Abstract
The discrimination of n-alkyl saturated aldehydes during the early stage of odorant recognition by the rat I7 olfactory receptor (OR-I7) is investigated. The concentrations of odorants necessary for 50% activation (or inhibition) of the OR-I7 are measured by calcium imaging recordings of dissociated rat olfactory sensory neurons, expressing recombinant OR-I7 from an adenoviral vector. These are correlated to the corresponding binding free energies computed for a homology structural model of OR-I7 built from the crystal structure of bovine visual rhodopsin at 2.2 Å resolution.
Advancing our understanding of ligand binding sites in transmembrane G-protein-coupled receptors (GPCRs) is a problem of great research interest (1). GPCRs are major targets in therapeutic applications since they mediate a wide range of signal transduction processes from the extracellular environment to the interior of every cell (2). In particular, the GPCRs that detect and distinguish odorant molecules in olfactory sensory neurons remain poorly understood. Here, we investigate the structure of the binding site of odorants in the rat I7 olfactory receptor (OR-I7). We focus on the molecular interactions that discriminate n-alkyl saturated aldehydes with 5-11 carbon atoms from those with shorter, or longer, chains. The analysis is based on calculations of binding free energies and direct comparisons with experimental data on the concentrations necessary for half maximal activation (or inhibition) of OR-I7. Experimental data are obtained by calcium imaging recordings of dissociated rat olfactory sensory neurons expressing recombinant OR-I7 from an adenoviral vector, as previously reported (3, 4). The calculations are based on free energy perturbation (FEP) methods (5) implemented in NAMD (6) and ONIOM quantum mechanics/ molecular mechanics (QM/MM) hybrid methods (7, 8) as implemented with Gaussian 03 (9).
Olfactory receptors (ORs) are expressed in the olfactory sensory neurons of the nasal epithelium (2), and constitute the largest superfamily of GPCRs in the genome (e.g., humans express ~350 functional ORs, while rodents express more than 1000) (10, 11). Their main biological function is the molecular recognition of small hydrophobic molecules that can easily disperse into the air. Upon binding to specific ligands, these ORs initiate the transduction of chemical recognition into a neural activity providing animals with the sense of smell so crucial for survival, health and reproduction (e.g., the recognition of specific odors associated with the presence and quality of food, the presence of toxins, predators, prey, mates and competitors). In spite of their central role in biology, the molecular structures of ORs remain unknown. In particular, the challenge of expressing ORs in large amounts (12-14) and the intrinsic difficulties in crystallizing these transmembrane proteins that are typically embedded in hydrophobic environments has so far defied the development of X-ray crystal models. It is, therefore, essential to approach their structural characterization with a combination of alternative methods, including biochemical studies and computational modeling techniques.
The OR-I7 (entry M64386 in GenBank) (15) is one of the most extensively investigated ORs. It has been cloned, expressed in neurons, functionally characterized as activated by odorants (16) and studied by computational modeling (17-19). Libraries of ligands have been screened for activation of OR-I7 expressed from olfactory sensory neurons, including homologous series of n-alkyl acids, aldehydes, ketones and alcohols (3, 4, 12, 20). At least 30 odorants were found to activate OR-I7, all of them aldehydes with hydrocarbon chains of 5-11 carbon atoms. These studies provided valuable insights on the structural properties of agonists that seem to be critical for binding, including the presence of the aldehyde functional group, a certain range of molecular length as well as certain conformational flexibility and level of unsaturation in the carbon chain. However, the interactions and mechanisms for molecular recognition and discrimination of agonists at the OR-I7 binding pocket remain poorly understood. In this paper, we focus on the response of OR-I7 to n-alkyl saturated aldehydes as correlated to the balance of electrostatic and hydrophobic interactions during the early stage of odorant recognition, with emphasis on the discrimination of aldehydes with 5-11 carbons from those with shorter, or longer chains.
Our experimental values of binding free energies of n-alkyl saturated aldehydes, relative to octanal, were estimated as ΔΔG† = RT ln([Cn]/[C8]) from the concentrations [Cn] necessary for half-maximal activation of the OR-I7 (EC50), for Cn=C6-C11, or the concentration at which 50% inhibition of activation by simultaneously applied octanal (IC50) is achieved for C5. In addition, we computed the corresponding binding free energies using a homology structural model of the OR-I7 (Figure 1). The model was built from the crystal structure of bovine visual rhodopsin at 2.2 Å resolution (21) by sequentially mutating nonconserved amino acid residues and reoptimizing the configuration, after alignment of the primary sequence of the I7 receptor (15) with the sequence of bovine rhodopsin (21). The correlation between calculated and experimental binding free energies (Figure 2) validates the homology model, yielding fundamental insight on the nature of interactions in the OR-I7 binding pocket. These results complement recent studies on a series of conformationally restricted octanal mimics (20) and earlier studies of odorant binding (17, 18) based on homology models built from an electron density map of rhodopsin at 7.5 Å resolution (22).
FIGURE 1.
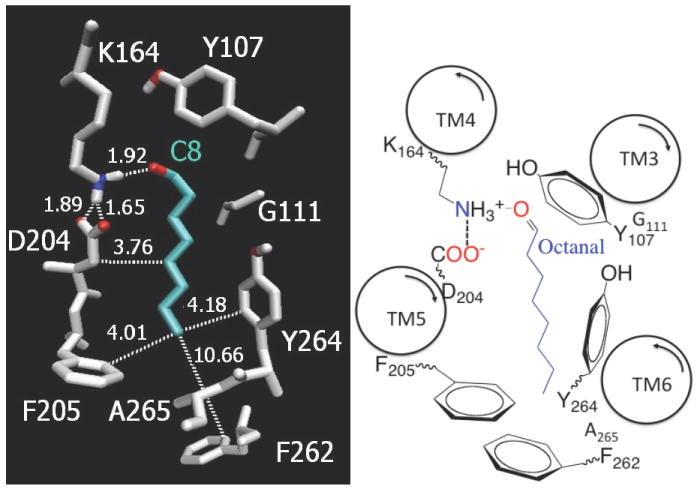
Left: Binding site of n-octanal (C8) in the rat OR-I7. Contact distances are indicated in Å. Color key: C (gray), O (red), N (blue) and aldehyde ligand (light blue). Right: Two-dimensional representation where the curved arrows inside each transmembrane (TM) α-helix indicate amino-carboxy polarity.
FIGURE 2.
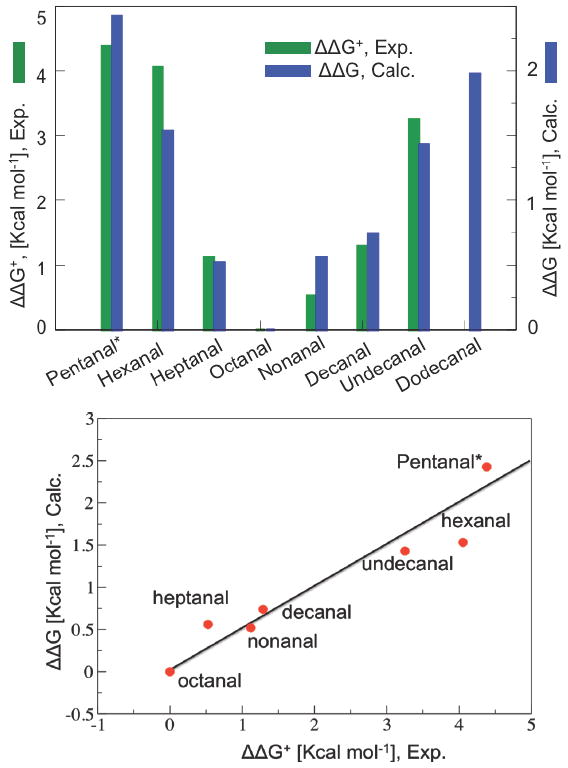
Correlation between calculated changes in binding free energies for saturated n-alkyl aldehydes, relative to octanal binding (ΔΔG in Kcal mol-1) and the experimental values ΔΔG† obtained from the EC50 for C6–C11, and the IC50 for C5 (marked *), including new data for nonanal (4.4±0.4μM); decanal: (16±1.7μM); and undecanal (446±16μM), and earlier data for hexanal (1736±440 μM), heptanal (12±3μM); and octanal 1(.9±0.2 μM).
Our analysis of binding energies for the homologous series of n-alkyl aldehydes indicates that odorants with saturated chains in the 5-11 carbon atom range are stabilized by hydrophobic interactions and hydrogen bonding to K164, a protonated amino acid residue forming a salt-bridge with the negatively charged counterion D204 (Figure 1). The dominant hydrophobic interactions involve contacts with several amino acid residues, including F205, Y107, A208, I209, G111, F262, Y264 and A265 (Figure 1). These results are consistent with the notion that functional groups and additional nonbonded interactions are determinants in ligand binding and activation of ORs (23). Hydrogen bonding with K164 is also consistent with earlier models (17, 18). However, the predicted localization of the binding site between TM5 and TM6 (supported by the strong correlation between experimental and predicted binding energies) is different from earlier proposals where the ligand was localized between TM6 and TM7 (e.g., with the hydrocarbon tail of octanal interacting with TM7).
All members of the homologous series of n-alkyl saturated aldehydes in the C5-C12 range exhibit the same binding motif with K164 (Figure 3), although with different binding energies due to differences in hydrophobic and steric interactions. The most stable member of the family is n-octanal (Figure 2) with a binding affinity of ~11 kcal mol-1. A difference in activation free energy of only 1.38 kcal mol-1 corresponds to an order of magnitude difference in bound lifetimes. Chains longer than octanal in this series (C9-C12) are penalized by steric interactions with F205 and Y264, due to the limited size of the binding pocket. For these larger odorants, conformational flexibility is critical to accommodate one or more kinks in the hydrocarbon chain and adopt bent conformations (Figure 3). Flexibility of the ligand, as well as relaxation of F205, are also essential aspects to maintain the hydrogen bond with K164 even when the ligand is buffeted by thermal fluctuations of amino acid residues (e.g., F205) with close contact interactions.
FIGURE 3.
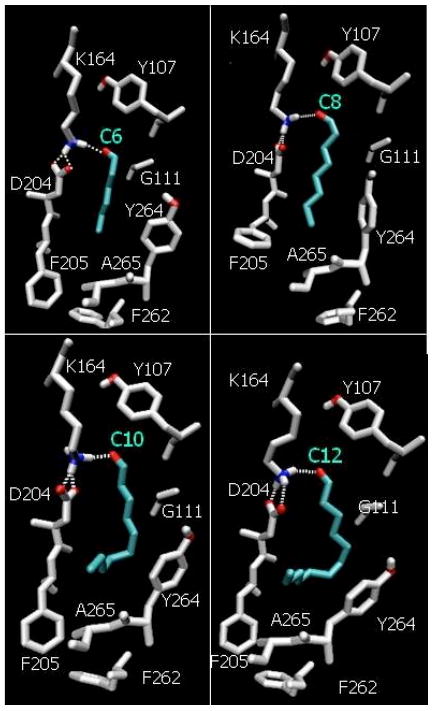
Binding of n-hexanal (C6), n-octanal (C8), n-decanal (C10), and n-dodecanal (C12) in our model of the rat OR-I7. Color key: C (gray), O (red), N (blue) and aldehyde ligand (light blue).
Chains shorter than octanal (C5-C7) are not penalized by steric interactions but they bind more weakly since they have less contact with amino acid residues (i.e., weaker hydrophobic interactions). In fact, n-alkyl saturated aldehydes smaller than n-pentanal have binding affinities comparable to room temperature thermal fluctuations. These results suggest that hydrogen-bonding and hydrophobic interactions in the OR-I7 can stabilize aldehydes with 5-11 carbon atoms so long as their hydrocarbon chains have sufficient conformational flexibility. The binding affinity is thus determined by the resulting balance of interactions, including hydrogen-bonding, hydrophobic interactions and steric repulsions.
It has been suggested that aldehyde odorants may covalently react with K164 in OR-I7 to form an imine (17, 18), which might explain the stringent aldehyde specificity of this OR. We performed a QM/MM analysis of the condensation reaction between n-octanal and K164 (see supporting information). Our calculations suggest that the resulting protonated Schiff-Base (pSB) would be thermodynamically favored when compared to its precursor hydrogen-bonded complex, and similarly stabilized by the negatively charged counterion D204. The calculated exothermicity of the condensation reaction (5 kcal mol-1) suggests that, if kinetically allowed, aldehydes could spontaneously form the imine pSB with K164 during the early stage of rat OR-I7 activation (17). These results are consistent with the correlated mutational analysis showing that K164 and D204 are critical to ligand binding, though the correlation does not necessarily discriminate between the covalent and non-covalent interactions (24). The resulting binding motif with K164 would be analogous to the pSB in rhodopsin where K296 (TM7) forms an imine with the aldehyde retinal (25). In rhodopsin, the pSB is also stabilized by a salt-bridge with a negatively charged counterion (i.e., E113) (26) that is crucial for the energetics of the primary activation step (27-29). A more definitive assessment of imine formation, however, awaits experimental evaluation possibly by cyanoborohydride reduction as performed on rhodopsin (30).
In summary, our homology model of the OR-I7 predicts binding free energies for n-alkyl saturated aldehydes in the C5-C11 range directly correlated with experimental data on activation of olfactory sensory neurons. We conclude that ligand binding, during the early stage of odorant recognition, is stabilized by hydrophobic interactions and hydrogen bonding to K164, a protonated amino acid residue that forms a salt-bridge with the negatively charged counterion D204. Saturated aldehydes shorter than pentanal do not bind since they are not sufficiently stabilized by hydrophobic interactions in the OR-I7. Aldehydes longer than octanal can bind when they have conformational flexibility to adopt bent conformations. Chains longer that undecanal, however, are too large to fit in the binding pocket.
Supplementary Material
Acknowledgments
V.S.B. acknowledges supercomputer time from NERSC and support from NSF CHE 0911520 and NIH 1R01-GM-084267-01. K.R. acknowledges support from NIH grants 1SC1GM083754 and G12 RR03060.
Footnotes
SUPPORTING INFORMATION AVAILABLE Description of computational methods and structural models. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Firestein S. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 3.Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. J Physiol (Lond) 2004;555:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araneda RC, Kini AD, Firestein S. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 5.Zwanzig RW. J Chem Phys. 1954;22:1420–1426. [Google Scholar]
- 6.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vreven T, Morokuma K. J Chem Phys. 2000;113:2969–2975. [Google Scholar]
- 8.Gascon JA, Leung SSF, Batista ER, Batista VS. J Chem Theory Comput. 2006;2:175–186. doi: 10.1021/ct050218h. [DOI] [PubMed] [Google Scholar]
- 9.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian, Inc. Wallingford, CT: 2004. [Google Scholar]
- 10.Niimura Y, Nei M. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niimura Y, Nei M. Proc Natl Acad Sci U S A. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krautwurst D, Yau KW, Reed RR. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 13.Saito H, Kubota M, Roberts RW, Chi QY, Matsunami H. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Von Dannecker LEC, Mercadante AF, Malnic B. Proc Natl Acad Sci U S A. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhao HQ, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- 17.Singer MS. Chem Senses. 2000;25:155–165. doi: 10.1093/chemse/25.2.155. [DOI] [PubMed] [Google Scholar]
- 18.Hall SE, Floriano WB, Vaidehi N, Goddard WA. Chem Senses. 2004;29:595–616. doi: 10.1093/chemse/bjh063. [DOI] [PubMed] [Google Scholar]
- 19.Lai PC, Singer MS, Crasto CJ. Chem Senses. 2005;30:781–792. doi: 10.1093/chemse/bji070. [DOI] [PubMed] [Google Scholar]
- 20.Peterlin Z, Li YD, Sun GX, Shah R, Firestein S, Ryan K. Chemistry & Biology. 2008;15:1317–1327. doi: 10.1016/j.chembiol.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 22.Schertler GFX. Eye. 1998;12:504–510. doi: 10.1038/eye.1998.138. [DOI] [PubMed] [Google Scholar]
- 23.Uchida N, Takahashi YK, Tanifuji M, Mori K. Nat Neurosci. 2000;3:1035–1043. doi: 10.1038/79857. [DOI] [PubMed] [Google Scholar]
- 24.Singer MS, Oliveira L, Vriend G, Shepherd GM. Receptors & Channels. 1995;3:89–95. [PubMed] [Google Scholar]
- 25.Nakayama TA, Khorana HG. J Biol Chem. 1991;266:4269–4275. [PubMed] [Google Scholar]
- 26.Sakmar TP, Franke RR, Khorana HG. Proc Natl Acad Sci U S A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gascon JA, Batista VS. Biophys J. 2004;87:2931–2941. doi: 10.1529/biophysj.104.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gascon JA, Sproviero EM, Batista VS. J Chem Theory Comput. 2005;1:674–685. doi: 10.1021/ct0500850. [DOI] [PubMed] [Google Scholar]
- 29.Gascon JA, Sproviero EM, Batista VS. Acc Chem Res. 2006;39:184–193. doi: 10.1021/ar050027t. [DOI] [PubMed] [Google Scholar]
- 30.Fager RS, Sejnowsk P, Abrahams Ew. Biochem Biophys Res Commun. 1972;47:1244–&. doi: 10.1016/0006-291x(72)90968-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


