Abstract
Background
LIN-12/Notch signaling is important for cell-cell interactions during development, and mutations resulting in constitutive LIN-12/Notch signaling can cause cancer. Loss of negative regulators of lin-12/Notch activity has the potential for influencing cell fate decisions during development and the genesis or aggressiveness of cancer.
Methodology/Principal Findings
We describe two negative modulators of lin-12 activity in C. elegans. One gene, sel-11, was initially defined as a suppressor of a lin-12 hypomorphic allele; the other gene, cdc-42, is a well-studied Rho GTPase. Here, we show that SEL-11 corresponds to yeast Hrd1p and mammalian Synoviolin. We also show that cdc-42 has the genetic properties consistent with negative regulation of lin-12 activity during vulval precursor cell fate specification.
Conclusions/Significance
Our results underscore the multiplicity of negative regulatory mechanisms that impact on lin-12/Notch activity and suggest novel mechanisms by which constitutive lin-12/Notch activity might be exacerbated in cancer.
Introduction
LIN-12/Notch signaling plays multiple roles in vulval development in C. elegans. Canonical roles for this signaling system in vulval development are the specification of a single anchor cell (AC), which is required for vulval induction, and the specification of the 2° vulval precursor cell (VPC) type. Numerous feedback loops and functional redundancies make vulval development relatively insensitive to small perturbations in signaling pathways. Thus, an individual regulator often does not cause an overt phenotype when it is removed in an otherwise wild-type genetic background, but its function may be revealed when it is removed in a sensitized genetic background. In C. elegans, a few core components of LIN-12/Notch signaling have been identified in phenotype-based genetic screens, but most core components and modulators have been identified as suppressors or enhancers in sensitized backgrounds (reviewed in [1]).
Mutations resulting in constitutive or elevated LIN-12/Notch signaling can cause cancer (reviewed in [2], [3]). In principle, abrogation of any system of negative regulation of lin-12/Notch activity has the potential for contributing to the development or aggressiveness of cancer. For example, there is some evidence that increased expression of the human SEL-1 ortholog, SEL1L, correlates with a decrease in tumor aggressiveness (reviewed in [4]), but how this correlation relates to effects on Notch activity is not clear. A compelling example is afforded by the ubiquitin ligase SEL-10/Fbw7, which targets LIN-12/Notch directly for proteasome-mediated degradation ([5]; reviewed in [6]). Point mutations in the extracellular domain of NOTCH1 are one cause of T cell acute lymphoblastic leukemia (T-ALL); when patients with these mutations relapse after chemotherapy, they generally have mutations in FBW7. Patients presenting with NOTCH1 mutations that remove the Fbw7 binding site do not have Fbw7 mutations upon relapse. These observations have implicated the loss of the negative regulation of NOTCH1 in patients whose T-ALL tumors are drug-resistant after relapse [7].
Here, we describe two negative modulators of lin-12 activity. One gene, sel-11, was initially defined as a suppressor of a lin-12 hypomorphic allele [8]. The other, cdc-42, is a well-studied Rho GTPase that we find has an effect on lin-12 activity that appears to be distinct from other roles of cdc-42 in vulval development described previously [9]. Our results underscore the multiplicity of mechanisms that impact on lin-12/Notch activity and suggest novel mechanisms by which aberrations in lin-12/Notch activity might be exacerbated in cancer.
Results
The genetic analysis of modulators of lin-12 activity involves combinations with different lin-12 alleles. Such alleles reduce or elevate lin-12 activity, and their effects on hallmark cell fate decisions are shown in Fig. 1. We first describe the genetic analysis that establishes hrd-1 as sel-11. We then describe genetic interactions between cdc-42 and lin-12.
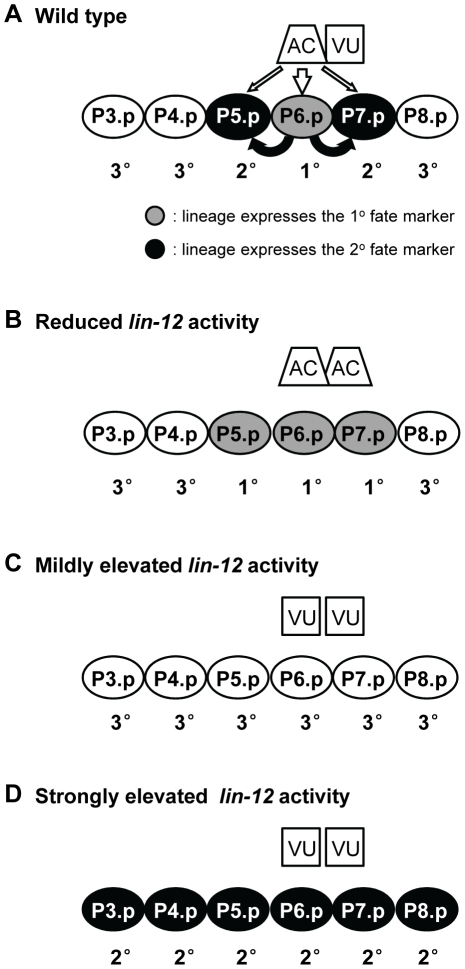
Figure 1. Signaling events that specify Vulval Precursor Cell (VPC) fates, and cell fate changes dependent on lin-12 activity levels.
(A) In wild-type C. elegans, vulval development is initiated in the early L3 stage when the anchor cell (AC) signals to the underlying VPCs via an inductive EGF signal, LIN-3 (white arrows). As a result, P6.p adopts the 1° cell fate. P6.p, in turn, sends a lateral signal (black arrows) to its neighboring VPCs, P5.p and P7.p, activating LIN-12 and causing them to adopt the 2° fate. During the AC/VU fate decision in the somatic gonad, the VU fate is determined by high lin-12 signaling. In wild-type animals, this results in 1 AC. (B) When lin-12 activity is reduced, a VU is often not specified, resulting in 2 ACs; in addition, as a result of increased inductive signaling from 2 ACs and/or reduced lateral signaling due to lower lin-12 activity, ectopic VPCs adopt the 1° fate at the expense of the 2° fate. The lin-12(n676n930) allele shows both aspects of this phenotype at 25°C [10]. (C) When lin-12 activity is mildly elevated, the AC fate is lost, and a VU is specified in its place. Since the inductive signal is lost, the underlying VPCs all adopt the 3° fate. lin-12(n379) and lin-12(n676) are examples of weak hypermorphic alleles, as is the lin-12(n676n930) allele at 15°C [10], [18]. (D) However, when lin-12 activity is strongly elevated, multiple VPCs are induced to adopt the 2° cell fate even though the AC is absent [18].
Demonstration that the negative regulator sel-11 encodes Hrd1p/Synoviolin
Mutations in sel-11 were originally identified in a genetic screen for negative regulators of lin-12 activity [8]. This screen was based on suppression of the egg-laying defective (Egl) phenotype caused by the hypomorphic allele, lin-12(n676n930) [10]; the properties of this allele are described further below. Extensive genetic analysis of the interactions between sel-11 and multiple lin-12 alleles in different cellular contexts established sel-11 as a negative regulator of lin-12 activity, but its molecular identity was not determined [8].
We molecularly identified sel-11 serendipitously because it is adjacent to the microRNA gene mir-61. A previous study in our laboratory concluded that mir-61 is expressed in response to LIN-12 activation and targets VAV-1, a negative regulator of lin-12 activity [11]. Our view of the role of mir-61 in vulval development was challenged by Miska et al. [12], who reported that a deletion of mir-61, nDf59, does not cause overt vulval lineage defects. We independently confirmed that nDf59 exhibits overtly normal vulval development by examining canonical 1°, 2°, and 3° cell fate markers (Table 1). However, as described below, our further analysis of nDf59 revealed that it removed sel-11, a negative regulator of lin-12 activity, and it is therefore problematic to use nDf59 to draw inferences about mir-61/250 function.
Table 1. Vulval cell fate marker expression in nDf59 is normal compared to control animals.
| 1° fate marker ( ayIs4 [egl-17::gfp] ): scored at Pn.px | ||||
| % P6.px only | % No expression | % P5.px and P6.px | N | |
| ayIs4 | 91.1 | 8.9 | 0 | 56 |
| ayIs4; nDf59 | 96.2* | 1.9 | 1.9 | 52 |
| 2° fate marker ( nIs106 [lin-11::gfp] ): scored at Pn.pxx | ||||
| % P5.pxx and/or P7.pxx | % P(5,6,7).pxx | N | ||
| nIs106 | 100 | 0 | 33 | |
| nDf59; nIs106 | 95.5 | 4.5# | 44 | |
| 3° fate marker ( arIs101 [K09H11.1::yfp] ): scored at Pn.px-Pn.pxx | ||||
| % Descendants of P3.p, P4.p, and P8.p | N | |||
| arIs101 | 100 | 53 | ||
| nDf59; arIs101 | 100& | 49 | ||
*1 animal had an anteriorly shifted AC above P5.px, and accordingly expression was seen in only P5.px.
By Fisher's exact test, not significantly different from nIs106 (P>0.5).
2/49 animals had an anteriorly shifted AC located above P5.p, and accordingly showed expression in descendants of P3.p, P7.p, and P8.p.
The possibility that nDf59 might affect a negative regulator of lin-12 activity arose when we examined the genetic interactions between nDf59 and lin-12(n676n930), a hypomorphic allele of lin-12 in which activity is compromised but not eliminated [10]. When lin-12(n676n930) hermaphrodites are grown at 25°C, they exhibit defects caused by reduced lin-12 activity: a highly penetrant egg-laying defect; a partially penetrant 2 AC defect; and, in hermaphrodites with 1 AC, a partially penetrant lateral signaling defect (P5.p and/or P7.p lose expression of a 2° fate marker) ([10]; Fig. 2B and 2C). At 15°C, lin-12(n676n930) retains mildly elevated lin-12 activity due to the presence of the n676 mutation. According to the proposed circuit, loss of mir-61 should decrease lin-12 activity; thus, we would expect that nDf59 might enhance phenotypes caused by lin-12(n676n930) at 25°C and suppress phenotypes of lin-12(n676n930) at 15°C. However, we obtained the opposite results, that nDf59 suppressed the loss-of-function phenotypes of lin-12(n676n930) at 25°C, and furthermore, enhanced the weak hypermorphic phenotype of this allele at 15°C (Fig. 2B and 2C). These results indicate that nDf59 increases lin-12 activity.
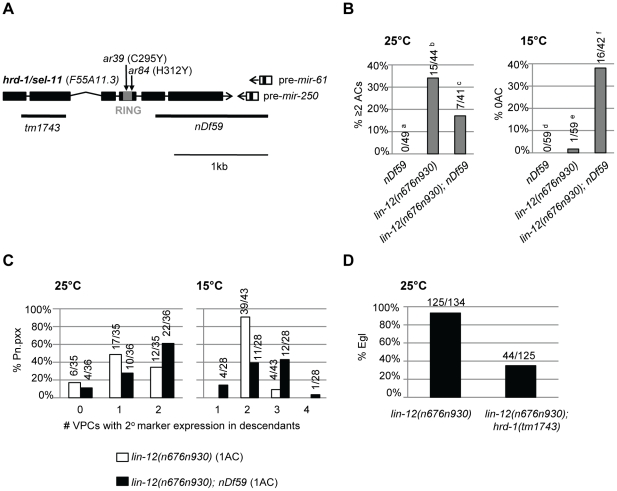
Figure 2. nDf59 enhancement of lin-12 activity and the identification of sel-11 as hrd-1.
(A) The mir-61/250 genomic region. The coding regions for the pre-microRNAs of mir-61 and mir-250 are separated by only 44bp, making it likely that these two miRNAs are cotranscribed. Expression of mir-61/250 in P5.p and P7.p depends on LIN-12 activation, mediated by LAG-1 Binding Sites in the 5′ flanking region [11]. The thick black lines indicate genomic regions deleted by tm1743 or nDf59 (see RESULTS). The RING domain of hrd-1/sel-11 is indicated by a grey box. Molecular lesions were found in the RING domain of hrd-1/sel-11 for two sel-11 alleles, ar39 and ar84, as indicated by arrows. These two alleles were found to contain point mutations in the RING finger domain of hrd-1, mutating either a histidine to a tyrosine (ar84, H312Y), or a cysteine to a tyrosine (ar39, C295Y), thereby mutating amino acids potentially important for the ubiquitination function of the RING domain. (B, C) lin-12(n676n930) has reduced activity at 25°C and mildly elevated activity at 15°C. The VU fate in the somatic gonad is determined by high lin-12 activity, as is the 2° vulval cell fate. (B) nDf59 decreases the number of anchor cells (AC) in the lin-12(n676n930) background at both 25°C and 15°C, which indicates an enhancement of lin-12 activity. The number of ACs observed for each strain is as follows: a 49/49 1 AC. b 29/44 1 AC, 13/44 2 ACs, 2/44 3 ACs. c 34/41 1 AC, 7/41 2 ACs. d 59/59 1 AC. e 56/59 1 AC, 2/59 2 ACs. f 25/42 1AC, 1/42 2 ACs. Full genotypes of animals scored for AC marker expression are unc-32(e189); arIs51[cdh-3::gfp]; nDf59 (GS5161), unc-32(e189)lin-12(n676n930); arIs51 (GS4894), and unc-32(e189)lin-12(n676n930); arIs51; nDf59 (GS5015). (C) nDf59 increases the number of VPCs expressing a 2° marker in the lin-12(n676n930) background at both 25°C and 15°C. When scoring for effects in the VPCs, we scored only animals with 1 AC to avoid the influence of altered numbers of ACs on VPC fate in the lin-12(n676n930) background. In a wild-type background, P5.p and/or P7.p express the 2° fate marker nIs106[lin-11::gfp] (Table 1, [11]). At 25°C, some lin-12(n676n930) animals have less than two VPCs that express a 2° fate marker; this phenotype is suppressed by nDf59. At 15°C, some lin-12(n676n930) animals have more than two; this phenotype is enhanced by nDf59. Full genotypes of animals scored for expression of the nIs106[lin-11::gfp] 2° fate marker are unc-32(e189)lin-12(n676n930); nIs106 (GS5014), and unc-32(e189)lin-12(n676n930); nDf59; nIs106 (GS5077). (D) hrd-1(tm1743), a null allele, suppresses the egg-laying defect (Egl) of lin-12(n676n930) at 25°C. Egl: egg-laying defective.
nDf59 had been reported to remove 1143 bases encompassing mir-61 and another microRNA, mir-250 [12]. We independently sequenced the nDf59 allele and confirmed the deletion breakpoints reported in WormBase (www.wormbase.org). During this process, we realized that, in addition to removing mir-61/250, nDf59 also removes part of the adjacent protein-coding gene, hrd-1 (Fig. 2A) (see MATERIALS AND METHODS). C. elegans hrd-1 [13] is the ortholog of yeast HRD1, which was identified in a genetic screen for genes that mediate HMG-CoA reductase degradation in Saccharomyces cerevisiae [14]. Another gene defined in the same screen, HRD3, corresponds to the C. elegans gene sel-1, the first molecularly characterized negative regulator of lin-12 [8], [15], [16].
In view of the functional relationship between HRD1 and HRD3 in yeast, we wondered if the genetic enhancement of lin-12(n676n930) by nDf59 may be explained if hrd-1, like HRD3/sel-1, is a negative regulator of lin-12 activity. This inference was confirmed by showing that hrd-1(tm1743), an apparent molecular null allele, suppresses the egg-laying defect (Egl) of lin-12(n676n930) at 25°C (Fig. 2D).
The map position of hrd-1 suggested that it might correspond to sel-11, previously defined as a negative regulator of lin-12 [8]. We therefore sequenced the two available alleles of sel-11, and found that they contain point mutations in the RING finger domain of HRD-1 (Fig. 2A). hrd-1 has been renamed sel-11, the first published name for the gene, in accordance with the accepted nomenclature convention in the field. The finding that two point mutations in the RING finger domain behave like a deletion of the gene suggests that the ubiquitin ligase activity of SEL-11 is important for negative regulation of lin-12 activity.
Genetic interactions implicate cdc-42 as a negative regulator of lin-12/Notch activity
As mentioned above, nDf59 deletes the microRNA mir-250, which appears likely to be cotranscribed with mir-61 in response to LIN-12 activation. Using the microRNA Registry release 2.0 prediction of the mir-250 sequence [17], we identified potential target genes computationally, using the same criteria that we used for mir-61 targets: we required that 3′ UTRs have at least 7 bases of perfect complementarity to the 5′ end of mir-250, with this putative “seed match” conserved in the C. briggsae orthologs of the C. elegans genes [11]. Our interest in cdc-42, a candidate identified in this way, was stimulated when we found that cdc-42(RNAi) enhanced lin-12 activity in sensitized backgrounds.
The backgrounds we used were afforded by the mild hypermorphs lin-12(n379) and lin-12(n676) (Fig. 3A). These mild hypermorphs lack an anchor cell and their VPCs generally behave as wild-type VPCs do in the absence of the inductive signal, i.e. they all adopt the 3° fate (Fig. 1). If lin-12(n379) or lin-12(n676) activity is increased, some or all of the VPCs adopt the 2° fate, resulting in a “Multivulva” phenotype that is visible in the dissecting microscope [8], [18], [19] (Fig. 3A). We verified that the Multivulva phenotype of cdc-42(RNAi); lin-12(n676) reflects an increased number of VPCs adopting the 2° fate using a transgenic marker (Fig. 3B). These observations are consistent with a role of cdc-42 as a negative regulator of lin-12 activity in the VPCs.
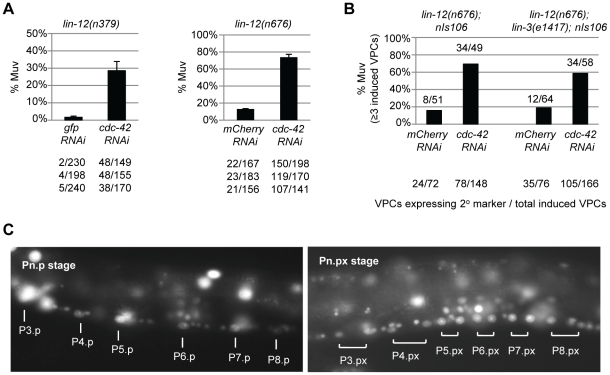
Figure 3. cdc-42(RNAi) enhances lin-12 activity in the VPCs.
(A) cdc-42(RNAi) enhances the Multivulva (Muv) phenotype of weak lin-12(d) alleles. Upon RNAi treatment, cdc-42 enhanced the Muv phenotype of lin-12(n379), as well as lin-12(n676). Muv is defined as three or more ventral protrusions. (B) The Multivulva phenotype of cdc-42(RNAi); lin-12(n676) hermaphrodites is not altered by lin-3(e1417), a mutation that reduces inductive signal expression. Animals were scored at the L3 Pn.pxx stage, where the VPCs have divided twice. The numbers of VPCs expressing the 2° fate marker nIs106[lin-11::yfp] are shown below. (C) A cdc-42 transcriptional reporter is expressed in the VPCs and their descendants. The transcriptional reporter was generated by fusing the 5′upstream region of cdc-42 to a 2nls-yfp reporter with an unc-54 3′UTR. Expression of the reporter was strong and uniform in all six VPCs at the Pn.p stage and was upregulated in the daughters of P5.p, P6.p and P7.p.
We note that in a lin-12(+) background, cdc-42(RNAi) had been shown to cause a supernumerary anchor cell, which induces an extra VPC to adopt the 1° fate [9]. However, the Multivulva phenotype of cdc-42(RNAi); lin-12(d) hermaphrodites appears to reflect enhanced lin-12(d) activity in the VPCs rather than supernumerary anchor cells or increased production of inductive signal. First, cdc-42(RNAi); lin-12(n676) hermaphrodites, like control lin-12(n676) hermaphrodites, do not have any anchor cells: in three independent experiments, cdc-42 RNAi performed on lin-12(n676); arIs51 animals resulted in 0/51, 0/80, 1/63 L3 animals that had AC marker expression, compared to 1/51, 0/73, 0/55 for control mCherry RNAi-treated animals. Second, the Multivulva phenotype of cdc-42(RNAi); lin-12(n676) hermaphrodites is not altered by lin-3(e1417), a mutation that reduces inductive signal expression (Fig. 3B). Third, as described above, in cdc-42(RNAi); lin-12(n676) hermaphrodites, extra VPCs adopt the 2° fate rather than the 1° fate.
We generated a transcriptional reporter containing the 5′ flanking region of cdc-42. This reporter drives strong, uniform expression in all six VPCs, and the level of expression appears to increase in the daughters of P5.p, P6.p and P7.p (Fig. 3C). The presence of cdc-42 in VPCs and their descendants is consistent with the multiple roles of cdc-42 in VPC development and specification inferred by Welchman [9] and the genetic interactions with lin-12 described here.
These observations are consistent with a role of cdc-42 as a negative regulator of lin-12 activity in the VPCs. Phenotypes caused by the expression of dominant-negative versions of Drosophila ortholog of cdc-42 have also suggested a role as a negative regulator of Notch signaling in wing development [20]. However, we were unable to obtain evidence that mir-250 negatively regulates cdc-42 in any assay (data not shown). When a revised mir-250 sequence was published (miRBase 10.1, [21]), it was evident that mir-250 has two additional adenines at its 5′ end that would preclude the appropriate seed match base pairing with cdc-42. Thus, although cdc-42 behaves genetically as a negative regulator of lin-12, it is not likely to be regulated by mir-250 in an analogous way to the LIN-12-mir-61-VAV-1 circuit.
Discussion
We have described here two negative regulators of lin-12/Notch activity, sel-11 and cdc-42. We discuss possible mechanisms by which these genes may influence lin-12/Notch activity in terms of our findings and relevant literature on their mammalian orthologs.
SEL-11/Hrd1p and a previously-described negative regulator, SEL-1/Hrd3p [15], [16], are both involved in the Hrd1p pathway of ER-associated degradation (ERAD). The Hrd1p pathway specifically targets proteins with misfolded lumenal domains for degradation. SEL-11/Hrd1p is the central E3 ubiquitin ligase, and SEL-1/Hrd3p acts in the recognition of terminally misfolded substrates [22], [23]. All lin-12 alleles that display genetic interactions with sel-1 and sel-11 [8], [15], [16] carry mutations that alter the extracellular domain, which would be positioned in the ER lumen during protein synthesis and folding [24], [25]. Loss of the Hrd1p quality control system likely increases the stability or export of these mutant LIN-12 proteins.
There is some evidence that increased expression of SEL1L, the human SEL-1 ortholog, correlates with a decrease in tumor aggressiveness (reviewed in [4]), but if this correlation reflects effects on Notch activity is not clear. In addition, the human SEL-11 ortholog, Synoviolin, is essential for development in mice and has been implicated in hyperproliferation of synovial tissues in rheumatoid arthritis and in the degradation of p53 and IRE1 [26], [27], [28]; furthermore, there appears to be multiple modes of crosstalk between Notch and p53 [29]. These observations are consistent with the possibility that loss of sel-11 might lead to elevated LIN-12/Notch activity in cancer.
C. elegans CDC-42 is a member of the Cdc42 subfamily of Rho family GTPases. Cdc42 has been implicated in many different cellular processes including polarity, cytoskeleton reorganization, vesicle trafficking, and signal transduction [30], [31]. The VPCs are polarized epithelial cells, with LIN-12 localized to the apical domain [32] and LET-23/EGFR localized to the basolateral domain [33]. Reciprocal negative regulation of endocytosis and trafficking of these receptors helps ensure proper pattern formation [11], [34], [35], [36], [37], [38], [39]. CDC-42 may contribute to this regulation through effects on VPC polarization and apical membrane organization, or through a general effect on endocytic traffic [40], [41], [42]. CDC-42 may affect LIN-12 signaling per se or through promoting the activity of the LET-23/EGFR-Ras-MAPK pathway: although genetic enhancement of lin-12(d) mutations was observed in the absence of the anchor cell, the normal source of inductive signal, it is possible that VAV-1 and CDC-42 affect a basal activity of LET-23 or another receptor tyrosine kinase that promotes Ras activity.
Cancer results from aberrations in cell-cell interactions and growth control, processes that are influenced by Notch activity. Depending on the cellular context, Notch can function as either a tumor suppressor (promoting differentiation) or proto-oncogene (promoting proliferation and/or suppressing apoptosis) [3]. In these roles, Notch crosstalks with p53 and Rho/CDC42 effectors [29], [43]. Our results raise the possibility that Cdc42 has effects on tumorigenesis through effects on Notch activity.
As mentioned in RESULTS, all of the lin-12 alleles used in this study carry missense mutations that alter the extracellular domain and cause constitutive LIN-12 activity; in some cases, the lin-12 alleles are composites of such activating mutations combined with second-site revertants that lower lin-12 activity. The missense activating alleles alter a negative regulatory domain [24] now known to be revealed under normal conditions by ligand binding (reviewed in [44]). Equivalent missense mutations in human Notch1 cause T-ALL and perhaps other cancers [45]. Whether SEL-11/Hrd1p/Synoviolin and CDC-42/Cdc42p act to promote the activity of wild-type LIN-12/Notch or only these missense mutant forms is not clear. Nevertheless, the clear effect on the LIN-12/Notch missense mutant forms associated with cancer suggest that the reduction in the activity of these negative regulators may be associated with cancer formation or progression.
Materials and Methods
Strains and genetic analysis
Caenorhabditis elegans var. Bristol strain N2 was the wild-type parent strain of all mutants and markers used. Key strains used herein were: GS4944 nDf59 (backcrossed to N2 4x), GS4420 ayIs4 [egl-17::gfp], GS5043 ayIs4; nDf59,
GS5775 nIs106 [lin-11p::gfp], GS5009 nDf59; nIs106, GS3804 pha-1(e2123); arIs101 [K09H11.1p::yfp], GS5010 nDf59; arIs101, GS5161 unc-32(e189); arIs51[cdh-3::gfp]; nDf59, GS4894 unc-32(e189)lin-12(n676n930); arIs51, GS5015 unc-32(e189)lin-12(n676n930); arIs51; nDf59, GS5014 unc-32(e189)lin-12(n676n930); nIs106, GS5077 unc-32(e189)lin-12(n676n930); nDf59; nIs106, GS3067 unc-32(e189)lin-12(n676n930), GS5186 unc-32(e189)lin-12(n676n930); hrd-1(tm1743) V, GS104 unc-32(e189)lin-12(n676n930); sel-11(ar39), GS257 unc-32(e189)lin-12(n676n930); sel-11(ar84), GS3196 lin-12(n379), GS23 lin-12(n676), GS4481 lin-12(n676); nIs106 [lin-11p::gfp], GS5098 lin-12(n676); lin-3(e1417); nIs106.
GE24 pha-1(e2123) was used as the recipient to create transgenes. All strains were grown using standard procedures at 20°C unless otherwise noted, except for strains with pha-1(+)-containing transgenes in the pha-1(e2123) background, which were maintained at 25°C. For strains that were scored at 25°C or 15°C, animals were maintained at the temperature of interest for at least two generations prior to scoring.
A strain carrying hrd-1(tm1743), backcrossed 4 times, was the gift of S. Arur and T. Schedl.
Transgenic lines
cdc-42p::2nls-yfp::unc-54 3′UTR reporter lines were made by injecting fusion PCR products [46] into pha-1(e2123) animals: pha-1(e2123); arEx843-847 [cdc-42p::2nls-yfp::unc-54 3′UTR (0.2 ng/ul), pBX (50 ng/ul)].
Fusion PCR products and plasmids
Fusion PCR was performed as reported [46]. ‘A’ primers and ‘B’ primers are forward and reverse primers, respectively, used to amplify promoter regions. ‘A*’ primers are nested forward primers used with the D* primer (see [46]) for the fusion step. The following primers were used to amplify the cdc-42 5′ region: AY56-A (CAATGGGCGATCAGGGTGTCTAT), AY56-A* (GCTAATAACCCGCACGGAGTAATG), AY56-B (agtcgacctgcaggcatgcaagctACTTGATCGTCTGCTTTTCGCCTG).
RNAi experiments
Feeding RNAi experiments were performed at 20°C as described [47], [48]. Briefly, gravid adults were bleached and the eggs were placed on plates seeded with HT115 cells expressing the dsRNA of interest. T7 polymerase expression in the HT115 cells had been induced with 6 mM IPTG for at least four hours at room temperature before plating the eggs. To score the number of pseudovulvae at the adult stage, animals were scored three days after eggs were placed on plates. To score at the L3 Pn.pxx stage, animals were scored roughly 45 hours after eggs were plated.
Imaging
All microscopy done on live animals was performed on a Zeiss Axioplan2 microscope, with a consistent exposure time used for each marker assayed.
Sequencing the hrd-1 genomic region of sel-11 alleles
The full genomic region of hrd-1, gene-to-gene, was amplified by PCR in two fragments for sequencing, using the primer pairs hrd-1-F1 (ATTGATATGGCACATTCAGAGCTTG)/hrd-1-R1 (GTTGTTGTGGAAATCCAAACTGATG), and hrd-1-F2 (CATTGTCTCCGCAGTTGGTTCC)/hrd-1-R2 (CATCGTCCTCTTTTTGTTCTGCTG). Two different alleles of sel-11, sel-11(ar39) and sel-11(ar84), were associated with two different alterations in the hrd-1 gene (see RESULTS). We note also that strains lacking sel-11 alleles but containing “sel(arX)”, a mutation that is in the background of some strains [8], did not contain any alterations in the hrd-1 gene.
Confirmation of deletion breakpoints for nDf59 and tm1743
Genomic DNA was extracted from backcrossed nDf59 and tm1743 strains, and the full genomic region of hrd-1 was amplified by PCR using the two primer pairs hrd-1-F1/hrd-1-R1, hrd-1-F2/hrd-1-R2 noted in the previous section. The deletion breakpoints for nDf59 and tm1743 reported in WormBase (www.wormbase.org) were verified by sequencing. In summary, nDf59 removes 1143 bp, and the deletion breakpoints for nDf59 are TGGATTTCCACAACAACCAGCTGGTGCC/GGAGGTGCTCAGCCTGG…GTTCTAGTCATTGCC/ATACGGAGGAAGGACTAAGC. tm1743 is a 473 bp deletion with a 38 bp insertion: the deletion breakpoints are TCATGTTCCAATTGCTCAAGTCT/ATTTTATTCGGAGATTTGAGAG… CTTAGGAATCCTCAATCTTGGGA/TAACAAAGCCGTTTACCTGCTCT, and CAAAGTTTCTTGTATATCTCATGTTCCAATTGCTCAAG is inserted. The published information about nDf59 only notes that mir-61/250 is deleted [12], but F55A11.3/hrd-1 is also affected (see WormBase and Fig. 2A).
Acknowledgments
We thank R. Ruiz and X. Zhou for technical assistance, S. Arur and T. Schedl for the backcrossed hrd-1(tm1743) strain and Maria Sallee and Marcus Vargas for critical reading of the manuscript. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center (CGC), which is supported by the NIH-National Center for Research Resources.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant to I.G. from the National Institutes of Health (R01CA095389). I.G. is an Investigator of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Greenwald I. LIN-12/Notch signaling in C. elegans. 2005. pp. 1–16. WormBook. [DOI] [PMC free article] [PubMed]
- 2.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biunno I, Cattaneo M, Orlandi R, Canton C, Biagiotti L, et al. SEL1L a multifaceted protein playing a role in tumor progression. J Cell Physiol. 2006;208:23–38. doi: 10.1002/jcp.20574. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard EJ, Wu G, Kitajewski J, Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 7.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundaram M, Greenwald I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics. 1993;135:765–783. doi: 10.1093/genetics/135.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welchman DP, Mathies LD, Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundaram M, Greenwald I. Genetic and phenotypic studies of hypomorphic lin-12 mutants in Caenorhabditis elegans. Genetics. 1993;135:755–763. doi: 10.1093/genetics/135.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasagawa Y, Yamanaka K, Ogura T. ER E3 ubiquitin ligase HRD-1 and its specific partner chaperone BiP play important roles in ERAD and developmental growth in Caenorhabditis elegans. Genes Cells. 2007;12:1063–1073. doi: 10.1111/j.1365-2443.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 14.Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant B, Greenwald I. The Caenorhabditis elegans sel-1 gene, a negative regulator of lin-12 and glp-1, encodes a predicted extracellular protein. Genetics. 1996;143:237–247. doi: 10.1093/genetics/143.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant B, Greenwald I. Structure, function, and expression of SEL-1, a negative regulator of LIN-12 and GLP-1 in C. elegans. Development. 1997;124:637–644. doi: 10.1242/dev.124.3.637. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- 19.de Souza N, Vallier LG, Fares H, Greenwald I. SEL-2, the C. elegans neurobeachin/LRBA homolog, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells. Development. 2007;134:691–702. doi: 10.1242/dev.02767. [DOI] [PubMed] [Google Scholar]
- 20.Baron M, O'Leary V, Evans DA, Hicks M, Hudson K. Multiple roles of the Dcdc42 GTPase during wing development in Drosophila melanogaster. Mol Gen Genet. 2000;264:98–104. doi: 10.1007/s004380000287. [DOI] [PubMed] [Google Scholar]
- 21.Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 23.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 24.Greenwald I, Seydoux G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature. 1990;346:197–199. doi: 10.1038/346197a0. [DOI] [PubMed] [Google Scholar]
- 25.Wen C, Greenwald I. p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans. J Cell Biol. 1999;145:1165–1175. doi: 10.1083/jcb.145.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao B, Lee SM, Chen A, Zhang J, Zhang DD, et al. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 2008;9:480–485. doi: 10.1038/embor.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamasaki S, Yagishita N, Sasaki T, Nakazawa M, Kato Y, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–122. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagishita N, Ohneda K, Amano T, Yamasaki S, Sugiura A, et al. Essential role of synoviolin in embryogenesis. J Biol Chem. 2005;280:7909–7916. doi: 10.1074/jbc.M410863200. [DOI] [PubMed] [Google Scholar]
- 29.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9:587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 31.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 32.Levitan D, Greenwald I. LIN-12 protein expression and localization during vulval development in C. elegans. Development. 1998;125:3101–3109. doi: 10.1242/dev.125.16.3101. [DOI] [PubMed] [Google Scholar]
- 33.Simske JS, Kaech SM, Harp SA, Kim SK. LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell. 1996;85:195–204. doi: 10.1016/s0092-8674(00)81096-x. [DOI] [PubMed] [Google Scholar]
- 34.Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–1839. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- 35.Shaye DD, Greenwald I. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature. 2002;420:686–690. doi: 10.1038/nature01234. [DOI] [PubMed] [Google Scholar]
- 36.Shaye DD, Greenwald I. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development. 2005;132:5081–5092. doi: 10.1242/dev.02076. [DOI] [PubMed] [Google Scholar]
- 37.Stetak A, Hoier EF, Croce A, Cassata G, Di Fiore PP, et al. Cell fate-specific regulation of EGF receptor trafficking during Caenorhabditis elegans vulval development. EMBO J. 2006;25:2347–2357. doi: 10.1038/sj.emboj.7601137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A. Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science. 2001;291:1055–1058. doi: 10.1126/science.1055642. [DOI] [PubMed] [Google Scholar]
- 39.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 40.Etienne-Manneville S. Cdc42—the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 41.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 42.Harris KP, Tepass U. Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J Cell Biol. 2008;183:1129–1143. doi: 10.1083/jcb.200807020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 46.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 47.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 48.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]