reagents
Ba/F3-gp130 cells were obtained from Immunex (Seattle, WA; Gearing et al., 1994), COS-7 cells were from the American Type Culture Collection (Rockville, MD/Manassas, VA) and Phoenix-Eco cells from U. Klingmüller (DKFZ, Heidelberg, Germany). All cells were grown in DMEM high-glucose culture medium (PAA Laboratories, Marburg, Germany) supplemented with 10% fetal calf serum, penicillin (60 mg/l), and streptomycin (100 mg/l) at 37°C with 5% CO2 in a water-saturated atmosphere. For cultivation of Ba/F3-gp130 cells the standard DMEM medium was supplemented with 10% FCS, penicillin (60 mg/l), streptomycin (100 mg/l), and 10 ng/ml Hyper-IL-6. Hyper-IL-6 is a fusion protein of IL-6 and the sIL-6R, which is a mimic of IL-6 trans-signaling (Fischer et al., 1997). Hyper-IL-6 was expressed and purified as described previously (Fischer et al., 1997). Recombinant IL-27 was a kind gift of Christoph Hölscher (Borstel, Germany). Mouse anti-IL-15 mAbs were purchased from Peprotech, Cell Concepts (Umkirch, Germany). Rabbit polyclonal anti-IL-15 Abs were from Morphosys AbD (Düsseldorf, Germany). Anti-Myc-tag (71D10), anti-phospho-tyrosine (P-Tyr-100), anti-STAT1/3, anti-phospho STAT1/3, anti-phospho-extracellular signal-regulated protein kinase 1/2 (ERK1/2), and anti-ERK1/2 Abs were purchased from Cell Signaling, New England Biolabs (Schwalbach, Germany). Anti-c-myc (9E10) and anti-gp130 (C-20) Abs were from Santa Cruz Biotechnology (Heidelberg, Germany). Anti-FLAG-tag (M2) mAbs were purchased from Sigma-Aldrich (Taufkirchen, Germany). Anti-GFP mAbs (clones 7.1 and 13.1) were purchased from Roche Applied Science (Mannheim, Germany). All restriction enzymes were obtained from Fermentas (St. Leon-Rot, Germany). The peroxidase conjugated secondary antibodies were purchased from Pierce (Thermo Scientific–Pierce, Perbio, Bonn, Germany). The allophycocyanin conjugated anti-mouse antibody was purchased from Dianova (Hamburg, Germany) and the Alexa Fluor 488–conjugated anti-rabbit antibody was purchased from Molecular Probes (Invitrogen, Karlsruhe, Germany). Murine IL-6 ELISA Duo Set was purchased from R&D (Wiesbaden-Nordenstadt, Germany). Recombinant IL-15 and IL-15Rα sushi domain were produced as previously described (Lorenzen et al., 2006). The plasmid pcDNA3-BACE was a kind gift of Andrea Rittger (Kiel, Germany).
Construction of IL-15-gp130, IL-15-WSX-1, IL-15-LIFR, and IL-15-OSMR and sushi-gp130, sushi-WSX-1, sushi-GPL, and FUSIO Expression Plasmids
The IL-15-fusion receptors pBSK-IL-15-gp130, pBSK-IL-15-WSX-1, pBSK-IL-15-LIFR, and pBSK-IL-15-OSMR were assembled as followed: bovine preprolactin (bPPL) signal peptide (MDSKGSSQKAGSRLLLLLVVSNLLLCQGVVSTTR), human IL-15 protein fragment (Bernard et al., 2004), and the N-terminally truncated receptor proteins (15 aa of the extracellular domain, transmembrane domain, and cytoplasmic domain). For construction of IL-15 fusion receptors, we used pBSK-PPL-IL-15, which was a kind gift of Yannick Jacques (Nantes, France; Bernard et al., 2004). pBSK-PPL-IL-15 encodes human IL-15, which consists of an alternative signal peptide from the bPPL and was used as a template to amplify the coding sequence by PCR (5′ primer: 5′-3′: GATCCTCGAGCCACCATGGACAGCAAAGGTTCG and 3′ primer: 5′-3′: GATCGAATTCAGAAGTGTTGATGAACATTTG). The purified PCR product was digested with XhoI and EcoRI and subcloned into pBSK-gp130 (ΔN-term). The resulting plasmid was named pBSK-IL-15-gp130, which was subsequently used to generate the remaining IL-15 fusion receptors by digestion of pBSK-IL-15-gp130 with EcoRI and BamHI and subcloning of PCR produced truncated WSX-1 (cDNA of Ba/F3 cells: 5′ primer 5′-3′: GACTGAATTCTCACTTCACCTACCAGATAATAGG and 3′ primer 5′-3′: GACTGGATCCTCAGACTAGAAGGCCCAGCTCCTC; amino acids 500-623), LIFR (template: p409-hLIFR; Kallen et al., 1999; 5′ primer: 5′-3′: GAATTCCCGGAGAAGAGTATGTATGTG and 3′ primer: 5′-3′: GGATCCTTAATCGTTTGGTT TGTTCTG; amino acids 820-1097), OSMR (template: p409-hOSMR; Cichy et al., 1998 5′ primer: 5′-3′: GAATTCCCCAGTGCTACGTTCACG and 3′ primer: 5′-3′: GGATCCTTAG CAGTAGTGTT CACC; amino acids 723-979) or receptor coding sequences (15 aa of extracellular domain, transmembrane domain, and cytoplasmic domain). The resulting plasmids were named pBSK-IL-15-WSX-1, pBSK-IL-15-LIFR, and pBSK-IL-15-OSMR. To determine the transmembrane regions of the receptors the programs DAS (Dense Alignment Surface; www.sbc.su.se/∼miklos) or TMHMM (Tied Mixture Hidden Markov Model; Technical University of Denmark, Lyngby, Denmark; www.cbs.dtu.dk/services/TMHMM) were used.
The IL-15Rα-sushi-fusion receptors IL-15Rα-sushi-gp130, IL-15Rα-sushi-WSX-1, and IL-15Rα-sushi-GPL were assembled as followed: gp130 signal peptide (MLTLQTWLVQALFIFLTTESTG), Myc-tag (EQKLISEEDL), human IL-15Rα-sushi protein fragment (Lorenzen et al., 2006) and the same N-terminally truncated receptor proteins. We used pQE30-IL-15Rα-sushi (Lorenzen et al., 2006) as a template for the PCR amplification of the cDNA coding for IL-15Rα-sushi (5′ primer: 5′-3′: AGCGAGGAGGACCTGATCACATGCCCTCCCCCC and 3′ primer: 5′-3′: GATCGAATTCGTCTCTAATGCATTTGAG). The coding sequence of the gp130 signal peptide and Myc-tag was amplified by using the plasmid pBSK- Jun-gp130 as a template (5′ primer: 5′-3′: GATCCTCGAGCCACCATGTTGACGTTGCAGACTTG and 3′ primer: 5′-3′ CAGGTCCTCCTCGCTGATCAGCTTCTGCTCACCTGTAGATTCAGTGG). Both PCR products were used for a subsequent PCR, because the coding sequence for IL-15Rα-sushi contains a 5′ overhang, which primes the coding sequence of the Myc-tag (5′ primer: 5′-3′: AGCGAGGAGGACCTGATCACATGCCCTCCCCCC and 3′ primer: 5′-3′ CAGGTCCTCCTCGCTGATCAGCTTCTGCTCACCTGTAGATTCAGTGG). The resulting PCR product was purified, digested with XhoI and EcoRI and ligated into pBSK-gp130(ΔN-term), also digested with XhoI and EcoRI. The resulting plasmid was named pBSK-IL-15Rα-sushi-gp130. To generate the plasmids pBSK-IL-15Rα-sushi-WSX-1, and pBSK-IL-15Rα-sushi-GPL, the gp130-receptor portion was exchanged by digestion of the plasmid pBSK-IL-15Rα-sushi-gp130 with EcoRI and BamHI and subcloning of the fragments coding for N-terminally truncated WSX-1 or GPL obtained from the plasmid pBSK-IL-15-WSX-1 or generated by PCR (digested, purified, and ligated) in the case of GPL (template: pcDNA3.1-GPL; 5′ primer: 5′-3′: GAATTCGGGACCAGCATAAATTTC and 3′ primer: 5′-3′: GGATCCTTAGACTTCTCCCTTGG; amino acid 500-727), respectively. The vector pcDNA3.1-GPL was a kind gift of Hugues Gascan (Unité Mixte INSERM, Angers, France). To determine the transmembrane regions of GPL the program TMHMM was used.
The FUSIO cDNA encoding signal peptide from bPPL, FLAG-tag, and β- and γ-receptor chains binding deficient hIL-15, murine WSX-1 (15 amino acids of the extracellular domain, transmembrane domain, and cytoplasmic domain), furin cleavage site, 2A peptide, signal peptide of human gp130, Myc-tag, and murine gp130 (15 amino acids of the extracellular domain, transmembrane domain, and cytoplasmic domain) was synthesized by GeneArt (Regensburg, Germany) and delivered in pMk plasmid.
The retroviral plasmid pMOWS encodes the puromycin resistance gene (Ketteler et al., 2002). For this study we modified the plasmid pMOWS by replacing the puromycin resistance gene with a hygromycin resistance gene. The hygromycin gene from the plasmid pCEP4 was amplified by PCR (5′ primer: 5′-3′: GATCAAGCTTCCACCATGAAAAAGCCTGAACTCACC and 3′ primer 5′-3′: GATCGTCGACTCAGTTAGCCTCCCCCATC-3′). The purified PCR product was digested with HindIII and SalI and subcloned into pMOWS. The resulting plasmid was named pMOWS-hygromycin. The IL-15 fusion receptors and IL-15Rα-sushi fusion receptors were subcloned into pMOWS and pMOWS-hygromycin as described in the Supplemental Materials and Methods. The resulting plasmids were named pMOWS-IL-15-gp130, pMOWS-IL-15-WSX-1, pMOWS-IL-15-LIFR, and pMOWS-IL-15-OSMR, as well as pMOWS-hygromycin-IL-15Rα-sushi-gp130, pMOWS-hygromycin-IL-15Rα-sushi-WSX-1, and pMOWS-hygromycin-IL-15Rα-sushi-GPL.
The FUSIO cDNA was subcloned into pMOWS. In brief pMOWS was digested with XagI and BamHI, and blunt ends were generated by Klenow fragment. The FUSIO cDNA was digested with EcoRV. Plasmid and FUSIO were purified and subcloned. The resulting plasmid was named pMOWS-FUSIO. The cDNAs coding for IL-15-WSX-1, IL-15-gp130 and sushi-gp130 were subcloned into p409 as described in the Supplemental Materials and Methods. The resulting plasmids were named p409-IL-15-WSX-1, p409-IL-15-gp130, and p409-sushi-gp130.
The cDNAs coding for IL-15-gp130 and sushi-gp130 were additionally subcloned into pEYFP-gp130. The resulting plasmids were named pEYFP-IL-15-gp130 and pEYFP-sushi-gp130.
Transfection, Transduction, and Selection of Murine Ba/F3-gp130 Cells
Ba/F3-gp130 cells were retrovirally transduced with the retroviral expression plasmids derivatives of pMOWS and pMOWS-hygromycin. pMOWS-IL-15-gp130, pMOWS-IL-15-WSX-1, pMOWS-IL-15-LIFR, pMOWS-IL-15-OSMR, as well as pMOWS-hygromycin-IL-15Rα-sushi-gp130, pMOWS-hygromycin-IL-15Rα-sushi-WSX-1 or pMOWS-hygromycin-IL-15Rα-sushi-GPL, pMOWS-FUSIO and the control vector pMOWS-GFP or pMOWS-hygromycin-GFP (5 μg each) were transiently transfected in 8 × 105 Phoenix-Eco cells using Lipofectamine 2000 according to manufacturer's instructions (Invitrogen). The transfection efficiency was typically ∼50%, which was estimated by green fluorescent protein (GFP) expression 24 h after transfection (Axiovert 200 microscope, Zeiss, Jena, Germany). Retroviral supernatants were produced as described (Ketteler et al., 2002). Retroviral supernatant, 250 μl, was applied to 1 × 105 Ba/F3-gp130 cells and mixed, and the solution was centrifuged at 1800 rpm for 2 h at 25°C. Transduced cells were grown in standard medium supplemented with 10 ng/ml Hyper-IL-6. Forty-eight hours after transduction, transduced Ba/F3-gp130 cells were selected in 1.5 μg/ml puromycin and/or 1 mg/ml hygromycin (PAA Laboratories) for at least 2 wk. After 2 wk of antibiotic selection, Hyper-IL-6 was washed away and the cells were screened for cytokine-independent growth.
Transfection of COS-7 Cells
COS-7 cells, 5 × 105, were transiently transfected with polymeric enhanced yellow fluorescent protein (pEYFP) and/or p409 expression plasmids using Turbofect (Fermentas) according to the manufacturer's instructions. The transfection efficiency was typically ∼50–70%, which was estimated by YFP expression 48 h after transfection (Axiovert 200 microscope, Zeiss). After 48 h, cells were lysed and coimmunoprecipitation, and/or Western blot experiments were performed.
Pervanadate Treatment of COS-7 Cells
Sodium orthovanadate solution, 500 μl at 100 mM, was incubated with 2.8 μl of 30% H2O2 to obtain pervanadate. Immediately after preparation of pervanadate COS-7 cells were treated with 1 mM pervanadate for 15 min at 37°C before lysis.
Preparation of Primary Murine Dendritic Cells
Murine bone marrow–derived dendritic cells were prepared as described (Lutz et al., 1999).
Coculture Experiments of Murine Dendritic Cells with Ba/F3-gp130 Cells
Murine dendritic cells were plated (2 × 105 cells) into a 12-well plate and stimulated with lipopolysaccharide (LPS; 10 ng/ml) overnight. The day after, the cells were washed three times with PBS to wash away LPS and Ba/F3-gp130 cells expressing either IL-15-gp130, IL-15-WSX-1 + sushi-gp130, Fos-gp130, or FUSIO were cocultured (2 × 106 cells) at a ratio of 1:10. After 24 h of coculturing the supernatants were analyzed for mIL-6 protein secretion. Murine IL-6 secretion was normalized to different Ba/F3-gp130 cell numbers obtained after 24 h by ELISA.
Detection of Target Gene Transcription by RT-PCR
For detection of target gene transcription (murine c-myc and murine Pim-1), retrovirally transduced Ba/F3-gp130 cells were washed three times in sterile PBS and starved for 6 h in serum-free DMEM. The cells were centrifuged, and total RNA was isolated using Nucleospin RNA II (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. RNA concentration was determined at 260 and 280 nm with the Nanodrop ND-1000 spectrophotometer (Peqlab, Erlangen, Germany) and cDNA was prepared using 2 μg of total RNA, oligo(dT) primer, dNTPs, and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas) following the manufacturer's instructions. The PCR was performed with 35 cycles of 95°C for 60 s, 55°C for 60 s, and 72°C for 60 s. Primer sequences to detect the target genes murine c-Myc and murine Pim-1 were described previously (Owaki et al., 2008).
Flow Cytometry Staining and Analysis
To detect the surface expression of the fusion receptors containing IL-15 and IL-15Rα-sushi, cells were washed with FACS buffer (PBS, 1% BSA) and incubated at 3 × 105 cells/100 μl FACS buffer containing 2 μg/ml anti-IL-15 mAb (Peprotech, Cell Concepts) or 1:100 diluted anti-Myc-tag (71D10) mAb (Cell Signaling) in fluorescent-activated cell sorting (FACS) buffer for 60 min on ice, respectively. After a single wash in FACS buffer, cells were incubated at 3 × 105 cells/100 μl FACS buffer containing a 1:100 dilution of allophycocyanin-conjugated anti-mouse mAb (Dianova) or Alexa Fluor 488–conjugated anti-rabbit mAb (Molecular Probes), respectively. Cells were washed once with FACS buffer, resuspended in 500 μl FACS buffer, and analyzed by flow cytometry (Becton-Dickinson, Heidelberg, Germany; FACS-Canto and FACS DIVA software).
Proliferation Assays
Ba/F3-gp130 cells expressing the leucine zipper, IL-15, and/or IL-15Rα-sushi fusion receptors were washed three times with sterile PBS and resuspended in DMEM containing 10% FCS at 5 × 103 cells per well of a 96-well plate. The cells were cultured for 3 d in a final volume of 100 μl with or without additional cytokines as indicated. For inhibition studies with soluble IL-15Rα-sushi domain, the recombinant protein was added as indicated. The CellTiter-Blue Cell Viability Assay (Promega, Mannheim, Germany) was used to determine the cell number following the manufacturer's instructions and measured on a Lambda Fluoro 320 fluorimeter (ex-filter 530/25, em-filter 590/35, sensitivity 75, Software KC4). Relative light units (RLU) values were normalized by subtractions of negative control values (not stimulated Ba/F3-gp130 cells) from all other values. All values were measured in quadruplicates.
Coimmunoprecipitation and Western Blotting
For coimmunoprecipitation transiently transfected COS-7 cells were washed with ice-cold PBS, collected by scraping, and subsequently lysed in 200 or 250 μl immunoprecipitation buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, and 1 mM Na3VO4). The immunoprecipitation buffer was supplemented with complete protease inhibitor cocktail tablets (Roche Diagnostics, Mannheim, Germany). Fifty microliters of each lysate was frozen and used as input controls if needed. The remaining 200 μl of the cell lysates was separated into two fresh tubes, which were filled up with immunoprecipitation buffer to a total volume of 200 μl. Anti-green fluorescent protein (GFP)-specific mAbs (2 μg) or anti-gp130–specific Abs (1 μg) were added to one reaction tube, and the mixture was incubated overnight at 4°C under gentle agitation. The second reaction tube was left untreated. Alternatively the complete 200 μl of lysate was used for immunoprecipitation. Fifty microliters of protein A or protein G agarose was added to each reaction tube, and the mixture was incubated for 4 h at 4°C under gentle agitation. The agarose was washed five times with 500 μl immunoprecipitation buffer and afterward was boiled in 50 μl 2.5× Laemmli buffer (92.5 mM Tris-HCl, pH 6.8, 25% glycerol, 5% SDS, 2.5% β-mercaptoethanol, and 0.025% bromphenol blue). The immunoprecipitation supernatant was separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane (GE Healthcare, Munich, Germany). The membrane was blocked with 5% skim milk powder in TBST (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 1% Tween 20) and probed with primary antibodies against GFP, Myc-tag, c-myc, gp130, IL-15, or phospho-tyrosine (PY) at 4°C overnight. After washing with TBST the membranes were incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (Thermo Scientific) and protein bands were visualized with the ECL detection system (GE Healthcare) according to the manufacturer's instructions.
For detection of Fos-gp130 dimers, COS-7 cells were washed with ice-cold PBS and collected by scraping and subsequently lysed in 300 μl mild lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, supplemented with the complete protease inhibitor cocktail tablets) The protein concentration of the cell lysates were determined by BCA Protein Assay kit (Thermo Scientific) according to the manufacturer's instructions. Equal amounts of proteins were separated by SDS-PAGE under reducing conditions. Western blotting and ECL detection were performed as described above.
For detection of phospho-STAT1/3 and phospho-ERK1/2, Ba/F3-gp130 cells, retrovirally transduced with cDNAs coding for the IL-15 and/or IL-15Rα-sushi fusion receptors, were washed three times with sterile PBS and starved for 6 h in serum-free DMEM. Cells were centrifuged, and the pellet was directly frozen in liquid nitrogen. Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM NaF, 1 mM Na3VO4, 1% IGEPAL [NP-40], and 1% Trition-X-100, supplemented with the complete protease inhibitor cocktail tablets; Roche Diagnostics). Determination of protein concentration, Western blotting, and ECL detection were performed as described above. The membranes were first probed with primary antibodies against phosphorylated STAT1, STAT3, and ERK1/2. After detection the membranes were stripped in stripping buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 0.1% β-mercaptoethanol for 30 min at 65°C, washed three times in TBST, blocked again, and probed for unphosphorylated STAT1, STAT3, and ERK1/2 proteins).
RESULTS
Ligand-independent IL-15/sIL-15Rα-sushi–induced gp130-Type Heterodimeric Receptors Are Constitutively Active
To achieve heterodimerization of gp130 with LIFR, OSMR, and WSX-1 and of GPL with OSMR, we decided to use the interaction of IL-15/IL-15Rα-sushi. The IL-15/sIL-15Rα-sushi proteins were chosen because of the high binding affinity of IL-15 to the IL-15Rα-sushi domain (hereafter referred to as sushi domain; Wang et al., 2005; Lorenzen et al., 2006). As shown in Supplemental Figure 1, A–F, we found that the Fos/Jun-leucine zippers (O'Shea et al., 1989) were not suitable to induce specific heterodimerization of gp130-type receptors. This was due to the unexpected finding that Fos-gp130-proteins are able to form biologically active, ligand-independent Fos-gp130-homodimers.
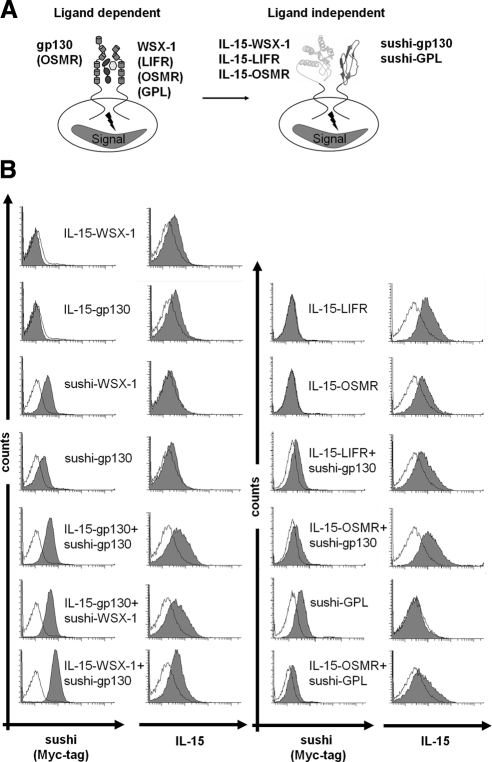
We substituted the entire extracellular portion of gp130 and WSX-1 and additionally of LIFR, and OSMR by the 148-amino acid sequence of IL-15 (PPL-IL-15) and the entire extracellular portion of gp130, WSX-1, and GPL by the 66-amino acid sequence of the sushi domain of the IL-15Rα as schematically illustrated in Figure 1A. To ease immunochemical detection of the chimeric proteins, a Myc-tag was placed immediately NH2-terminal of the sushi domain sequence. No immunotag was used in case of IL-15. The transmembrane and cytoplasmic domains of the receptors were left intact. The resulting chimeric proteins were named IL-15-gp130, IL-15-WSX-1, IL-15-LIFR, and IL-15-OSMR, as well as sushi-gp130, sushi-WSX-1, and sushi-GPL. The cDNAs coding for all receptors were transduced in Ba/F3-gp130 cells. Additionally double transduced Ba/F3-gp130 cells were generated for IL-15-gp130 + sushi-gp130, IL-15-gp130 + sushi-WSX-1, IL-15-WSX-1 + sushi-gp130, IL-15-LIFR + sushi-gp130, IL-15-OSMR + sushi-gp130, and IL-15-OSMR + sushi-GPL. Retrovirally transduced cell lines were analyzed by RT-PCR and FACS analysis, indicating expression of the respective mRNAs and surface expression of all receptor chimeras (Supplemental Figure 2 and Figure 1B). Ba/F3 cell were described not to be responsive to IL-15, because of lack of IL-2Rβ expression. Ba/F3 cells genetically complemented with IL2Rβ were able to proliferate in the presence of IL-15 (Demirci and Li, 2004). However, we stimulated Ba/F3-gp130 cells with Hyper-IL-6 (fusion protein of IL-6 and sIL-6R, 10 ng/ml), IL-15, or IL-15 in combination with soluble IL-15Rα-sushi domain, which has been shown to act agonistically on cells that express only IL-2Rβ and IL-2Rγ (Mortier et al., 2006).
Figure 1.
Schematic organization, idealized activation and surface expression of gp130/WSX-1, gp130/LIFR, gp130/OSMR wild type and gp130/WSX-1, gp130/LIFR, gp130/OSMR, and OSMR/GPL. (A) Scheme of the gp130 and WSX-1 receptors with the extracellular domains comprising immunglobulin-like domain (Ig, light green), cytokine binding domain (CBD, dark green), three fibronectin-like domains (FNIII, green), transmembrane domain (TM) and cytoplasmic domain (CD, black line). Dimerization is induced after binding of IL-27. All receptors were truncated 15 amino acids above the transmembrane domain and replaced by IL-15 or the sushi domain of IL-15Rα. (B) Cell surface expression of gp130, WSX-1, LIFR, OSMR, and GPL chimeric receptor in various stably transduced Ba/F3-gp130 cell lines analyzed by flow cytometry. The cell lines were labeled as described in Materials and Methods. Untransduced Ba/F3-gp130 cells were used as negative controls (unfilled histograms).
As depicted in Supplemental Figure 3A only Hyper-IL-6, but not IL-15 or IL-15 in combination with soluble IL-15Rα-sushi domain induced proliferation of Ba/F3-gp130 cells. Additionally, we performed coculture experiments using Ba/F3-gp130 cells transduced with a cDNA coding for GFP and Ba/F3-gp130 cells transduced with IL-15-gp130, IL-15-WSX-1 + sushi-gp130 or Fos-gp130. Depletion of Hyper-IL6 in our coculture experiments led to the death of the Ba/F3-gp130-GFP cells within 96 h, which could not be rescued by coculture with IL-15–presenting Ba/F3-gp130 cells (Supplemental Figure 3B). We conclude from these control experiments that Ba/F3-gp130 cells cannot be self-stimulated with IL-15 as part of our chimeric receptor chains.
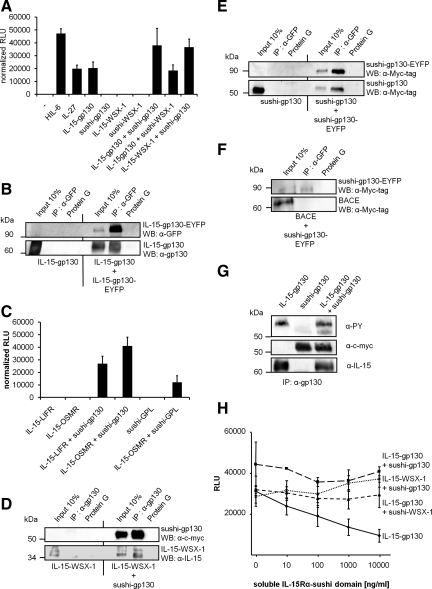
Next, we asked whether IL-15/IL-15Rα-sushi heterodimerization of chimeric receptors is sufficient to induce ligand-independent cell growth. Therefore, all retrovirally transduced Ba/F3-gp130 cell lines were cultured in medium lacking Hyper-IL-6. Surprisingly, we found that cells transduced with IL-15-gp130 alone were able to proliferate in the absence of Hyper-IL-6, indicating that this chimeric receptor forms active IL-15-gp130 homodimeric complexes (Figure 2A). To demonstrate the physical interaction of two IL-15-gp130 proteins, we performed coimmunoprecipitation experiments using lysates containing IL-15-gp130 and IL-15-gp130-EYFP. We used single- or double-transfected cells (IL-15-gp130 or IL-15-gp130 + IL-15-gp130-EYFP) and anti-GFP-antibodies for the coimmunoprecipitation of IL-15-gp130. As indicated in the left panel of Figure 2B, IL-15-gp130 was not detected after coimmunoprecipitation from lysates of single-transfected cells, indicating that IL-15-gp130 did not unspecifically interacted with anti-GFP-antibodies or protein G agarose. However IL-15-gp130 was coimmunoprecipitated with IL-15-gp130-EYFP as indicated on the right panel of Figure 2B. We conclude that IL-15-gp130 can form homodimers on the cell surface that induce gp130-dependent but cytokine-independent signaling. The unexpected IL-15-gp130 receptor activation could be explained by the tendency of IL-15 to form dimers and higher molecular aggregates as shown by size exclusion chromatography (Supplemental Figure 4A). This tendency to form dimers of IL-15 was also recently observed by others (Hanick et al., 2008).
Figure 2.
Biological activity of IL-15 and IL-15Rα-sushi gp130, WSX-1, LIFR, OSMR, and GPL chimeric receptor proteins. (A) Equal numbers of Ba/F3-gp130 cells stably transduced with IL-15-gp130, sushi-gp130, IL-15-WSX-1, sushi-WSX-1, IL-15-gp130 + sushi-gp130, IL-15-gp130 + sushi-WSX-1, and IL-15-WSX-1 + sushi-gp130 were cultured for 3 d in the absence of Hyper-IL-6. As a control Ba/F3-gp130 were cultured in the presence or absence of 10 ng/ml Hyper-IL-6 or IL-27. Proliferation was measured as indicated in Materials and Methods. (B) Detection of IL-15-gp130 homodimers by coimmunoprecipitation. COS-7 cells were transiently transfected with p409-IL-15-gp130 or p409-IL-15-gp130 and pEYFP-IL-15-gp130. Forty-eight hours after transfection cells were lysed and IL-15-gp130-EYFP was immunoprecipitated with anti-GFP mAbs. As a control lysates were incubated only with protein G agarose. Input, immunoprecipitation supernatant and protein G agarose control supernatant were separated by SDS-PAGE. Proteins were detected with anti-GFP mAbs and visualized by ECL detection. The membrane was stripped and probed with anti-gp130 Abs, and proteins were visualized by ECL detection. (C) Equal numbers of Ba/F3-gp130 cells stably transduced with IL-15-LIFR, IL-15-LIFR + sushi-gp130, IL-15-OSMR, IL-15-OSMR + sushi-gp130, sushi-GPL, and IL-15-OSMR + sushi-GPL were cultured for 3 d in the absence of Hyper-IL-6. Proliferation was measured as indicated in Materials and Methods. (D) Detection of heterodimeric interaction between IL-15-WSX-1 and sushi-gp130 by coimmunoprecipitation. COS-7 cells were transiently transfected with p409-IL-15-WSX-1 or p409-IL-15-WSX-1 and p409-sushi-gp130. Forty-eight hours after transfection cells were lysed and sushi-gp130 was immunoprecipitated with anti-gp130 Abs. As a control lysates were incubated only with protein G agarose. Input, immunoprecipitation supernatant and protein A agarose control supernatant were separated by SDS-PAGE. Proteins were detected with anti-c-myc mAbs and visualized by ECL detection. The membrane was stripped and probed with IL-15-gp130 Abs, and proteins were visualized by ECL detection. Detection of sushi-gp130 homodimers by coimmunoprecipitation. (E) COS-7 cells were transiently transfected with p409-sushi-gp130 or p409-sushi-gp130 and pEYFP-sushi-gp130. Forty-eight hours after transfection cells were lysed and sushi-gp130-EYFP was immunoprecipitated with anti-GFP mAbs. As a control, lysates were incubated only with protein G agarose. Input, immunoprecipitation supernatant and protein G agarose control supernatant were separated by SDS-PAGE. Proteins were detected with anti-GFP mAbs and visualized by ECL detection. The membrane was stripped and probed with anti-gp130 Abs, and proteins were visualized by ECL detection. (F) Specificity of coimmunoprecipitation analysis. COS-7 cells were transiently transfected with pcDNA3-BACE and pEYFP-sushi-gp130. Forty-eight hours after transfection cells were lysed, and sushi-gp130-EYFP was immunoprecipitated with anti-GFP mAbs. As a control lysates were incubated with only protein G agarose. Input, immunoprecipitation supernatant and protein G agarose control supernatant were separated by SDS-PAGE. Proteins were detected with an anti-Myc-tag mAbs and visualized by ECL detection. (G) Tyrosine-phosphorylation of IL-15-gp130 and sushi-gp130. COS-7 cells were transiently transfected with p409-IL-15-gp130 or p409-sushi-gp130 or p409-IL-15-gp130 and p409-sushi-gp130 and starved for the last 20 h before lysis. Forty-eight hours after transfection, COS-7 cells were treated with pervanadate as described in Materials and Methods and lysed rapidly. The gp130 chimeric proteins were immunoprecipitated with anti-gp130 Abs. Immunoprecipitation supernatant was separated by SDS-PAGE. Proteins were transferred onto PVDF membrane and were detected with an anti-phosphotyrosine (PY) mAbs and visualized by ECL detection. To confirm immunoprecipitation of IL-15-gp130 and sushi-gp130, Western blots against IL-15 or c-myc were also performed. (H) Equal numbers of Ba/F3-gp130 cells stably transduced with IL-15-gp130, IL-15-gp130 + sushi-gp130, IL-15-gp130 + sushi-WSX-1, and IL-15-WSX-1 + sushi-gp130 were cultured for 3 d in the absence of Hyper-IL-6 and increasing amounts of the soluble sushi domain. Proliferation was measured as indicated in Materials and Methods.
Importantly, Ba/F3-gp130 cells retrovirally transduced with sushi-gp130 or the IL-15 chimeras IL-15-WSX-1, IL-15-OSMR, and IL-15-LIFR showed no cytokine-independent proliferation (Figure 2A and C). These results may indicate that sushi-gp130 on its own does not form homodimers or ′inactive′ homodimers and that homodimers of WSX-1, OSMR, and LIFR induced by IL-15 homodimerization are not functionally active in terms of cytokine-independent induction of proliferation and STAT phosphorylation of the respective stably transduced Ba/F3-gp130 cells. Therefore we used Ba/F3-gp130 cells retrovirally transduced with sushi-gp130 and IL-15-WSX-1. Ba/F3-gp130-IL-15-WSX-1 + sushi-gp130 cells were able to proliferate cytokine-independently, indicating the formation of a biologically functional IL-15-WSX-1 and sushi-gp130 receptor complex. As expected, cells stably transfected with IL-15-gp130 and sushi-gp130 or with IL-15-gp130 and sushi-WSX-1 also proliferated cytokine-independently. Because the IL-15-gp130 receptor chimera was already able to induce cytokine-independent growth, we were not able to decide if the proliferation of these cells is dependent on homo- or heterodimerization of the fusion receptors. However, we concluded that the proliferation of Ba/F3-gp130-IL-15-WSX-1+sushi-gp130 was based on the formation of a functional ligand-independent receptor heterodimer consisting of IL-15-WSX-1 and sushi-gp130, because none of the Ba/F3-gp130 cell lines neither transduced with sushi-gp130 nor IL-15-WSX-1 showed cytokine-independent proliferation (Figure 2A).
Ba/F3-gp130 cells retrovirally transduced with sushi-gp130 were used to introduce the cDNAs coding for IL-15-LIFR or IL-15-OSMR, respectively. Importantly, only the double- but not the single-transduced cells lines were able to grow cytokine-independently (Figure 2C). IL-31 was shown to induce the formation of a GPL/OSMR heterodimer (Dillon et al., 2004). Here we show that sushi-GPL and IL-15-OSMR alone were biologically inactive, indicating that the cytokine-independent proliferation of Ba/F3-gp130 cells retrovirally transduced with both cDNAs coding for sushi-GPL and IL-15-OSMR was due to functional heterodimerization of sushi-GPL and IL-15-OSMR (Figure 2C).
To demonstrate the physical interaction of sushi-gp130 and IL-15-WSX-1 proteins, we performed coimmunoprecipitation experiments using lysates containing sushi-gp130 and IL-15-WSX-1. Again, we used single- or double-transfected cells (IL-15-WSX-1 or sushi-gp130+IL-15-WSX-1) and anti-gp130 antibodies for coimmunoprecipitation of sushi-gp130. As indicated in the left panel of Figure 2D, IL-15-WSX-1 was not detected after coimmunoprecipitation from lysates of single-transfected cells, indicating that IL-15-WSX-1 did not unspecifically interact with anti-gp130 antibodies or protein A agarose. IL-15-WSX-1 was coimmunoprecipitated with sushi-gp130 as indicated on the right panel of Figure 2D. However, sushi-gp130 could also be coimmunoprecipitated with sushi-gp130-EYFP (Figure 2E), even though this chimeric receptor combination does not induce cytokine-independent cell proliferation (Figure 2A) and STAT-phosphorylation (Figure 3A) and was not expected to interact with each other. We have performed a series of additional coimmunoprecipitation experiments with sushi-gp130 and Δcys-Fos-gp130 or IL-15-gp130 and Δcys-Fos-gp130. These proteins were also expected not to interact with each other. However, in all combinations applied, we could coimmunoprecipitate the other gp130 chimeric receptor chain (Supplemental Figure 4, B–E). Finally, we chose the transmembrane protein BACE, which does not belong to the gp130-superfamily, for coimmunoprecipitation experiments (Vassar et al., 2009). As depicted in Figure 2F, BACE does not interact with sushi-gp130. We conclude that in our experimental setting, coimmunoprecipitation of gp130 chimeric receptor proteins is specific and is not caused by overexpression, because overexpression does not lead to coimmunoprecipitation of sushi-gp130 and BACE. Previously, it has been shown that gp130 can form preformed dimers with gp130 and other receptors of the gp130-superfamily such as LIFR (Tenhumberg et al., 2006). The current view is that dimerization of the preformed gp130-receptor molecules is mediated by extracellular interaction of the receptors. Our data also point out that the intracellular part or the transmembrane domain of gp130 receptor chains can interact with each other.
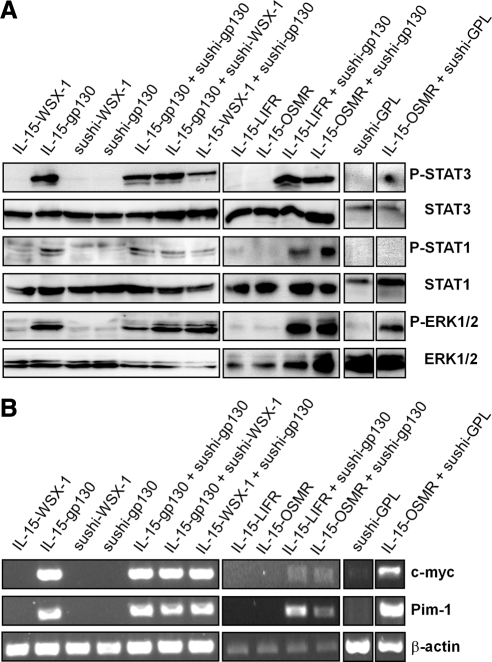
Figure 3.
Activation of STAT and ERK proteins and transcription of the STAT3 target genes c-myc and Pim-1 in Ba/F3-gp130 cells stably transduced with chimeric gp130-type receptors. (A) After 6-h serum starvation Ba/F3-gp130 cells stably transduced with IL-15-WSX-1, IL-15-gp130, sushi-WSX-1, sushi-gp130, IL-15-gp130 + sushi-gp130, IL-15-gp130 + sushi-WSX-1, IL-15-WSX-1 + sushi-gp130, IL-15-LIFR, IL-15-LIFR + sushi-gp130, IL-15-OSMR, IL-15-OSMR + sushi-gp130, sushi-GPL, and IL-15-OSMR + sushi-GPL were left untreated. Subsequently, cells were lysed and separated by SDS/PAGE. Proteins were transferred onto PVDF membranes. Proteins were detected with anti-phospho-STAT1, anti-STAT3, or anti-ERK1/2 Abs and visualized by ECL detection. The membranes were stripped, probed with anti-STAT1, anti-STAT3, or anti-ERK1/2 Abs, and proteins were visualized by ECL detection. (B) Analysis of STAT3 target gene expression of c-myc and Pim-1 in Ba/F3-gp130 cells stably transduced with IL-15-WSX-1, IL-15-gp130, sushi-WSX-1, sushi-gp130, IL-15-gp130 + sushi-gp130, IL-15-gp130 + sushi-WSX-1, IL-15-WSX-1 + sushi-gp130, IL-15-LIFR, IL-15-LIFR + sushi-gp130, IL-15-OSMR, IL-15-OSMR + sushi-gp130, sushi-GPL, and IL-15-OSMR + sushi-GPL by RT-PCR.
We analyzed the phosphorylation status of gp130-receptor chains in “nonactive” (sushi-gp130/sushi-gp130) versus “active” (IL-15-gp130/sushi-gp130 or IL-15-gp130) receptor combinations. We define active receptor chain combinations that induce cytokine-independent proliferation and STAT activation. We precipitated the gp130 chimeric proteins with anti-gp130 antibodies and analyzed tyrosine phosphorylation of gp130-chimeric receptor chains after pervanadate treatment, which blocks dephosphorylation. Our results clearly showed that only active receptor combinations lead to phosphorylation of gp130-receptor chains (Figure 2G). Only a faint phosphorylation signal was found in nonactive receptor combination, which also did not lead to subsequent STAT-phosphorylation (Figure 3A). We conclude that all chimeric gp130 receptor chains can form dimers/multimers independent of the extracellular part but dependent on the intracellular or transmembrane domain. Importantly, simple interaction of the intracellular or transmembrane domain does not lead to sustained receptor activation. This is illustrated by the cases of sushi-gp130 homodimers, which are inactive receptor chains, and of extracellular IL-15/sushi or IL-15/IL-15 dimerization, which leads to active receptor chains.
Next, we asked, whether the soluble IL-15Rα-sushi domain was able to inhibit the proliferation of Ba/F3-gp130 cells stably transduced with IL-15-gp130, IL-15-gp130 + sushi-gp130, IL-15-gp130 + sushi-WSX-1, or IL-15-WSX-1 + sushi-gp130. As shown in Figure 2H, the proliferation of cells stably transduced with IL-15-gp130 alone was inhibited by soluble IL-15Rα-sushi domain in a concentration-dependent manner. The proliferation of the other tested cell lines was not inhibited by soluble IL-15Rα-sushi domain. These data indicated that the proliferation of Ba/F3-gp130 cells stably transduced with IL-15-gp130 is mediated by IL-15 homodimerization, which was competed by soluble IL-15Rα-sushi to abrogate gp130 signaling. Interestingly, we could not inhibit the growth of cells stably transduced with a combination of IL-15- and sushi-receptor chimeras (gp130/gp130 and gp130/WSX-1). Duitman et al. (2008) showed that IL-15 formed complexes with the IL-15Rα already within the cell. The high-affinity constant of an IL-15/IL-15Rα-complex (Kd: 38 pM) is achieved by an electrostatic interface characterized by a very low dissociation rate (Koff: 1.4 × 10−5 s−1) (Mortier et al., 2006). Therefore it can be assumed that once formed complexes between IL-15 and IL-15Rα-sushi cannot be dissociated even by high molar excess of soluble IL-15Rα-sushi domain.
We tested whether the coexpression of the IL-15-sushi chimeras resulted in activation of the signaling proteins STAT1/3 and ERK1/2 in stably transduced Ba/F3-gp130 cells. For this purpose, cell lysates were analyzed by Western blot with anti-phospho STAT1, anti-phospho STAT3, and anti-phospho ERK1/2 specific antibodies. As depicted in Figure 3A, expression showed STAT3 and ERK1/2-phosphorylation and no STAT1-phosphorylation (Chattopadhyay et al., 2007). In contrast, cells only expressing IL-15-WSX-1, sushi-WSX-1, sushi-gp130, IL-15-LIFR, or IL-15-OSMR showed no STAT1, STAT3, and ERK1/2 phosphorylation (Figure 3A).
The transcription of the genes c-myc and Pim-1 has been shown to be dependent on STAT3 activation (Owaki et al., 2008). To ask whether the homo- and heterodimeric receptor complexes could activate target genes of IL-6-type cytokines, the cell lines were analyzed for c-myc and Pim-1 transcription by RT-PCR. As depicted in Figure 3B, only those cell lines that were able to proliferate in a cytokine-independent manner and that showed STAT1 and/or STAT3 and ERK1/2 phosphorylation also transcribed measurable amounts of c-myc and Pim-1 mRNAs.
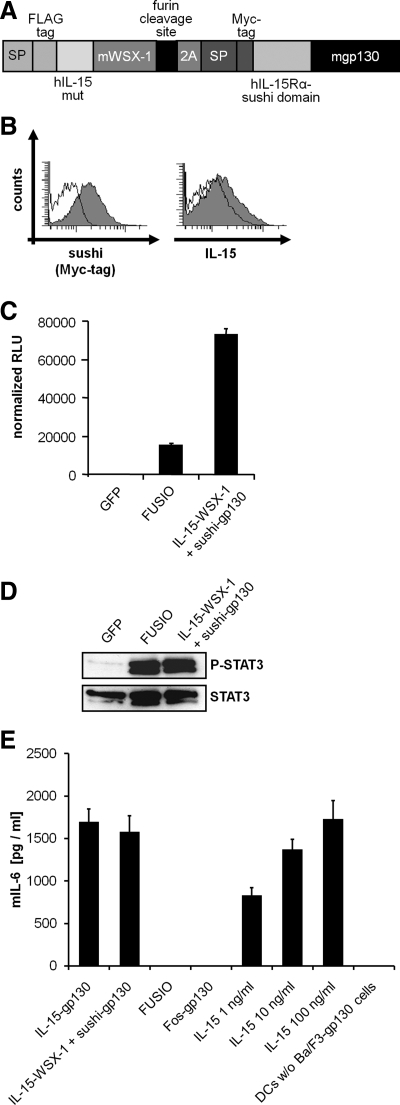
To facilitate the application of our system in vivo, we generated a second generation expression cDNA coding for human-IL-15/murine-WSX-1 and human-sushi/murine gp130. First of all we mutated the cDNA coding for human IL-15 at two sites, which led to two amino acid exchanges at IL-15/D8S and IL-15/Q108S, resulting in a lack of binding to the β- and γ-receptor subunits, respectively (Pettit et al., 1997). Moreover, both cDNAs coding for IL-15-WSX-1 and sushi-gp130 were combined in one transcript, separated by a 2A peptide coding sequence and a furin cleavage site. The 2A peptide from the foot-and-mouth-disease virus is a self-processing sequence to achieve expression of at least two separate proteins from a single open reading frame (Fang et al., 2005). The cleavage of the two proteins is thought to happen in a cotranslational process. We called the resulting single reading frame cDNA FUSIO, which consisted of a FLAG-tagged human IL-15 mutant, transmembrane and cytoplasmic region of murine WSX-1, furin cleavage site, 2A peptide, c-myc–tagged human IL-15Rα-sushi domain, and transmembrane and cytoplasmic region of murine gp130 (Figure 4A). Stably transduced Ba/F3-gp130 cells with the cDNA coding for FUSIO expressed both chimeric receptor chains on the cell surface (Figure 4B) and showed cytokine-independent proliferation and STAT3 phosphorylation, whereas control cells stably transduced with a cDNA coding for GFP were not able to proliferate in the absence of Hyper-IL-6 and did not show any STAT3 phosphorylation (Figure 4, C and D), demonstrating the functionality of our approach. However, stably transduced Ba/F3-gp130 cells with the cDNAs coding for IL-15-WSX1 and sushi-gp130 from separate genes showed stronger cytokine-independent proliferation than cells stably transduced with the combined IL15-WSX-1/sushi-gp130-FUSIO cDNA, even though intensity of STAT3 phosphorylation was comparable (Figure 4, C and D).
Figure 4.
Biological Activity of FUSIO. (A) Scheme of the FUSIO protein containing signal peptide from bovine preprolactin (bPPL), FLAG-tag, β- and γ-receptor-binding–deficient hIL-15, murine WSX-1 (15 amino acids of the extracellular domain, transmembrane domain, and cytoplasmic domain), furin cleavage site, 2A peptide, signal peptide of human gp130, Myc-tag, and murine gp130 (15 amino acids of the extracellular domain, transmembrane domain, and cytoplasmic domain). (B) Cell surface expression of gp130 and WSX-1 chimeric receptor encoded by FUSIO in stably transduced Ba/F3-gp130 cells analyzed by flow cytometry. The cell lines were labeled as described in Materials and Methods. Untransduced Ba/F3-gp130 cells were used as negative controls (unfilled histograms). (C) Equal numbers of Ba/F3-gp130 cells stably transduced with GFP, FUSIO, or IL-15-WSX-1 + sushi-gp130 from two cDNAs were cultured for 3 d in the absence of Hyper-IL-6. Proliferation was measured as indicated in Materials and Methods. (D) Activation of STAT-3 proteins in BaF/3-gp130 cells stably transduced with GFP, FUSIO, or IL-15-WSX-1 + sushi-gp130 from two cDNAs. After 6-h serum starvation Ba/F3-gp130 cells stably transduced with GFP, FUSIO, or IL-15-WSX-1 + sushi-gp130 were left untreated. Subsequently, cells were lysed, and 50 μg was separated by SDS/PAGE. Proteins were transferred onto a PVDF membrane and were detected with anti-phospho-STAT3 mAbs and visualized by ECL detection. The membrane was stripped and probed with anti-STAT3 mAbs, and proteins were visualized by ECL detection. (E) Coculture experiment of murine dendritic cells with Ba/F3-gp130 cell lines expressing different fusion receptors and quantification of IL-15–mediated murine IL-6 secretion. Murine dendritic cells were stimulated with LPS (10 ng/ml) overnight. The day after murine DCs were washed and cocultured with Ba/F-gp130 cells expressing either IL-15-gp130, IL-15-WSX-1 + sushi-gp130, FUSIO, or Fos-gp130 for 24 h. As a control murine dendritic cells were stimulated with 1, 10, and 100 ng/ml recombinant IL-15 or left untreated. After 24 h supernatants were analyzed for mIL-6 secretion.
Primary dendritic cells were described to produce IL-6 after stimulation with IL-15 (Brandt et al., 2003). Therefore, we analyzed whether our cell membrane–located IL-15-chimeric receptors would induce IL-6 production in primary murine dendritic cells through IL-15-trans-signaling via direct cell–cell contacts. Here we used the cell lines Ba/ F3-gp130-IL-15-gp130, Ba/F3-gp130-IL-15-WSX-1 + sushi-gp130, Ba/F3-gp130-Fos-gp130, and Ba/F3-gp130-FUSIO. As depicted in Figure 4E cocultures of primary murine dendritic cells with Ba/F3-gp130-IL-15-gp130, Ba/F3-gp130-IL-15-WSX-1 + sushi-gp130 cells but not with Ba/F3-gp130-Fos-gp130, induced a strong induction of IL-6 protein expression, indicating that the membrane located IL-15 protein is biologically active and induces IL-15-trans-signaling. Interestingly Ba/F3-gp130-FUSIO cells, which expressed an IL-15 mutant, which cannot bind to the β- and γ-receptor subunits, did not induce IL-6 protein expression via IL-15-trans-signaling. These results indicate that the FUSIO cDNA could potentially be used for the analysis of in vivo consequences of cytokine-independent, constitutive gp130/WSX-1 receptor activation.
We concluded from these experiments that the presence of the IL-15/IL-15Rα-sushi domain system is useful for the ligand-independent heterodimerization of all known gp130-type receptor complexes, leading to long-term cytokine-independent cell proliferation and constitutive activation of associated intracellular gp130-type receptor signaling pathways and target gene activation.
DISCUSSION
Here we describe a novel strategy to enforce cytokine-independent, constitutive signaling of heterodimeric gp130 receptor complexes. Our study reveals three major findings. First, replacing the extracellular domain of gp130, LIFR, OSMR, WSX-1, and GPL by IL-15/IL-15Rα-sushi domain is sufficient to heterodimerize gp130 with LIFR, OSMR, and WSX-1 as well as GPL with OSMR and as a consequence leading to sustained STAT1 and/or STAT3 and ERK1/2 phosphorylation and transcriptional activation of the gp130-type receptor target genes c-myc and Pim-1. Second, introduction of functional combinations of the chimeric receptor proteins into IL-6/sIL-6R–dependent Ba/F3-gp130 pre-B-cells confers sustained and long-term factor-independent growth of these cells, suggesting permanent activation of the heterodimeric receptor complexes. Third, exploiting the higher stability of an IL-15/sushi-IL-15Rα heterodimer when compared with IL-15 homodimers alone, we demonstrate that soluble sushi protein can be used to destabilize homodimeric IL-15-gp130 complexes, thereby interfering with the constitutive signaling activity of IL-15-gp130 but not with IL-15/IL-15Rα-sushi domain complexes, again demonstrating the exceptionally high affinity between IL-15 and IL-15Rα-sushi domain. These findings indicate that once formed IL-15/sushi complexes could not be dissociated by soluble IL-15Rα-sushi domain. Interestingly, IL-15 was recently described as a cytokine that can only leave the cell in complex with its IL-15Rα receptor (Duitman et al., 2008). Here, surface expression of IL-15 was detected in the absence of IL-15Rα (e.g., IL-15-gp130), demonstrating that IL-15 fused to a truncated gp130 can also leave the cell and that the driving force of IL-15 secretion is not its specific association with IL-15Rα but its fusion or association with a membrane receptor.
The approach taken assumes that the activation of gp130-, WSX-1–, LIFR-, OSMR-, and GPL-associated JAK kinases are triggered by close juxtapositioning of the cytoplasmic tails within gp130-type heterodimeric receptor complexes. gp130, like other cytokine receptors has two cytokine-binding domains (CBD) and additional subdomains such as an immunglobulin-like domain and fibronectin-like (FNIII) domains. It is believed that during activation, cytokine ligands reorganize their receptor complexes in such a way that the JAKs are correctly juxtaposed allowing their activation by cross-phosphorylation. There is increasing evidence that many cytokine receptors appear to exist as inactive, preassembled complexes at the cellular surface. This has been demonstrated for the receptors for erythropoietin (Livnah et al., 1999; Remy et al., 1999; Constantinescu et al., 2001), growth hormone (Frank, 2002), interferon γ (Krause et al., 2002), the βc signaling component in the receptors for IL-3, IL-5, and granulocyte-macrophage colony-stimulating factor (Carr et al., 2001), IL-6R (Schuster et al., 2003), and also gp130 (Giese et al., 2005; Tenhumberg et al., 2006). These data implied that ligand binding induces a local reorganization within the receptor complex, thereby triggering intracellular signaling.
The contact between IL-6-type cytokines and their gp130-type β-receptors occurs via the site II and site III on the respective cytokine. IL-6, IL-11, IL-27 (p28-subunit), CLC, CNTF, and NNT-1 require additional α-receptors such as IL-6R, IL11-R, EBI3, and CNTFR to mediate gp130-type β-receptor homo- or heterodimerization. This interaction occurs via site I of the cytokine. gp130-type receptors contact the sites II and III via cytokine-binding domains and/or the immunglobulin-like domains, which are separated from the plasma membrane by three fibronectin-like domains. At least for gp130 it is clear that deletion of one or more of the fibronectin-like domains leads to receptor molecules, which can still bind their ligands but fail to activate intracellular signal transduction cascades (Kurth et al., 2000). Single particle electron microscopy was used to visualize the entire extracellular hexameric IL-6/IL-6R/gp130 complex, containing the immunglobulin-like domain, the cytokine-binding domain and the three fibronectin-like domains (Skiniotis et al., 2005). The authors showed that the COOH terminal portions of the cytokine binding domain are ∼100 Å apart after ligand binding. At the same time the COOH terminal portions of the membrane proximal fibronectin-like domains are dimerized, which would lead to close juxtaposition of the transmembrane domains of the cell expressed receptor. In this view, the functional role of the fibronectin-like domains would be to assemble the transmembrane domains in close proximity. However, the molecular mechanism how cytokine binding is coupled to JAK and STAT activation is not understood. Currently no structural information for full-length cytokine receptors containing transmembrane or intracellular domains is available. Preformed receptor dimerization can be mediated by extracellular, transmembrane, or intracellular domains. Here, we present evidence that inactive dimerization of gp130 can be mediated by transmembrane or intracellular domains, which formally did not exclude that preformed dimer formation is mediated by the extracellular domains in vivo. Here, we have used gp130 chimeric proteins Δcys-Fos-gp130, IL15-gp130, and sushi-gp130, because a completely extracellular truncated gp130 molecule was not transported to the cell surface (Stuhlmann-Laeisz et al., 2006). Importantly, in our experimental setting, simple interaction of the intracellular or transmembrane gp130 receptor domains of does not lead to sustained receptor activation. Although we have not addressed this point in our study directly, our results imply that a functional active gp130-type receptor dimer needs to be positioned not only in close proximity to allow for full activation of the receptor, but also to be aligned in a specific manner (Greiser et al., 2002).
Recently, a molecular model based on crystal structure and electron microscopy data of the functional CNTF receptor has shown that most likely this receptor complex consists of a tetramer of CNTF, CNTF-R, gp130, and LIFR (Skiniotis et al., 2008). In contrast, the functional IL-6 receptor complex is believed to be formed of two molecules of IL-6, two molecules of IL-6R, and two molecules of gp130, thus forming a hexamer (Skiniotis et al., 2008). These two receptor complex set-ups are geometrically very different and it is therefore remarkable that the forced dimerization of all possible gp130 family receptor complexes leads to activation of intracellular signaling.
Our approach to heterodimerize gp130-type receptor complexes with the help of the IL-15/IL-15Rα-sushi domain is the first successful strategy to confer constitutive heterodimeric gp130-type receptor activation in the complete absence of any extracellular stimulation. Our findings add support to the current view that all IL-6-type cytokines induce conformational changes within the extracellular domains of heterodimeric gp130 receptor complexes that is necessary for optimal alignment and binding of the amino acids in the heterodimerization domain. Finally, this rearrangement stabilizes the heterodimerized receptor in a conformation that holds gp130-type receptor–associated JAK kinases in close proximity thereby facilitating signaling.
Constitutive activation of STAT3, the gp130-dependent transcription factor has been implicated in many human neoplastic malignancies including multiple myeloma (Catlett-Falcone et al., 1999; Rawat et al., 2000), prostate cancer, melanoma, ovarian cancer, renal carcinoma (Bromberg, 2002), and gastric cancer (Jenkins et al., 2005). Artificially dimerized and therefore constitutively active STAT3 was shown to possess oncogenic potential, and STAT3 was therefore designated as an oncogene (Bromberg et al., 1999). So far it is not understood which processes lead to the observed constitutive activation of STAT3. It is not unlikely that in such malignancies, constitutive activation of homodimeric gp130 receptor complex or a heterodimeric gp130/WSX-1, gp130/LIFR, gp130/OSMR, or GPL/OSMR receptor complex either by an autocrine or paracrine loop of stimulating cytokines or by genetic alterations causes the activation of STAT3 (Selander et al., 2004). In fact, recently, Rebouissou et al. (2008) described the discovery of marked activation of the IL-6 signaling pathway in inflammatory hepatocellular adenomas, which was directly caused by gain-of-function somatic mutations within the gp130 receptor chain. The gain-of-function gp130 variants were active in a ligand-independent manner, which is highly comparable to our homo- and heterodimeric receptor complexes. These data may directly connect the long-time observed STAT3 activation in various cancers with constitutive gp130-receptor signaling (Rebouissou et al., 2008). Our study might be the starting point to study the oncogenic potential of permanently activated heterodimeric gp130 receptor complexes resulting in constitutively activated STAT1/3 and ERK1/2, which in turn activate the RAS/RAF/mitogen-activated protein kinase (MAPK) pathway in vivo. Moreover, constitutively active heterodimeric gp130-type receptor complexes would provide novel molecular tools to establish animal models of diseases in which activated STAT1/3 is believed to play a role. To avoid IL-15–mediated responses of our receptor chimeras in vivo, we have used an IL-15 mutant, which cannot bind to the β- and γ-receptor chains of the IL-15 receptor signaling complex, but was still able to bind the IL-15Rα-sushi domain to induce cytokine-independent signaling of the chimeric receptor proteins (Pettit et al., 1997). Therefore, our constitutively active heterodimeric receptor molecules in combination with the 2A peptide technology (Fang et al., 2005) may be used to generate animal models to assess the consequences of constitutive pathophysiologic activation of gp130 heterodimeric receptor complexes at a cell autonomous level.
There are several potential advantages of using models based on ligand-independent, constitutive heterodimeric gp130-type receptor complexes rather than to rely on a constitutively active STAT3 mutant or to transgenic mice overexpressing the respective cytokines. First, activation of STAT3 does not reflect the activation pattern after heterodimeric gp130-type receptor activation because the RAS/MAPK pathway and the PI3K pathway is not activated. Second, systemic cytokine supply does not reflect situations in which chronic activation of the receptors is achieved only in certain tissues or areas of the body. Our receptor chimeras could now be used to induce gp130-type receptor signaling in a cell autonomous manner and if combined with tissue specific promoters in tissue specific transgenic mouse models. The use of the receptor chimeras therefore will provide novel insights into the consequences of uncontrolled activation of heterodimeric gp130-type receptor complexes and the resulting signaling cascades in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stefanie Schnell and Britta Hansen for excellent technical assistance and H. Gascan (INSERM, Angers, France) for the kind gift of a GPL cDNA. This work was supported by the Deutsche Forschungsgemeinschaft to S.R.-J. (SFB415, Project C6) and to J.S. (SFB415, Project B5) and by the German cluster of excellence “Inflammation at Interfaces.”
Abbreviations used:
- ERK1/2
extracellular signal-regulated protein kinase 1/2
- gp130
glycoprotein 130 kDa
- GPL
gp130-like receptor
- IL-6
interleukin 6
- IL-15
interleukin 15
- IL-15Rα
interleukin 15 receptor alpha
- LIFR
leukemia inhibitory factor receptor
- OSMR
oncostatin M receptor
- STAT
signal transducer and activator of transcription
- WSX-1
interleukin 27 receptor with a WSX signature motif (W-S-X-W-S).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-03-0240) on June 16, 2010.
REFERENCES
- Bernard J., et al. Identification of an interleukin-15alpha receptor-binding site on human interleukin-15. J. Biol. Chem. 2004;279:24313–24322. doi: 10.1074/jbc.M312458200. [DOI] [PubMed] [Google Scholar]
- Boulanger M. J., Chow D.-C., .revnova E. E., Garcia K. C. Hexameric structure and assembly of the interleukin-6/IL-6 α receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- Brandt K., Bulfone-Paus S., Foster D. C., Rückert R. Interleukin-21 inhibits dendritic cell activation and maturation. Blood. 2003;102:4090–4098. doi: 10.1182/blood-2003-03-0669. [DOI] [PubMed] [Google Scholar]
- Bromberg J. Stat proteins and oncogenesis. J. Clin. Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Bulfone-Paus S., Bulanova E., Budagian V., Paus R. The interleukin-15/interleukin-15 receptor system as a model for juxtacrine and reverse signaling. BioEssays. 2006;28:362–377. doi: 10.1002/bies.20380. [DOI] [PubMed] [Google Scholar]
- Carr P. D., Gustin S. E., Church A. P., Murphy J. M., Ford S. C., Mann D. A., Woltring D. M., Walker I., Ollis D. L., Young I. G. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. doi: 10.1016/s0092-8674(01)00213-6. [DOI] [PubMed] [Google Scholar]
- Catlett-Falcone R., et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Tracy E., Liang P., Robledo O., Rose-John S., Baumann H. Interleukin-31 and oncostatin-M mediate distinct signaling reactions and response patterns in lung epithelial cells. J. Biol. Chem. 2007;282:3014–3026. doi: 10.1074/jbc.M609655200. [DOI] [PubMed] [Google Scholar]
- Cichy J., Rose-John S., Pure E. Regulation of the Type II OSM receptor expression in human lung-derived epithelial cells. FEBS Lett. 1998;429:412–416. doi: 10.1016/s0014-5793(98)00643-7. [DOI] [PubMed] [Google Scholar]
- Constantinescu S. N., Keren T., Socolovsky M., Nam H., Henis Y. I., Lodish H. F. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc. Natl. Acad. Sci. USA. 2001;98:4379–4384. doi: 10.1073/pnas.081069198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci G., Li X. C. IL-2 and IL-15 exhibit opposing effects on Fas mediated apoptosis. Cell. Mol. Immunol. 2004;1:123–128. [PubMed] [Google Scholar]
- Dillon S. R., et al. Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat. Immunol. 2004;5:752–760. doi: 10.1038/ni1084. [DOI] [PubMed] [Google Scholar]
- Duitman E. H., Orinska Z., Bulanova E., Paus R., Bulfone-Paus S. How a cytokine is chaperoned through the secretory pathway by complexing with its own receptor: lessons from interleukin-15 (IL-15)/IL-15 receptor alpha. Mol. Cell. Biol. 2008;28:4851–4861. doi: 10.1128/MCB.02178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Qian J. J., Yi S., Harding T. C., Tu G. H., VanRoey M., Jooss K. Stable antibody expression at therapeutic levels using the 2A peptide. Nat. Biotechnol. 2005;23:584–590. doi: 10.1038/nbt1087. [DOI] [PubMed] [Google Scholar]
- Fischer M., Goldschmitt J., Peschel C., Brakenhoff J. P., Kallen K. J., Wollmer A., Grotzinger J., Rose-John S. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat. Biotechnol. 1997;15:145–145. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- Frank S. J. Receptor dimerization in GH and erythropoietin action–it takes two to tango, but how? Endocrinology. 2002;143:2–10. doi: 10.1210/endo.143.1.8607. [DOI] [PubMed] [Google Scholar]
- Gearing D. P., Ziegler S. F., Comeau M. R., Friend D., Thoma B., Cosman D., Park L., Mosley B. Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor. Proc. Natl. Acad. Sci. USA. 1994;91:1119–1123. doi: 10.1073/pnas.91.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese B., Roderburg C., Sommerauer M., Wortmann S. B., Metz S., Heinrich P. C., Muller-Newen G. Dimerization of the cytokine receptors gp130 and LIFR analysed in single cells. J. Cell Sci. 2005;118:5129–5140. doi: 10.1242/jcs.02628. [DOI] [PubMed] [Google Scholar]
- Greiser J. S., Stross C., Heinrich P. C., Behrmann I., Hermanns H. M. Orientational constraints of the gp130 intracellular juxtamembrane domain for signaling. J. Biol. Chem. 2002;277:26959–26965. doi: 10.1074/jbc.M204113200. [DOI] [PubMed] [Google Scholar]
- Hanick N. A., Rickert M., Varani L., Bankovich A. J., Cochran J. R., Kim D. M., Surh C. D., Garcia K. C. Elucidation of the interleukin-15 binding site on its alpha receptor by NMR. Biochemistry. 2008;46:9453–9461. doi: 10.1021/bi700652f. [DOI] [PubMed] [Google Scholar]
- Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M., Nakajima K., Hirano T. IL-6 cytokine family and signal transduction: a model of the cytokine system. J. Mol. Med. 1996;74:1–12. doi: 10.1007/BF00202068. [DOI] [PubMed] [Google Scholar]
- Jenkins B. J., et al. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling. Nat. Med. 2005;11:845–852. doi: 10.1038/nm1282. [DOI] [PubMed] [Google Scholar]
- Kallen K.-J., Grötzinger J., Lelièvre E., Vollmer P., Aasland D., Renné C., Müllberg J., Meyer zum Büschenfelde K.-H., Gascan H., Rose-John S. Receptor recognition sites of cytokines are organized as exchangeable modules: transfer of the LIFR binding site from CNTF to IL-6. J. Biol. Chem. 1999;274:11859–11867. doi: 10.1074/jbc.274.17.11859. [DOI] [PubMed] [Google Scholar]
- Ketteler R., Glaser S., Sandra O., Martens U. M., Klingmuller U. Enhanced transgene expression in primitive hematopoietic progenitor cells and embryonic stem cells efficiently transduced by optimized retroviral hybrid vectors. Gene Ther. 2002;9:477–487. doi: 10.1038/sj.gt.3301653. [DOI] [PubMed] [Google Scholar]
- Krause C. D., Mei E., Xie J., Jia Y., Bopp M. A., Hochstrasser R. M., Pestka S. Seeing the light: preassembly and ligand-induced changes of the interferon gamma receptor complex in cells. Mol. Cell. Proteom. 2002;1:805–815. doi: 10.1074/mcp.m200065-mcp200. [DOI] [PubMed] [Google Scholar]
- Kurth I., Horsten U., Pflanz S., Timmermann A., Kuster A., Dahmen H., Tacken I., Heinrich P. C., Muller-Newen G. Importance of the membrane-proximal extracellular domains for activation of the signal transducer glycoprotein 130. J. Immunol. 2000;164:273–282. doi: 10.4049/jimmunol.164.1.273. [DOI] [PubMed] [Google Scholar]
- Livnah O., Stura E. A., Middleton S. A., Johnson D. L., Jolliffe L. K., Wilson I. A. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- Lorenzen I., Dingley A. J., Jacques Y., Grotzinger J. The structure of the interleukin-15 alpha receptor and its implications for ligand binding. J. Biol. Chem. 2006;281:6642–6647. doi: 10.1074/jbc.M513118200. [DOI] [PubMed] [Google Scholar]
- Lutz M. B., Kukutsch N., Ogilvie A. L., Rössner S., Koch F., Romani N., Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- Mortier E., Quemener A., Vusio P., Lorenzen I., Boublik Y., Grotzinger J., Plet A., Jacques Y. Soluble interleukin-15 receptor alpha (IL-15R alpha)-sushi as a selective and potent agonist of IL-15 action through IL-15R beta/gamma hyperagonist IL-15 x IL-15R alpha fusion proteins. J. Biol. Chem. 2006;281:1612–1619. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Stafford W. F., Kim P. S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245:646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- Owaki T., Asakawa M., Morishima N., Mizoguchi I., Fukai F., Takeda K., Mizuguchi J., Yoshimoto T. STAT3 is indispensable to IL-27-mediated cell proliferation but not to IL-27-induced Th1 differentiation and suppression of proinflammatory cytokine production. J. Immunol. 2008;180:2903–2911. doi: 10.4049/jimmunol.180.5.2903. [DOI] [PubMed] [Google Scholar]
- Patel N., Herrman J. M., Timans J. C., Katelein R. A. Functional replacement of cytokine receptor extracellular domains by leucine zippers. J. Biol. Chem. 1996;271:30386–30391. doi: 10.1074/jbc.271.48.30386. [DOI] [PubMed] [Google Scholar]
- Pettit D. K., Bonnert T. P., Eisenman J., Srinivasan S., Paxton R., Beers C., Lynch D., Miller B., Yost J., Grabstein K. H., Gombotz W. R. Structure-function studies of interleukin 15 using site-specific mutagenesis, polyethylene glycol conjugation, and homology modeling. J. Biol. Chem. 1997;272:2312–2318. doi: 10.1074/jbc.272.4.2312. [DOI] [PubMed] [Google Scholar]
- Pflanz S., Hibbert L., Mattson J., D., Rosales R., Vaisberg E., Bazan J. F., Phillips J. H., McClanahan T. K., de Waal Malefyt R., Kastelein R. A. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J. Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- Rawat R., Rainey G. J., Thompson C. D., Frazier-Jessen M. R., Brown R. T., Nordan R. P. Constitutive activation of STAT3 is associated with the acquisition of an interleukin 6 independent phenotype by murine plasmacytomas and hybridomas. Blood. 2000;96:3514–3521. [PubMed] [Google Scholar]
- Rebouissou S., Amessou M., Couchy G., Poussin K., Imbeaud S., Pilati C., Izard T., Balabaud C., Bioulac-Sage P., Zucman-Rossi J. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2008;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I., Wilson I. A., Michnick S. W. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- Scheller J., Grötzinger J., Rose-John S. Updating IL-6 classic- and trans-signaling. Signal Transduction. 2006;6:240–259. [Google Scholar]
- Schuster B., Meinert W., Rose-John S., Kallen K.-J. The human interleukin-6 (IL-6) receptor exists as a preformed dimer in the plasma membrane. FEBS Lett. 2003;538:113–116. doi: 10.1016/s0014-5793(03)00154-6. [DOI] [PubMed] [Google Scholar]
- Selander K. S., Li L., Watson L., Merrell M., Dahmen H., Heinrich P. C., Muller-Newen G., Harris K. W. Inhibition of gp130 signaling in breast cancer blocks constitutive activation of Stat3 and inhibits in vivo malignancy. Cancer Res. 2004;64:6924–6933. doi: 10.1158/0008-5472.CAN-03-2516. [DOI] [PubMed] [Google Scholar]
- Skiniotis G., Boulanger M. J., Garcia K. C., Walz T. Signaling conformations of the tall cytokine receptor gp130 when in complex with IL-6 and IL-6 receptor. Nat. Struct. Mol. Biol. 2005;12:545–551. doi: 10.1038/nsmb941. [DOI] [PubMed] [Google Scholar]
- Skiniotis G., Lupardus P. J., Martick M., Walz T., Garcia K. C. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol. Cell. 2008;31:737–748. doi: 10.1016/j.molcel.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmann-Laeisz C., Lang S., Chalaris A., Paliga K., Sudarman E., Eichler J., Klingmüller U., Samuel M., Ernst M., Rose-John S., Scheller J. Forced dimerization of gp130 leads to constitutive STAT3 activation, cytokine independent growth and blockade of differentiation of embryonic stem cells. Mol. Biol. Cell. 2006;17:2986–2995. doi: 10.1091/mbc.E05-12-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga T., Kishimoto T. gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Tenhumberg S., Schuster B., Zhu L., Kovaleva M., Scheller J., Kallen K. J., Rose-John S. gp130 dimerization in the absence of ligand: preformed cytokine receptor complexes. Biochem. Biophys. Res. Commun. 2006;346:649–657. doi: 10.1016/j.bbrc.2006.05.173. [DOI] [PubMed] [Google Scholar]
- Vassar R., Kovacs D. M., Yan R., Wong P. C. The beta-secretase enzyme BACE in health and Alzheimer's disease: regulation, cell biology, function, and therapeutic potential. J. Neurosci. 2009;29:12787–12794. doi: 10.1523/JNEUROSCI.3657-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rickert M., Garcia K. C. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.