Abstract
Nuclear receptors act as ligand-modulated transcription factors and orchestrate a plethora of cellular functions central to health and disease. Although studied for more than half a century, many mysteries surrounding the mechanism of action of nuclear receptors remain unresolved. Herein, using isothermal titration calorimetry (ITC) in conjunction with macromolecular modeling (MM), we provide evidence that the binding of ERα nuclear receptor to its DNA response element is coupled to proton uptake by two ionizable residues, H196 and E203, located at the protein-DNA interface. Alanine substitution of these ionizable residues decouples protonation and hampers the binding of ERα to DNA by nearly an order of magnitude. Remarkably, H196 and E203 are predominantly conserved across ~50 members of the nuclear receptor family, implying that proton-coupled equilibrium may serve as a key regulatory switch for modulating protein-DNA interactions central to nuclear receptor function and regulation. Taken together, our findings unearth an unexpected but a critical step in the molecular action of nuclear receptors and suggest that they may act as sensors of intracellular pH.
Keywords: Nuclear receptors, Estrogen receptor α, Estrogen response element, DNA-binding, Proton-coupled equilibrium, Isothermal titration calorimetry
Estrogen receptor α (ERα) is a member of a family of ligand-modulated transcription factors that have come to be known as nuclear receptors (NRs) (1–4). All members of NR family are evolutionarily related and share a core modular architecture comprised of a central DNA-binding (DB) domain flanked between an N-terminal trans-activation (TA) domain and a C-terminal ligand-binding (LB) domain (5–7). A typical scenario for the activation of nuclear receptors involves the secretion of lipophilic messengers such as hormones and vitamins by appropriate tissues. Upon their diffusion through the cell membrane, these ligands bind to the LB domain of nuclear receptors and allow their translocation into the nucleus to modulate gene expression (8–10). While the DB domain recognizes specific promoter elements, the LB domain additionally serves as a platform for the recruitment of a multitude of cellular proteins, such as transcription factors, co-activators and co-repressors, to the site of DNA transcription and thereby allowing nuclear receptors to exert their action at genomic level in a concerted fashion (11, 12). The TA domain is believed to be responsive to growth factors acting through the MAPK signaling and may further synergize the action of various co-activators and co-repressors recruited by the LB domain at the site of DNA transcription (13, 14). In this manner, nuclear receptors orchestrate a diverse array of cellular functions from embryonic development to metabolic homeostasis and their malfunction has been widely implicated in disease (5, 15–19).
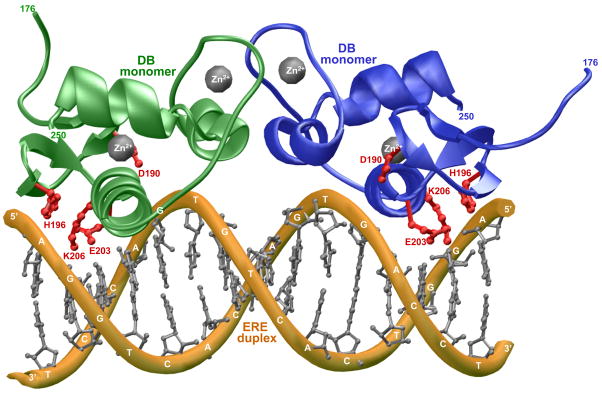
Discovered more than half a century ago, ERα mediates the action of estrogens such as estradiol and its hyperactivation leads to the genesis of large fractions of breast cancer (20–26). The DB domain of ERα binds as a homodimer to the AGGTCAnnnTGACCT consensus motif, termed estrogen response element (ERE), located within the promoters of target genes (27). DNA-binding is accomplished through a pair of tandem C4-type Zinc fingers, with each finger containing a Zn2+ ion coordinated in a tetrahedral arrangement by four highly conserved cysteine residues (28, 29). The first Zinc finger (ZF-I) within each monomer of DB domain recognizes the hexanucleotide sequence 5′-AGGTCA-3′ within the major groove at each end of the ERE duplex, whilst the second Zinc finger (ZF-II) is responsible for the homodimerization of DB domain upon DNA binding. Close scrutiny of 3D structure of the DB domain of ERα in complex with ERE duplex reveals that a triplet of ionizable residues — D190, H196 and E203 — either protrude deep into the comfort of the major groove at the protein-DNA interface or appear to reside within touching distance of DNA backbone (28) (Figure 1). Given that the placement of these residues in close proximity to the negatively charged phosphate backbone of DNA would be energetically unfavorable due to electrostatic repulsions, we hypothesized that the sidechain moieties of D190, H196 and E203 may become protonated upon the binding of ERα to DNA so as to neutralize the intermolecular repulsions and further enhance the favorable role of electrostatic forces central to driving protein-DNA interactions. This notion is further corroborated by the fact that the imidazole sidechain of H196 stacks against the highly basic sidechain of K206 — a scenario that could reduce the sidechain pKa of H196 and thereby rendering it more amenable to protonation upon the binding of DB domain to DNA. It is also of worthy note that the acidic sidechains of D190 and E203 are positioned in close proximity to each other within the DB domain. It is thus conceivable that the more acidic sidechain of D190 may be able to increase the pKa value of the sidechain of E203 allowing it to become protonated more easily upon the binding of DB domain to DNA at its own expense. Although both D190 and E203 are located at the protein-DNA interface, protonation of E203 would be more desirable as it directly inserts into the major groove of DNA.
Figure 1.
3D atomic model of the DB domain of human ERα in complex with ERE duplex containing the AGGTCAcagTGACCT consensus sequence. Note that the DB domain binds to DNA as a homodimer. One monomer of the DB domain is shown in green and the other in blue. The Zn2+ divalent ions are depicted as gray spheres and the sidechain moieties of D190, H196, E203 and K206 within the DB monomers are colored red. The DNA backbone is shown in yellow and the bases are colored gray for clarity. The numerals at the termini of DB monomers indicate the boundaries of DB domain within the amino acid sequence of human ERα.
In an effort to test our hypothesis, we have employed here isothermal titration calorimetry (ITC) in conjunction with macromolecular modeling (MM) to analyze the binding of DB domain of ERα to a 21-mer dsDNA oligo containing the ERE motif, hereinafter referred to as ERE duplex. Our data reveal that H196 and E203, but not D190, indeed become protonated upon the binding of ERα to DNA. Furthermore, alanine substitution of these ionizable residues decouples protonation and hampers the binding of ERα to DNA by nearly an order of magnitude. Our study suggests that the proton-coupled equilibrium observed here may be a general feature of the nuclear receptor family.
MATERIALS and METHODS
Protein preparation
The DB domain (residues 176–250) of human ERα (Expasy# P03372) was cloned into pET102 bacterial expression vector — with an N-terminal thioredoxin (Trx)-tag and a C-terminal polyhistidine (His)-tag — using Invitrogen TOPO technology. Trx-tag was included to maximize protein expression in soluble fraction, while the His-tag was added to aid in protein purification through Ni-NTA affinity chromatography. Additionally, thrombin protease sites were introduced at both the N- and C-termini of the DB domain to aid in the removal of tags after purification. The protein was subsequently expressed in Escherichia coli BL21*(DE3) bacterial strain (Invitrogen) and purified on a Ni-NTA affinity column using standard procedures. Briefly, bacterial cells were grown at 20°C in LB media supplemented with 50μM ZnCl2 to an optical density of 0.5 at 600nm prior to induction with 0.5mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial culture was further grown overnight at 20°C and the cells were subsequently harvested and disrupted using a BeadBeater (Biospec). After separation of cell debris at high-speed centrifugation, the cell lysate was loaded onto a Ni-NTA column and washed extensively with 20mM imidazole to remove non-specific binding of bacterial proteins to the column. The recombinant protein was subsequently eluted with 200mM imidazole and dialyzed against an appropriate buffer to remove excess imidazole. Further treatment on a Hiload Superdex 200 size-exclusion chromatography (SEC) column coupled in-line with GE Akta FPLC system led to purification of recombinant DB domain to apparent homogeneity as judged by SDS-PAGE analysis. The identity of recombinant protein was confirmed by MALDI-TOF mass spectrometry analysis. Final yield was typically between 5–10mg protein of apparent homogeneity per liter of bacterial culture. Treatment with thrombin protease significantly destabilized the recombinant DB domain and it appeared to be proteolytically unstable. For this reason, except for control experiments to ensure that the tags had no effect on the binding of DB domain to DNA, all experiments reported herein were carried out on recombinant fusion DB domain containing a Trx-tag at the N-terminus and a His-tag at the C-terminus. Protein concentration was determined by the fluorescence-based Quant-It assay (Invitrogen) and spectrophotometrically using an extinction coefficient of 29,045 M−1cm−1 calculated for the recombinant fusion DB domain using the online software ProtParam at ExPasy Server (30). Results from both methods were in an excellent agreement.
Site-directed mutagenesis
pET102 bacterial expression vector expressing wildtype DB domain of ERα was subjected to QuikChange Lightening kit (Stratagene) to generate single mutants D190A (DB_D190A), S193A (DB_S193A), H196A (DB_H196A), Y197A (DB_Y197A), S201A (DB_S201A), E203A (DB_E203A), K206A (DB_K206A), K210A (DB_K210A), R211A (DB_R211A), and the double mutant H196A/E203A (DB_AA). All mutant DB domains were expressed, purified and characterized as described above. When analyzed by size-exclusion chromatography (SEC) using a Hiload Superdex 200 column, all mutant DB domains exhibited virtually indistinguishable elution volumes to those observed for the wildtype DB domain, implying that the point substitution of specific residues did not lead to protein unfolding and that the mutant DB domains retained the compact globular fold characteristic of wildtype DB domain. These observations were further confirmed by circular dichroism (CD) analysis.
DNA synthesis
21-mer DNA oligos containing the ERE consensus site (AGGTCAnnnTGACCT) were commercially obtained from Sigma Genosys. The complete nucleotide sequences of the sense and antisense oligos constituting the ERE duplex is presented below:
5′-cccAGGTCAcagTGACCTgcg-3′
3′-gggTCCAGTgtcACTGGAcgc-5′
Oligo concentrations were determined spectrophotometrically on the basis of their extinction coefficients derived from their nucleotide sequences using the online software OligoAnalyzer 3.0 (Integrated DNA Technologies) based on the nearest-neighbor model (31). To obtain double-stranded DNA (dsDNA) annealed oligos to generate the ERE duplex, equimolar amounts of sense and antisense oligos were mixed together and heated at 95°C for 10min and then allowed to cool to room temperature. The efficiency of oligo annealing to generate dsDNA was close to 100% as judged by Native-PAGE and circular dichroism (CD) analysis.
ITC measurements
Isothermal titration calorimetry (ITC) experiments were performed on a Microcal VP-ITC instrument and data were acquired and processed using fully automized features in Microcal ORIGIN software. Measurements were repeated 3–4 times in Phosphate, Hepes, Tricine or Tris buffer. All buffers were made up to a final concentration of 50mM containing 5mM β-mercaptoethanol at pH 7.0. Additionally, 0–105mM NaCl was added to adjust the ionic strength of all buffers to 110mM. This ionic strength was high enough to prevent non-specific binding of DB domain of ERα to DNA, yet low enough to allow ITC analysis to be conducted with a high signal-to-noise ratio. Various constructs of the DB domain and the ERE duplex were prepared in an appropriate buffer and de-gassed using the ThermoVac accessory for 5min. The experiments were initiated by injecting 25 × 10μl aliquots of 50–100μM of ERE duplex from the syringe into the calorimetric cell containing 1.8ml of 5–10μM of DB domain solution at 25°C. The change in thermal power as a function of each injection was automatically recorded using Microcal ORIGIN software and the raw data were further processed to yield binding isotherms of heat release per injection as a function of molar ratio of ERE duplex to dimer-equivalent DB domain. The heats of mixing and dilution were subtracted from the heat of binding per injection by carrying out a control experiment in which the same buffer in the calorimetric cell was titrated against the ERE duplex in an identical manner. Control experiments with scrambled dsDNA oligos generated similar thermal power to that obtained for the buffer alone, implying that there was no non-specific binding of DB domain to non-cognate DNA. Experiments on the binding of thrombin-cleaved DB domain to DNA gave similar results to those conducted on recombinant fusion protein, implying that the tags had no effect on DNA-binding. However, due to poor stability and low yield of thrombin-cleaved DB domain and particularly in the case of mutant DB domains, all experiments reported herein were carried out on recombinant fusion DB domain containing a Trx-tag at the N-terminus and a His-tag at the C-terminus. Additionally, titration of a protein construct containing thioredoxin with a C-terminal His-tag (Trx-His) in the calorimetric cell with ERE duplex in the syringe produced no observable signal, implying that the tags do not interact with ERE duplex. In a similar manner, titration of wildtype or mutant DB domains in the calorimetric cell with Trx-His construct in the syringe produced no observable signal, implying that the tags do not interact with any of the wildtype or mutant DB domains. To extract observed affinity (Kobs) and observed enthalpy (ΔHobs), the binding isotherms were iteratively fit to the following built-in function by non-linear least squares regression analysis using the integrated Microcal ORIGIN software:
| [1] |
where q(i) is the heat release (kcal/mol) for the ith injection, n is the binding stoichiometry, V is the effective volume of protein solution in the calorimetric cell (1.46ml), P is the total dimer-equivalent concentration of DB domain in the calorimetric cell (μM) and L is the concentration of ERE duplex added (μM). The above equation is derived from the binding of a ligand to a macromolecule using the law of mass action assuming one-site model (32). Observed free energy of binding (ΔGobs) was calculated from the relationship:
| [2] |
where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (298 K). Observed entropic contribution (TΔSobs) to binding was calculated from the relationship:
| [3] |
The net change in the number of protons (Δm) absorbed or released per DB monomer upon binding to DNA and the intrinsic binding enthalpy (ΔHint) due to direct protein-DNA interactions and protonation of ionizable moities were calculated from the slope and y-intercept of ΔHobs-ΔHion plots by linear fits of data to the equation:
| [4] |
where ΔHobs is the observed binding enthalpy and ΔHion is the ionization enthalpy of each buffer. The ΔHion values of various buffers used were +1.22 kcal/mol (Phosphate), +5.02 kcal/mol (Hepes), +7.64 kcal/mol (Ticine) and +11.35 kcal/mol (Tris) (33–35).
Macromolecular modeling
Macromolecular modeling (MM) was employed to generate a 3D atomic model of the DB domain of ERα in complex with the ERE duplex using the MODELLER software based on homology modeling (36). The X-ray structure of DB domain of ERα in complex with a dsDNA oligo containing the ERE motif but with varying flanking sequences was used as a template (with a PDB code of 1HCQ). A total of 100 atomic models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The atomic model was rendered using RIBBONS (37) and the electrostatic surface potentials were generated using MOLMOL (38).
RESULTS and DISCUSSION
Binding of the DB domain of ERα to DNA is coupled to proton uptake
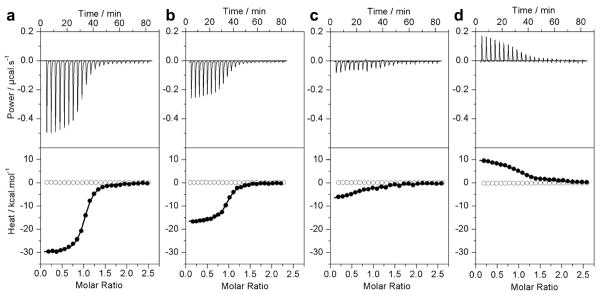
To test our hypothesis that the binding of ERα to DNA is coupled to proton uptake, we measured the binding of DB domain of ERα to ERE duplex in buffers of varying ionization enthalpies using ITC. Figure 2 shows representative ITC isotherms obtained from such measurements, while detailed thermodynamic parameters are reported in Table 1. It should be noted here that a classical test for ligand binding coupled to proton exchange is the dependence of observed enthalpy (ΔHobs) on ionization enthalpy (ΔHion) of the reaction buffer. Since different buffers are characterized by distinct ionization entahlpies, the observed enthalpy of ligand binding displays sharp dependence on the buffer employed due to varying contributions from coupled protonation/deprotonation. Our data indeed suggest that the ΔHobs for the binding of DB domain of ERα to DNA is highly dependent on the nature of buffer conditions employed (Figure 2). Thus, the ΔHobs of binding goes from being highly exothermic (−30.52 kcal/mol) in Phosphate buffer to being endothermic (+9.82 kcal/mol) in Tris buffer and thereby mirrors the ΔHion of the respective buffers ranging from +1.22 kcal/mol to +11.35 kcal/mol (33–35). This salient observation demonstrates that the binding of DB domain of ERα to DNA is directly coupled to proton uptake. Although such coupled equilibrium could result from the protonation of DNA bases, the sidechain moieties of D190, H196 and E203 within the DB domain must be considered as the major suspects for proton uptake due to their close proximity to the negatively charged phosphate backbone of DNA — a situation that would be inconceivable on thermodynamic grounds bar their protonation.
Figure 2.
Representative ITC isotherms for the binding of ERE duplex to the wildtype DB domain of ERα in Phosphate (a), Hepes (b), Tricine (c) and Tris (d) buffers at pH 7.0 and 25°C. The upper panels show the raw ITC data expressed as change in thermal power with respect to time over the period of titration. In the lower panels, change in molar heat is expressed as a function of molar ratio of ERE duplex to dimer-equivalent DB domain (●). The solid lines in the lower panels show the fit of data to a one-site model, as embodied in Eq [1], using Microcal Origin software. Note also that the DB domain shows no non-specific binding to scrambled dsDNA oligos (○).
Table 1.
Observed thermodynamic parameters for the binding of ERE duplex to the wildtype DB domain of ERα in various buffers at pH 7.0 and 25°C
| Kobs/nM | ΔHobs/kcal.mol−1 | TΔSobs/kcal.mol−1 | ΔGobs/kcal.mol−1 | |
|---|---|---|---|---|
| Phosphate | 43 ± 13 | −30.52 ± 0.27 | −20.44 ± 0.38 | −10.08 ± 0.20 |
| Hepes | 59 ± 6 | −17.22 ± 0.50 | −7.34 ± 0.53 | −9.88 ± 0.07 |
| Tricine | 238 ± 84 | −6.06 ± 1.54 | +3.02 ± 1.68 | −9.08 ± 0.23 |
| Tris | 336 ± 6 | +9.82 ± 0.64 | +19.10 ± 0.52 | −8.94 ± 0.12 |
The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from 3–4 independent measurements. All errors are given to one standard deviation.
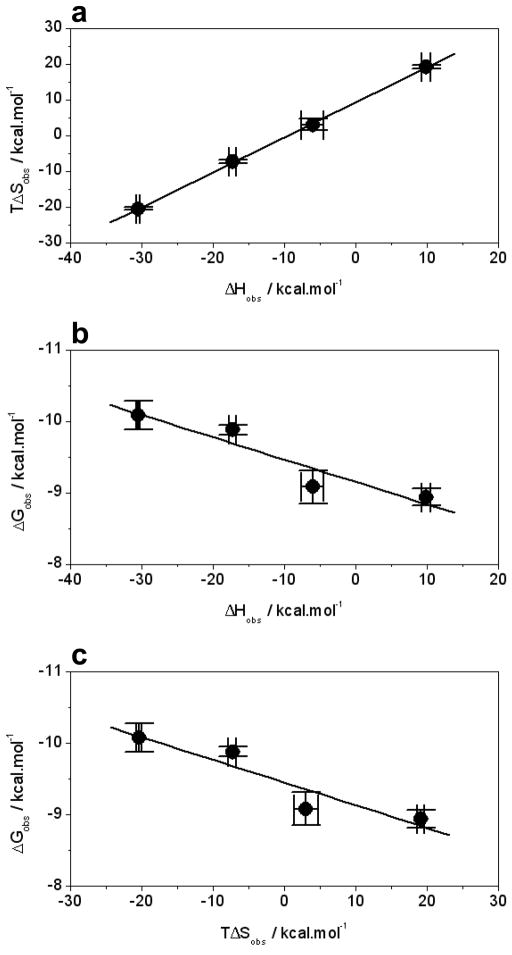
It is also of worthy note that while the enthalpy is favorable for the binding of ERα to DNA in Phosphate buffer, it contributes a substantial energetic penalty in Tris buffer (Table 1). In fact, close scrutinization of thermodynamic parameters observed for the binding of ERα to DNA in various buffers suggests that while enthalpy solely drives this protein-DNA interaction in Phosphate and Hepes buffers of low ionization enthalpies, entropy plays a major role in Tris and Tricine buffers of high ionization enthalpies. In particular, in the case of Tris buffer, it is the entropy that drives binding against the backdrop of enthalpic penalty. These observations imply that physiological settings with low ionization enthalpies are likely to favor the binding of ERα to DNA while the opposing conditions may be somewhat inhibitory. This is indeed corroborated by the fact that the binding affinity for ERα-DNA complexation drops by nearly an order of magnitude from 43nM in Phosphate buffer to 336nM in Tris buffer (Table 1). It should be borne in mind that the favorable enthalpic contributions to binding largely result from the release of heat upon the formation of tight electrostatic interactions, hydrogen bonding and hydrophobic contacts between protein and DNA. Thus, in buffers of low ionization enthalpy, the favorable enthalpic contribution is only slightly offset to compensate for proton-coupled equilibrium rendering enthalpy as the sole driving force accompanied by entropic penalty. In contrast, in buffers of high ionization enthalpy, the favorable enthalpic contribution is largely offset and even overridden by the enthalpic penalty due to the proton-coupled equilibrium with entropy either contributing favorably or serving as the sole driving force at the expense of enthalpy.
This reciprocal relationship between enthalpy and entropy lies in the enthalpy-entropy compensation phenomenon (39–43) — macromolecular interactions are compensated by equal but opposing entropic changes such that there is little or no net gain in the overall free energy. Thus, buffers of high ionization enthalpy gain a substantial increase in entropy upon the release of a proton, presumably due to an increase in the degrees of freedom that become available to water molecules after being freed from their hydration shell surrounding the exchangeable proton prior to its release. However, in the case of buffers of low ionization enthalpy, the exchangeable proton would be expected to be more “economically” hydrated such that the release of water molecules from the rather small hydration shell contributes relatively little to the overall entropy gain but at the same time draws less heat to be removed. Such enthalpy-entropy compensations for the binding of DB domain to ERE duplex in various buffers are illustrated in Figure 3a. Consistent with the foregoing arguments, it should also be noted that while the increase in the binding affinity of the DB domain to DNA correlates with overall favorable enthalpy change in various buffers, the increase in favorable entropy change seems to oppose such protein-DNA interactions (Figures 3b and 3c).
Figure 3.
Inter-dependence of observed enthalpic change (ΔHobs), entropic change (TΔSobs) and free energy change (ΔGobs) for the binding of ERE duplex to the wildtype DB domain of ERα in various buffers. (a) ΔHobs-TΔSobs plot. (b) ΔHobs-ΔGobs plot. (c) TΔSobs-ΔGobs plot. Note that the solid lines represent linear fits to the data in all plots. All error bars were calculated from 3–4 independent measurements and are given to one standard deviation.
Residues H196 and E203 serve as sole proton acceptors upon the binding of ERα to DNA
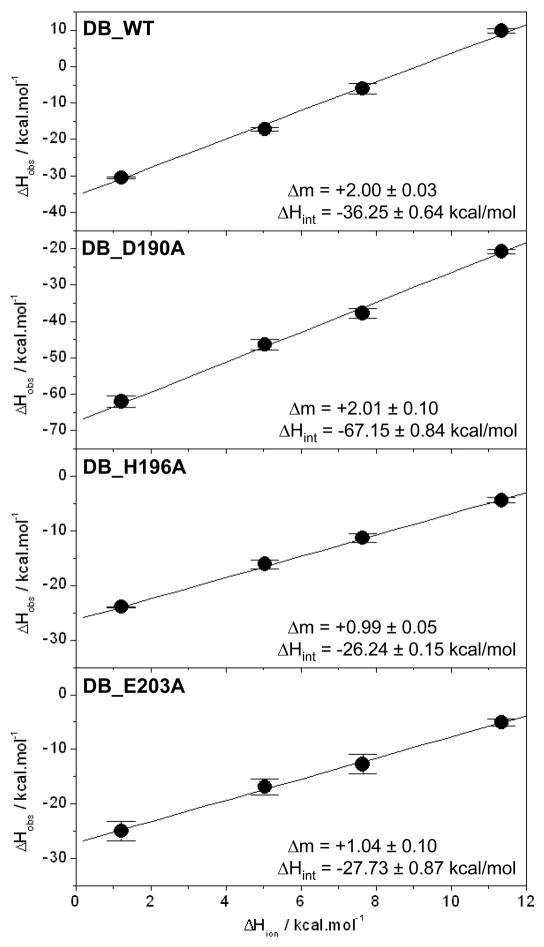
For processes in which ligand binding is coupled to proton exchange, the observed enthalpy (ΔHobs) is related to the ionization enthalpy (ΔHion) by the relationship ΔHobs=2ΔmΔHion+ΔHint, where Δm is the net change in the number of protons absorbed or released per DB monomer upon binding to DNA and ΔHint is the intrinsic binding enthalpy due to direct protein-DNA interactions and protonation of ionizable moities. A plot of ΔHobs versus ΔHion should thus yield a linear curve with the slope 2Δm and y-intercept equal to ΔHint. As shown in Figure 4, such analysis reveals that the binding of wildtype DB domain (DB_WT) of ERα to DNA results in the uptake of two protons per DB monomer. It should be noted here that a positive slope equates to proton uptake and a negative slope to proton release in this analysis. The fact that the binding of each monomer of DB domain to DNA is coupled to a net uptake of two protons implies that at least two of the three possible residues in D190, H196 and E203 may serve as proton acceptors. Could it be possible that only two of these residues are involved in proton uptake, or do all three residues fractionally contribute to a net uptake of two protons?
Figure 4.
Dependence of observed enthalpy (ΔHobs) as a function of ionization enthalpy (ΔHion) of various buffers upon the binding of ERE duplex to the wildtype DB domain (DB_WT), the D190A single mutant of DB domain (DB_D190A), the H196A single mutant of DB domain (DB_H196A) and the E203A single mutant of DB domain (DB_E203A) of ERα at pH 7.0 and 25°C. The ΔHion of various buffers used were +1.22 kcal/mol (Phosphate), +5.02 kcal/mol (Hepes), +7.64 kcal/mol (Ticine) and +11.35 kcal/mol (Tris) (33–35). The solid lines within each panel represent fit of data points to Eq [4]. Note that the net change in the number of protons (Δm) absorbed or released per DB monomer upon binding to DNA and the intrinsic binding enthalpy (ΔHint) due to direct protein-DNA interactions and protonation of ionizable moieties for each DB construct are provided within the corresponding panels. Error bars were calculated from 3–4 independent measurements. All errors are given to one standard deviation.
To address this question, we introduced single alanine substitutions at positions D190, H196 and E203 within the DB domain and then conducted the binding of these mutant domains to ERE duplex using ITC. The ΔHobs-ΔHion plot for the binding of D190A mutant of DB domain (DB_D190A) to DNA reveals that there is no net change in the number of protons exchanged relative to DB_WT domain (Figure 4), implying that the residue D190 is most likely not responsible for the proton-coupled equilibrium observed here. In striking contrast, the ΔHobs-ΔHion plots for the binding of H196A (DB_H196A) and E203A (DB_E203A) mutants of the DB domain to DNA reveal that only one proton is exchanged in each case (Figure 4), arguing strongly that the residues H196 and E203 are the sole sites of protonation.
Table 2 provides complete thermodynamic parameters for the binding of wildtype and various mutants of the DB domain to ERE duplex in Phosphate buffer. It is clearly evident from these data that while the D190A mutation has little effect on the binding affinity of DB domain to DNA, H196A and E203A mutations both reduce the binding affinity by several folds. Remarkably, the binding of double mutant H196A/E203A of DB domain (DB_AA) to DNA is about an order of magnitude weaker relative to the wildtype DB domain (DB_WT), arguing further that both H196 and E203 residues are likely involved in proton uptake upon the binding of ERα to DNA. It should however be noted that the poor stability of the DB_AA construct made measurements feasible only in Phosphate buffer and no reliable analysis could be carried out in other buffers for direct comparison with the DB_WT construct. It is also of interest that while the H196A and E203A mutations result in the reduction of enthalpy for the binding of DB domain to DNA by about 5–7 kcal/mol due to removal of enthalpic contribution of protonation at H196 or E203 and corresponding protein-DNA interactions at these positions, the enthalpy change for the D190A mutation is nearly two-fold more favorable relative to the wildtype DB domain (Table 2). It is thus conceivable that the D190A mutation results in local secondary and tertiary structural changes within the DB domain and that the partial folding of DB_D190A mutant domain upon binding to DNA also favorably contributes to the binding enthalpy. However, such enhancement in favorable enthalpy does not translate into higher binding affinity of D190A mutant domain to DNA due to an equally compensating entropic contribution as discussed in the previous section.
Table 2.
Observed thermodynamic parameters for the binding of ERE duplex to wildtype and various mutant constructs of the DB domain of ERα in Phosphate buffer at pH 7.0 and 25°C
| Kobs/nM | ΔHobs/kcal.mol−1 | TΔSobs/kcal.mol−1 | ΔGobs/kcal.mol−1 | |
|---|---|---|---|---|
| DB_WT | 43 ± 13 | −30.52 ± 0.27 | −20.44 ± 0.38 | −10.08 ± 0.20 |
| DB_D190A | 67 ± 6 | −62.05 ± 1.64 | −52.33 ± 1.48 | −9.80 ± 0.06 |
| DB_H196A | 190 ± 24 | −23.95 ± 0.08 | −14.76 ± 0.14 | −9.18 ± 0.07 |
| DB_E203A | 172 ± 48 | −24.97 ± 1.81 | −15.66 ± 1.97 | −9.26 ± 0.17 |
| DB_AA | 387 ± 90 | −23.77 ± 2.29 | −15.00 ± 2.36 | −8.77 ± 0.14 |
| DB_S193A | 102 ± 10 | −45.10 ± 1.29 | −35.54 ± 1.25 | −9.55 ± 0.04 |
| DB_Y197A | 316 ± 7 | −31.12 ± 0.41 | −22.24 ± 0.37 | −8.88 ± 0.02 |
| DB_S201A | 119 ± 9 | −28.70 ± 0.69 | −19.23 ± 0.64 | −9.46 ± 0.05 |
| DB_K206A | 313 ± 6 | −30.86 ± 0.86 | −21.97 ± 0.85 | −8.89 ± 0.07 |
| DB_K210A | 326 ± 11 | −25.96 ± 0.17 | −17.09 ± 0.18 | −8.86 ± 0.09 |
| DB_R211A | 745 ± 76 | −8.21 ± 0.09 | +0.17 ± 0.10 | −8.38 ± 0.11 |
The various constructs of the DB domain are the wildtype consruct (DB_WT), the single mutant constructs (DB_D190A, DB_H196A, DB_E203A, DB_S193A, DB_Y197A, DB_S201A, DB_K206A, DB_K210A, DB_R211A) and the H196A/E203A double mutant construct (DB_AA). The binding stoichiometries to the fits agreed to within ±10%. Errors were calculated from 3–4 independent measurements. All errors are given to one standard deviation.
pH tightly regulates the binding of DB domain of ERα to DNA
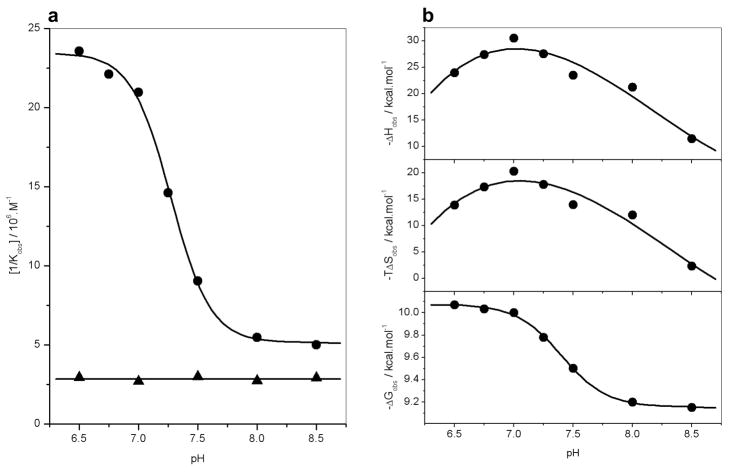
In an effort to further support the notion that the residues H196 and E203 serve as the sole sites of protonation upon the binding of ERα to DNA, we analyzed the binding of wildtype (DB_WT) and double mutant H196A/E203A (DB_AA) constructs of DB domain to ERE duplex as a function of solution pH (Figure 5). Our data reveal that while the binding affinity of DB_WT construct to DNA is sharply dependent on solution pH in a sigmoidal fashion, binding affinity of DB_AA construct to DNA is independent of solution pH (Figure 5a). Taken collectively, our data suggest strongly that the binding of DB domain of ERα is coupled to proton uptake and that the sidechain moieties of H196 and E203 serve as sole proton acceptors in this capacity.
Figure 5.
Dependence of thermodynamics on pH for the binding of ERE duplex to the wildtype DB domain (DB_WT) and the H196A/E203A double mutant of DB domain (DB_AA) of ERα in Phosphate buffer at 25°C. (a) Representative [1/Kobs]-pH plots for the DB_WT (●) and DB_AA (▲) domains. The solid lines respectively indicate sigmoidal and linear fits to data points for clarity. (b) Representative ΔHobs-pH (top panel), TΔSobs-pH (middle panel) and ΔGobs-pH (bottom panel) plots for the DB_WT domain. In the top and middle panels, the solid lines indicate polynomial fits to data points for clarity. In the bottom panel, the solid line indicates sigmoidal fit to data points for clarity.
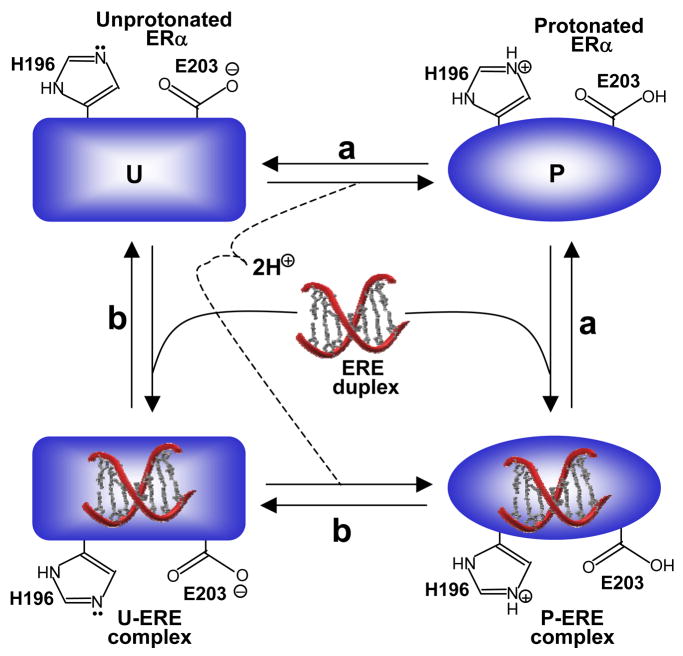
It is however of worthy note that the protonation of H196 and E203 within ERα could precede or follow the subsequent binding of DNA. In this manner, ERα could bind to DNA in both the protonated and unprotonated forms. Figure 6 provides a thermodynamic cycle for the various equilibria linked to the binding of ERα to DNA. It is clearly evident from such a cycle that the binding of ERα to DNA may or may not be coupled to proton uptake depending on solution pH. Thus, under low pH values, ERα may become fully protonated prior to binding DNA. On the other hand, under high pH values, ERα may become fully unprotonated and the proton uptake may be decoupled for its binding to DNA. That ERα can bind to DNA both in the protonated and unprotonated forms is further supported by the sigmoidal response of binding affinity (Kobs) of DB_WT to DNA as a function of pH (Figure 5a). Thus, the plateau values of [1/Kobs]-pH plot at low and high pH values correspond to the intrinsic binding affinities of the protonated and unprotonated forms of DB_WT domain to DNA, respectively.
Figure 6.
A thermodynamic cycle for the various equilibria linked to the binding of ERα to DNA. (a) ERα becomes protonated in the free form and the resulting protonated form (P) binds to DNA. (b) ERα binds to DNA in the unprotonated form (U) and the resulting liganded form becomes protonated.
It should also be noted that the ΔHobs-pH and TΔSobs-pH plots for the binding of DB_WT domain to DNA display bell-shaped curves characteristic of a proton-coupled ligand-binding event (Figure 5b, top and middle panels). Such behavior arises due to the fact that the enthalpy of protonation of free form is different from that of liganded form. Since the ratio of free and liganded forms of the protein varies as a function of pH in going from a low pH value to high, the enthalpic contribution due to their protonation to the observed enthalpy varies accordingly, reaching zero at the extreme values and a maximum in between these extremes where the ratio of the protonation of free form to liganded form equals unity. Expectedly, the ΔGobs-pH plot for the binding of DB_WT to DNA follows sigmoidal behavior in agreement with the ability of both the protonated and unprotonated forms to bind to DNA with distinct affinities (Figure 5b, bottom panel).
Electrostatic surface potentials reveal that the protonation of H196 and E203 optimizes thermodynamic constraints
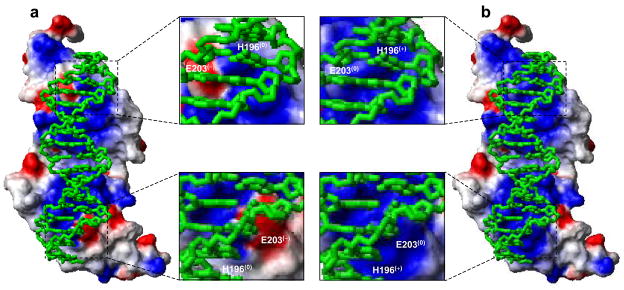
In an attempt to rationalize the effect of protonation of H196 and E203 on electrostatics at the protein-DNA interface, we generated molecular surfaces of the DB domain of ERα in complex with the ERE duplex depicting protein electrostatic potentials (Figure 7). Our data reveal how such protonation switches polarization of protein surface at residues H196 and E203 so as to render it thermodynamically more favorable for coming into contact with DNA. In the free conformation, H196 and E203 occupy what appear to be respectively neutral and negatively charged spots on the protein surface that is destined to come in close contact with DNA. It is further evident that while the presence of neutral charge at H196 may not in any way compromise the subsequent binding of DNA, protonation at this position could bring about favorable energetic contributions as a direct result of favorable electrostatic interactions with the negatively charged phosphate backbone. In contrast, the build up of negative charge at E203 would hamper the subsequent binding of DNA due to electrostatic repulsions with the negatively charged phosphate backbone suggesting that protonation at this position would relieve such energetic barriers. Taken together, the electrostatic surface potentials of the DB domain of ERα alone and in complex with DNA argue strongly that the protonation of H196 and E203 would optimize thermodynamic constraints so to allow the two molecular surfaces to come in close proximity to attain a tight molecular fit worthy of the rather high affinity that this DNA-protein complex displays.
Figure 7.
Molecular surfaces depicting electrostatic potentials of the DB domain of ERα containing H196 and E203 in unprotonated forms (a) and protonated forms (b) in complex with the ERE duplex. The blue and red colors respectively denote the density of positive and negative charges, while the apolar and polar surfaces are indicated by white/gray color on the molecular surfaces. In the expanded views, the locations of H196 and E203 are clearly marked on the molecular surfaces with the parenthesis indicating the overall charge on each residue under unprotonated and protonated forms. The ERE duplex is displayed as a stick model and colored green for clarity.
Proton-coupled binding to DNA appears to be a hallmark of nuclear receptor family
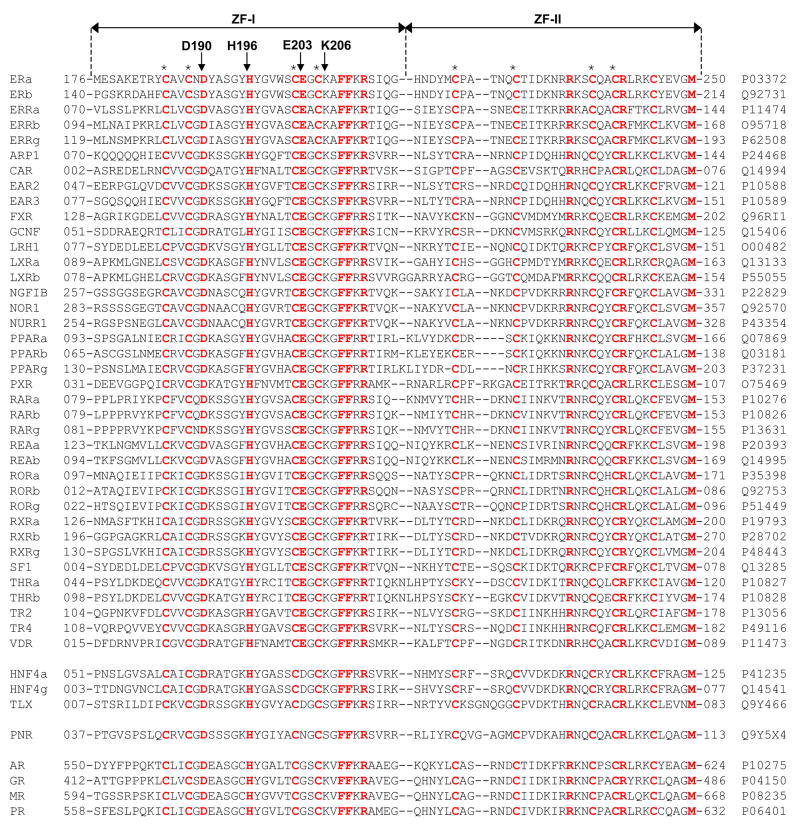
In an attempt to analyze the extent to which the ability of ERα to become protonated upon binding to DNA is shared by other members, we generated amino acid sequence alignment of the DB domains of the entire human NR family (Figure 8). It should be noted that the DB domains of NR family are poorly conserved and display less than 15% sequence identity outside the quartet of cysteine residues involved in coordinating the Zn2+ divalent ion within each of the two Zinc fingers of the DB domain. Thus, the residues conserved among the various DB domains bear a significant importance and must have co-evolved for a common physiological function.
Figure 8.
Amino acid sequence alignment of DB domains of all known members of human NR family. Absolutely conserved residues are shown in red, while all other residues are depicted in black. Each member is denoted by its acronym in the left column with the corresponding Expasy code provided in the right column for access to complete proteomic details on each member. The numerals hyphenated to amino acid sequence at each end denote the boundaries of DB domains for each member. The cysteine residues within each of the two Zinc fingers of DB domains, denoted ZF-I and ZF-II, that coordinate the Zn2+ ion in a tetrahedral arrangement are marked by asterisks. Residues D190, H196, E203 and K206, located within the DB domain of ERα, are indicated by vertical arrows.
Remarkably, H196 and E203 rank among these conserved residues within the DB domains of human NR family. Thus, while H196 is absolutely conserved within all members of human NR family, E203 is predominantly conserved in most members, with notable exceptions being the androgen receptor (AR), glucocorticoid receptor (GR), mineralocorticoid receptor (MR) and progesterone receptor (PR), which all have a glycine substitution for E203. Interestingly, E203 is substituted by an asparagine in photoreceptor-specific nuclear receptor (PNR), implying that hydrogen bonding at this position may play a critical role in protein-DNA interaction pertinent to the physiological function of this nuclear receptor. Finally, E203 is substituted by a related acidic and ionizable aspartate residue in hepatocyte nuclear factor 4a (HNF4a), hepatocyte nuclear factor 4g (HNF4g) and the tail-less orphan receptor (TLX), indicating that protonation at this position upon binding to DNA may also be critical for these nuclear receptors. Thus, the highly conserved nature of H196 and E203 among the functionally-diverse members of the human NR family argues strongly that the proton-coupled binding to DNA may have evolved as a general mechanism for nuclear receptor function and regulation.
It is also of worthy note that although the residue D190 is absolutely conserved among all members of the human NR family, implying that although it does not serve as a proton acceptor, it must also play a pivotal role in protein-DNA interactions pertinent to nuclear receptors. Additionally, our thermodynamic data indicate that the D190A substitution has no bearable effect on the binding affinity of DB domain of ERα to DNA (Table 2) — an observation that is in conflict with evolutionary constraints being placed upon this residue in the human NR family. Further scrutiny of the binding of wildtype DB domain (DB_WT) versus the D190A mutant (DB_D190A) to DNA suggests that although they bind with virtually indistinguishable affinities, the underlying thermodynamic forces display distinct features. Thus, while the binding of both domains is driven by favorable enthalpic factors accompanied by entropic penalties, DB_D190A generates twice as much heat relative to DB_WT implying that this residue may be critical for the folding and stability of ERα and that the more favorable heat likely results from the partial folding of DB_D190A mutant domain upon binding to DNA as noted earlier.
In an effort to further decipher the molecular basis of how nuclear receptors bind to their cognate DNA promoter elements with specificity, we also analyzed and compared the thermodynamics of binding of ERE duplex to DB domain of ERα containing alanine substitutions for a number of additional amino acid residues located at the protein-DNA interface (Table 2). These point mutations include S193A (DB_S193A), Y197A (DB_Y197A), S201A (DB_S201A), K206A (DB_K206A), K210A (DB_K210A) and R211A (DB_R211A). As shown in Table 2, alanine substitution of these residues reduces the binding of DB domain to DNA by as little as 2-fold in the case of S193A mutation to as large as 17-fold in the case of R211A mutation relative to the wildtype construct, implying that these residues contribute differentially to the free energy of binding. Of these six residues at the protein-DNA interface, only R211 is absolutely conserved within the DB domains of all nuclear receptors (Figure 8). This salient observation suggests strongly that in addition to H196 and E203, R211 is also likely to be a critical residue involved in the binding of all nuclear receptors to their cognate DNA sequences. However, the fact that the residues S193, Y197, S201, K206 and K210 show variability within the DB domains of nuclear receptors argues strongly in favor of their role in determining the specificity of binding of nuclear receptors to DNA. Nonetheless, it should be borne in mind that complete understanding of molecular basis of DNA-specificity of nuclear receptors awaits detailed thermodynamic analysis coupled with site-directed mutagenesis of specific amino acid residues within DB domains of other nuclear receptors that we hope to accomplish in our future studies.
CONCLUSIONS
Nuclear receptor function is tightly regulated by a multitude of post-translational modifications such as phosphorylation, acetylation, sumoylation, ubiquitination and glycosylation (44–47). However, such modifications usually occur in regions outside the DB domain. The fact that the DB domain of ERα is directly regulated via proton-coupled equilibrium of two critical residues, H196 and E203, located at the protein-DNA interface not only adds to the repertoire of tricks and treats employed by nuclear receptors but also bears significant implications for furthering our understanding of this important family of transcription factors.
Although the binding of DB domain of ERα to DNA appears to be coupled to proton uptake, it is not clear from our data as to how such coupled equilibrium might dictate the physiological role of this important nuclear receptor. Changes in intracellular pH regulate a multitude of cellular processes such as metabolic homeostasis and apoptosis (48). Furthermore, it is believed that ionizable residues within proteins sense such changes and activate a variety of proton pumps and ion transporters that in turn mediate extracellular transport of protons and anions to regulate intracellular pH (49–51). It is thus conceivable that changes in intracellular pH may also tightly regulate the transcriptional activity of ERα through direct modulation of two ionizable residues, H196 and E203, located at the protein-DNA interface. Protonation of such residues would clearly enhance intermolecular hydrogen bonding and electrostatic interactions critical to driving this key protein-DNA interaction and vice versa. Although pKa values of sidechains of histidine and glutamate within proteins are respectively around 6 and 4 (52), these are likely to be influenced by the neighboring ionizable amino acid residues in the DB domain as noted earlier (Figure 1). Thus, protonation/deprotonation of H196 and E203 may not necessarily require large changes but may be mediated by small changes in intracellular pH. Whatever the exact physiological role of proton-coupled equilibrium observed here, our current study clearly warrants further investigating the role of pH in physiological processes governed by ERα and other nuclear receptors.
In the crystal structure of the DB domain of ERα in complex with ERE duplex solved nearly two decades ago (28), it was proposed that the negative charge on E203 was largely neutralized through the formation of a salt bridge with the neighboring K206. On the contrary, our study here shows that the negative charge on E203 is rather neutralized through its protonation allowing it to participate in the formation of hydrogen bonding with DNA in a more harmonious manner. Additionally, the crystal structural analysis also suggested the involvement of H196 in dictating protein-DNA interactions through hydrogen bonding with the phosphate backbone. The fact that H196 acquires a net positive charge through protonation upon the binding of DNA suggests that H196 is more likely to engage in the formation of a salt bridge with the phosphate backbone. Taken together, our study exquisitely reveals how a combined approach involving site-directed mutagenesis in conjunction with thermodynamics can complement structural data and further define key residues involved in protein-DNA interactions.
In short, our present study demonstrates that the protonation of H196 and E203 in ERα is coupled to the binding of DNA and that such protonation is required for high-affinity protein-DNA interaction through thermodynamic optimization of intermolecular contacts. Given that H196 and E203 are conserved in a vast majority of ~50 members of the nuclear receptor family, our findings suggest that the nuclear receptors may act as sensors of intracellular pH and bear important consequences for a paradigm shift of their molecular action. Finally, the proton-coupled equilibrium characterized here may serve as a novel target for therapeutic intervention of nuclear receptors.
Acknowledgments
The authors are deeply indebted to Thomas Harris, Marius Sudol and Vineet Gupta for their critical reading of the manuscript and many helpful suggestions.
ABBREVIATIONS
- DB
DNA-binding
- ERα
Estrogen receptor α
- ERE
Estrogen response element
- ITC
Isothermal titration calorimetry
- LB
Ligand-binding
- MALDI-TOF
Matrix-assisted laser desorpton/ionization time of flight
- MAPK
Mitogen-activated protein kinase
- NMR
Nuclear magnetic resonance
- NR
Nuclear receptor
- SEC
Size-exclusion chromatography
- TA
Trans-activation
- Trx
Thioredoxin
- ZF
Zinc finger
Footnotes
This work was supported by funds from the National Institutes of Health (Grant# R01-GM083897) and the USylvester Braman Family Breast Cancer Institute to AF. CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929). BJD and AF are members of the Sheila and David Fuente Graduate Program in Cancer Biology at the Sylvester Comprehensive Cancer Center of the University of Miami.
References
- 1.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- 3.Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, Lazar MA, Mangelsdorf DJ, Moore DD, Qin J, Steffen DL, Tsai MJ, Tsai SY, Yu R, Margolis RN, Evans RM, O’Malley BW. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–746. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEwan IJ. Nuclear receptors: one big family. Methods Mol Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- 6.Barnett P, Tabak HF, Hettema EH. Nuclear receptors arose from pre-existing protein modules during evolution. Trends Biochem Sci. 2000;25:227–228. doi: 10.1016/s0968-0004(00)01579-6. [DOI] [PubMed] [Google Scholar]
- 7.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64:310–319. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 8.Egea PF, Klaholz BP, Moras D. Ligand-protein interactions in nuclear receptors of hormones. FEBS Lett. 2000;476:62–67. doi: 10.1016/s0014-5793(00)01672-0. [DOI] [PubMed] [Google Scholar]
- 9.Claessens F, Gewirth DT. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- 10.Green S, Chambon P. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet. 1988;4:309–314. doi: 10.1016/0168-9525(88)90108-4. [DOI] [PubMed] [Google Scholar]
- 11.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ham J, Parker MG. Regulation of gene expression by nuclear hormone receptors. Curr Opin Cell Biol. 1989;1:503–511. doi: 10.1016/0955-0674(89)90012-4. [DOI] [PubMed] [Google Scholar]
- 13.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 14.Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- 15.Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46:13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb B, Beitel LK, Wu J, Elhaji YA, Trifiro M. Nuclear receptors and disease: androgen receptor. Essays Biochem. 2004;40:121–136. doi: 10.1042/bse0400121. [DOI] [PubMed] [Google Scholar]
- 17.Gurnell M, Chatterjee VK. Nuclear receptors in disease: thyroid receptor beta, peroxisome-proliferator-activated receptor gamma and orphan receptors. Essays Biochem. 2004;40:169–189. doi: 10.1042/bse0400169. [DOI] [PubMed] [Google Scholar]
- 18.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 19.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen EV, Jacobson H. Basic guides to the mechanism of estrogen action. Recent Prog Horm Res. 1962;18:318–414. [Google Scholar]
- 21.Jensen EV. On the mechanism of estrogen action. Perspect Biol Med. 1962;6:47–59. doi: 10.1353/pbm.1963.0005. [DOI] [PubMed] [Google Scholar]
- 22.Toft D, Gorski J. A receptor molecule for estrogens: isolation from the rat uterus and preliminary characterization. Proc Natl Acad Sci U S A. 1966;55:1574–1581. doi: 10.1073/pnas.55.6.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toft D, Shyamala G, Gorski J. A receptor molecule for estrogens: studies using a cell-free system. Proc Natl Acad Sci U S A. 1967;57:1740–1743. doi: 10.1073/pnas.57.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980–1989. [PubMed] [Google Scholar]
- 25.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 26.Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 27.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 29.Schwabe JW, Neuhaus D, Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- 30.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker JM, editor. Humana Press; Totowa, New Jersey, USA: 2005. pp. 571–607. [Google Scholar]
- 31.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 32.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 33.Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 34.Kozlov AG, Lohman TM. Large contributions of coupled protonation equilibria to the observed enthalpy and heat capacity changes for ssDNA binding to Escherichia coli SSB protein. Proteins Suppl. 2000;4:8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz-Salmeron E, Yassin Z, Clemente-Jimenez MJ, Las Heras-Vazquez FJ, Rodriguez-Vico F, Baron C, Garcia-Fuentes L. Thermodynamic analysis of the binding of glutathione to glutathione S-transferase over a range of temperatures. Eur J Biochem. 2001;268:4307–4314. doi: 10.1046/j.1432-1327.2001.02349.x. [DOI] [PubMed] [Google Scholar]
- 36.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 37.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 38.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 39.Lumry R, Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- 40.Eftink MR, Anusiem AC, Biltonen RL. Enthalpy-entropy compensation and heat capacity changes for protein-ligand interactions: general thermodynamic models and data for the binding of nucleotides to ribonuclease A. Biochemistry. 1983;22:3884–3896. doi: 10.1021/bi00285a025. [DOI] [PubMed] [Google Scholar]
- 41.Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 42.Sharp K. Entropy-enthalpy compensation: fact or artifact? Protein Sci. 2001;10:661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starikov EB, Norden B. Enthalpy-entropy compensation: a phantom or something useful? J Phys Chem B. 2007;111:14431–14435. doi: 10.1021/jp075784i. [DOI] [PubMed] [Google Scholar]
- 44.Chen YX, Du JT, Zhou LX, Liu XH, Zhao YF, Nakanishi H, Li YM. Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta. Chem Biol. 2006;13:937–944. doi: 10.1016/j.chembiol.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Faus H, Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60:520–528. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- 46.Popov VM, Wang C, Shirley LA, Rosenberg A, Li S, Nevalainen M, Fu M, Pestell RG. The functional significance of nuclear receptor acetylation. Steroids. 2007;72:221–230. doi: 10.1016/j.steroids.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weigel NL, Moore NL. Steroid receptor phosphorylation: a key modulator of multiple receptor functions. Mol Endocrinol. 2007;21:2311–2319. doi: 10.1210/me.2007-0101. [DOI] [PubMed] [Google Scholar]
- 48.Boron WF. Regulation of intracellular pH. Adv Physiol Educ. 2004;28:160–179. doi: 10.1152/advan.00045.2004. [DOI] [PubMed] [Google Scholar]
- 49.Khaled AR, Moor AN, Li A, Kim K, Ferris DK, Muegge K, Fisher RJ, Fliegel L, Durum SK. Trophic factor withdrawal: p38 mitogen-activated protein kinase activates NHE1, which induces intracellular alkalinization. Mol Cell Biol. 2001;21:7545–7557. doi: 10.1128/MCB.21.22.7545-7557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khaled AR, Reynolds DA, Young HA, Thompson CB, Muegge K, Durum SK. Interleukin-3 withdrawal induces an early increase in mitochondrial membrane potential unrelated to the Bcl-2 family. Roles of intracellular pH, ADP transport, and F(0)F(1)-ATPase. J Biol Chem. 2001;276:6453–6462. doi: 10.1074/jbc.M006391200. [DOI] [PubMed] [Google Scholar]
- 51.Puceat M, Roche S, Vassort G. Src family tyrosine kinase regulates intracellular pH in cardiomyocytes. J Cell Biol. 1998;141:1637–1646. doi: 10.1083/jcb.141.7.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]