Abstract
The P2Y1 receptor is a prothrombotic G protein-coupled receptor (GPCR) activated by ADP. Preference for the North (N) ring conformation of the ribose moiety of adenine nucleotide 3′,5′-bisphosphate antagonists of the P2Y1 receptor was established by using a ring-constrained methanocarba (a bicyclo[3.1.0]hexane) ring as a ribose substitute. A series of covalently linkable N6-methyl-(N)-methanocarba-2′-deoxyadenosine-3′,5′-bisphosphates containing extended 2-alkynyl chains was designed and binding affinity at the human (h) P2Y1 receptor determined. The chain of these functionalized congeners contained hydrophilic moieties, a reactive substituent, or biotin, linked via an amide. Variation of the chain length and position of an intermediate amide group revealed high affinity of carboxylic congener 8 (Ki 23 nM) and extended amine congener 15 (Ki 132 nM), both having a 2-(1-pentynoyl) group. A biotin conjugate 18 containing an extended ε-aminocaproyl spacer chain exhibited higher affinity than a shorter biotinylated analogue. Alternatively, click coupling of terminal alkynes of homologous 2-dialkynyl nucleotide derivatives to alkyl azido groups produced triazole derivatives that bound to the P2Y1 receptor following deprotection of the bisphosphate groups. The preservation of receptor affinity of the functionalized congeners was consistent with new P2Y1 receptor modeling and ligand docking. Attempted P2Y1 antagonist conjugation to PAMAM dendrimer carriers by amide formation or palladium-catalyzed reaction between an alkyne on the dendrimer and a 2-iodopurine-derivatized nucleotide was unsuccessful. A dialkynyl intermediate containing the chain length favored in receptor binding was conjugated to an azide-derivatized dendrimer, and the conjugate inhibited ADP-promoted human platelet aggregation. This is the first example of attaching a strategically functionalized P2Y receptor antagonist to a PAMAM dendrimer to produce a multivalent conjugate exhibiting a desired biological effect, i.e. antithrombotic action.
INTRODUCTION
The family of P2Y receptors consists of eight subtypes of G protein-coupled receptors (GPCRs) that respond to extracellular purine and pyrimidine nucleotides. Native ligands of these receptors include ATP, ADP, UTP, UDP, and UDP-glucose (1,2). Two subfamilies have been delineated based on second messengers and sequence homology, i.e., a cluster of P2Y1, P2Y2, P2Y4, P2Y6, and P2Y11 receptors, which activate phospholipase C-β via Gq, and a smaller cluster of P2Y12, P2Y13, and P2Y14, which inhibit adenylyl cyclase via Gi.
P2Y1 receptors are involved in activation of platelets and other hematopoietic cells (3–6), vascular function (7), apoptotic events (8), inflammation, glial cell differentiation and function (9–11), and development of vision (12). Activation of a P2Y1 receptor expressed in brain astrocytes participates in cytoprotection, as indicated in a forebrain model using hydrogen peroxide, one of the main reactive oxygen species generated by traumatic brain injury, ischemic stroke, and various neurodegenerative diseases (10,11). On the surface of platelets, simultaneous activation of the P2Y1 and P2Y12 receptors by ADP induces aggregation (3,4). The P2Y1–mediated response is associated with the initial shape change and rapid aggregation, and the P2Y12 receptor is associated with amplification of aggregation. P2Y12 receptor antagonists are used extensively as antithrombotic agents, and new antagonists are currently under development (13).
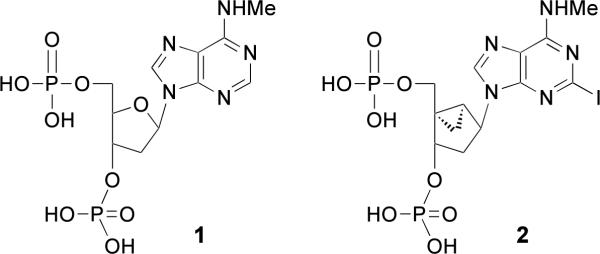
A prototypical competitive antagonist of the P2Y1 receptor, bisphosphate derivative MRS2179 1 (Chart 1) widely-used as a pharmacological probe, has a KB of 102 nM (14). Newer and more potent and selective P2Y1 receptor antagonists, such as the conformationally locked North (N)-methanocarba nucleotide MRS2500 2 ((1′R,2′S,4′S,5′S)-4-(2-iodo-6-methylamino-purin-9-yl)-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane, Ki 0.79 nM), which contain a bicyclo[3.1.0]hexane ring system in place of ribose, have been designed and shown to have promise in preclinical studies as antithrombotic agents (5,15–18). This novel drug concept is also supported by studies of mice in which the P2Y1 receptor has been genetically deleted (19, 20).
Chart 1.
Prototypical nucleotide antagonists of the P2Y1 receptor.
While most substitutions at positions other than C2 of adenine in this family of (N)-methanocarba nucleotides related to MRS2500 decrease antagonist potency at the P2Y1 receptor, 2-alkynyl substitution preserves or enhances antagonist potency (18). Structure activity relationship (SAR) analysis indicated that hydrophobic substituents at the C2 position are favored. In the present study, we have extended our search for antagonists of the P2Y1 receptor by modifying the C2 position with novel alkynyl substituents. We have generated new P2Y1 antagonists using a conceptual approach for covalently linking GPCR ligands to carrier molecules while preserving the receptor interactions, i.e., the functionalized congener approach (21). Carrier molecules may include small molecules, biopolymers, or synthetic polymers that are biocompatible. The linkage used between the nucleotide pharmacophore and a carrier moiety was explored systematically in a series of small model compounds that contain typical linking moieties that may be used for tethering to the polymer, and thus can be evaluated for effects on receptor recognition. These model compounds incorporated groups such as a carboxylic acid, an amine, or a terminal alkyne that allow introduction of hydrophilic or chemically reactive substituents or link to a carrier. A terminal alkyne-derivatized nucleotide was coupled by a “click” reaction (22) to a polymeric carrier, a polyamidoamine (PAMAM) dendrimer that had been derivatized with terminal azide groups. The most biologically favorable chain length among the resulting triazole derivatives was incorporated in a covalent conjugate with the dendrimer. The resulting multivalent conjugate displayed antithrombotic activity characteristic of P2Y1 antagonists in a human platelet preparation (5,16). Attempted conjugation of the nucleotides to dendrimers by alternate routes, e.g. amide formation or palladium-catalyzed reaction between an alkyne on the dendrimer and an iodoaryl-derivatized nucleotide, was unsuccessful.
EXPERIMENTAL PROCEDURES
Chemical synthesis
Materials and instrumentation
Methylamine, PAMAM dendrimer (G3, 20 wt% solution in methanol; G4, 5 wt% solution in methanol) with an ethylenediamine core, and other reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO). Dialysis membranes (Spectra/Pore Membrane, MWCO 3500, flat width 18 mm) were purchased from Spectrum Laboratories, Inc. (Rancho Dominguez, CA). Compound 25 was synthesized as reported from the starting material 24 (Organomed Corp., Coventry, RI) (15). Azido dendrimer derivative 51 was prepared as described (23). 1H NMR spectra of monomeric derivatives were obtained with a Varian Gemini 300 spectrometer using CDCl3 and CD3OD as solvents. Chemical shifts are expressed in δ values (ppm) with tetramethylsilane (δ = 0.00) for CDCl3 and water (δ 3.30) for CD3OD. TLC analysis was performed using aluminum sheets precoated with silica gel F254 (0.2 mm) from Aldrich. The nucleotide derivatives were purified by HPLC before biological testing, and the purity of each was shown to be >95%. The mobile phases consisted of System A: linear gradient solvent system of CH3CN/triethyl ammonium acetate from 5/95 to 60/40 in 20 min, flow rate 1.0 mL/min; System B: linear gradient solvent system of CH3CN/tetrabutyl ammonium phosphate from 20/80 to 60/40 in 20 min, flow rate 1.0 mL/min; System C: linear gradient solvent system of CH3CN/tetrabutyl ammonium phosphate from 20/80 to 60/40 in 13 min, flow rate 0.5 mL/min; System D: with a Luna 5 l RP-C18(2) semipreparative column (250 × 10.0 mm; Phenomenex, Torrance, CA) and using the following conditions: flow rate of 2 mL/min; 10 mM triethylammonium acetate (TEAA)-CH3CN from 100:0 to 0:100 in 10 min and held at 0:100 for 10 min; and System E: a Luna 5 l RP-C18(2) semipreparative column (250 × 10.0 mm; Phenomenex, Torrance, CA) using the following conditions: flow rate of 2 mL/min; 10 mM triethylammonium acetate (TEAA)-CH3CN from 100:0 to 60:40 in 30 min. High resolution mass spectroscopic (HRMS) measurements were performed on a proteomics optimized Q-TOF-2 (Micromass-Waters) using external calibration with polyalanine, unless noted. Observed mass accuracies are those expected, based on the known performance of the instrument as well as trends in masses of standard compounds observed at intervals during the series of measurements. Reported masses are observed masses uncorrected for this time-dependent drift in mass accuracy.
General procedure for 2-(methoxycarbonylalkynyl) derivatives 26–28
A solution of 25 (30 mg, 0.038 mmol) in DMF (5 mL) containing Et3N (7.6 μL) was treated with (PPh3)2PdCl2 (2.63 mg, 0.0038 mmol) and CuI (1.5 mg, 0.0076 mmol) (13). Then, the appropriate acetylene derivative (0.076 mmol, 2 equiv) was subsequently added dropwise to the reaction mixture, which was then stirred under argon at room temperature for 7 h. The solvent was removed under reduced pressure, and the residue was purified by flash chromatography to obtain the desired compounds as solids.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(methoxycarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (26)
Yield: 28.40 (90%). 1H NMR (CDC13) δ 8.10 (s, 1H), 5.26 (dd, 1H, J = 14.2, 7.9 Hz), 5.10 (d, 1H, J = 6.9 Hz), 4.58 (dd, 1H, J = 11.1, 4.7 Hz), 3.80 (dd, 1H, J = 11.1, 6.3 Hz), 3.64 (s, 3H), 3.15 (c, 3H, J= 7.5 Hz), 2.67 (m, 4H), 2.23 (dd, 1H, J = 15.3, 8.4 Hz), 2.02 (m, 1H), 1.70 (m, 1H), 1.43, 1.41, 1.40, 1.39 (4s, 36H), 1.04 (m, 1H), 0.90 (m, 1H); HRMS (ESI MS m/z) calculated for C35H58N5O10P2+ (M+H)+, 770.3653; found, 770.3658.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(methoxycarbonyl)-1-pentynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (27)
Yield: 34 mg (85%). 1H NMR (CDC13) δ 8.10 (s, 1H), 5.26 (dd, 1H, J = 8.7, 2.4 Hz), 5.12 (d, 1H, J = 7.2 Hz), 4.59 (dd, 1H, J = 11.4, 4.8 Hz), 3.77 (dd, 1H, J = 11.7, 6.3 Hz), 3.61 (s, 3H), 3.15 (m, 3H), 2.47 (m. 2H), 2.38 (m, 1H), 2.24 (m, 1H), 1.92 (m,1H), 1.82 (m, 3H),1.42, 1.41, 1.40, 1.38 (4s, 36 H), 1.04 (m, 1H), 0.90 (m, 1H); HRMS (ESI MS m/z) calculated for C36H60N5O10P2+ (M+H)+, 784.3815; found, 784.3794.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(methoxycarbonyl)-1-hexynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (28)
Yield: 29 mg (95%). 1H NMR (CDC13) δ 8.07 (s, 1H), 5.26 (dd, 1H, J = 14.7, 8.7 Hz), 5.12 (d, 1H, J = 6.9 Hz), 4.57 (dd, 1H, J = 11.1, 4.6 Hz), 3.76 (dd, 1H, J = 11.4, 6.3 Hz), 3.60 (s, 3H), 3.14 (c, 3H, J = 7.2 Hz), 2.40 (t, 2H, J = 7.2 Hz), 2.29 (t, 2H, J = 7.2 Hz), 2.18 (m, 1H), 2.02 (m, 1H), 1.68 (m, 4H), 1.41, 1.39, 1.37 (3s, 36H), 1.04 (m, 1H), 0.88 (m, 1H); HRMS (ESI MS m/z) calculated for C37H62N5O10P2+ (M+H)+, 798.3966; found, 798.3953.
General procedure for 2-(methoxycarbonylalkynyl) derivatives 5–7
A mixture of the appropriate ester 26–28 (0.012 mmol) in CH2C12 (1 mL) was treated with trifluoroacetic acid (TFA, 29 μL), and the reaction mixture was stirred at room temperature for 3 h. After removal of the solvent, the crude product was purified by HPLC with System E and isolated in the triethylammonium salt form to obtain the desired compounds as solid.
(1'R,2'S,4'S,5'S)-4-{2-[(Methoxycarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (5)
Yield: 8.3 mg (73%). 1H NMR (D2O) δ 8.52 (s, 1H), 5.23 (dd, 1H, J = 13.8, 6.8 Hz), 5.02 (d, 1H, J = 6.0 Hz), 4.59 (dd, 1H, J = 11.7, 5.1 Hz), 3.79 (s, 3H), 3.71 (dd, 1H, J = 11.4, 4.5 Hz), 3.12 (s, 3H), 2.82 (m, 4H), 2.29 (dd, 1H, J = 15.1, 7.9 Hz), 2.04 (m, 1H), 1.70 (m, 1H), 1.24 (m, 1H), 1.04 (m, 1H); 31P NMR (D2O) δ 0.90 (s), 0.40 (s); HRMS (ESI MS m/z) calculated for C19H24N5O10P2− (M–H)−, 544.1004; found, 544.0998; HPLC RT 6.9 min (98%) in solvent System A, 15.1 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(Methoxycarbonyl)-1-pentynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (6)
Yield: 9.3 mg (82%). 1H NMR (D2O) δ 8.53 (s, 1H), 5.22 (m, 1H), 5.05 (m, 1H), 4.59 (m, 1H), 3.79 (s, 3H), 3.68 (m, 1H), 3.12 (s, 3H), 2.64 (t, 4H, J = 7.5 Hz), 2.58 (t, 1H, J = 6.9 Hz), 2.29 (m, 1H), 1.99 (m, 2H), 1.24 (m, 1H), 1.04 (m, 1H); 31P NMR (D2O) δ 0.87 (s), 0.23 (s); HRMS (ESI MS m/z) calculated for C20H26N5O10P2− (M–H)−, 558.1160; found, 558.1135; HPLC RT 7.5 min (98%) in solvent System A, 15.7 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(Methoxycarbonyl)-1-hexynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (7)
Yield: 8.3 mg (71%). 1H NMR (D2O) δ 8.55 (s, 1H), 5.21 (m, 1H), 5.05 (m, 1H), 4.60 (m, 1H), 3.73 (s, 3H), 3.68 (m, 1H), 3.13 (s, 3H), 2.55 (t, 4H, J = 6.8 Hz), 2.52 (t, 1H, J = 7.3 Hz), 2.26 (m, 1H), 1.95 (m, 2H), 1.54 (m, 1H), 1.72 (m, 1H), 1.26 (m, 1H), 1.05 (m, 1H); 31P NMR (D2O) δ 0.92 (s), 0.41 (s); HRMS (ESI MS m/z) calculated for C21H28N5O10P2− (M–H)−, 572.1317; found, 572.1286; HPLC RT 8.2 min (98%) in solvent System A, 16.4 min (99%) in System B.
General procedure for 2-(carboxyalkynyl) derivatives 8–10
Aqueous KOH (0.5 mL of a 0.25 M solution) was added to a solution of the appropriate ester 5 – 7 (0.008 mmol) in water (1 mL), and the mixture was stirred overnight at room temperature. After removal of the solvent, the crude product was purified by HPLC with System E and isolated in the triethylammonium salt form to obtain the desired compounds as solids.
(1'R,2'S,4'S,5'S)-4-{2-[Hydroxycarbonyl]-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (8)
Yield: 5.2 mg (69%). 1H NMR (D2O) δ 8.49 (s, 1H), 5.16 (dd, 1H, J = 15.0, 9.0 Hz), 5.01 (d, 1H, J = 6.6 Hz), 4.55 (dd, 1H, J = 10.2, 5.1 Hz), 3.70 (dd, 1H, J = 11.4, 4.7 Hz), 3.09 (s, 3H), 2.70 (dd, 4H, J = 18.1, 5.8 Hz), 2.25 (m. 1H), 1.94 (m, 2H), 1.08 (t, 1H, J = 7.5 Hz); 31P NMR (D2O) δ 0.63 (s), 0.16 (s); HRMS (ESI MS m/z) calculated for C18H22N5O10P2− (M–H)−, 530.0847; found, 530.0347; HPLC RT 5.5 min (98%) in solvent System A, 15.6 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[Hydroxycarbonyl]-1-pentynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (9)
Yield: 5.0 mg (64%). 1H NMR (D2O) δ 8.48 (s, 1H), 5.19 (m, 1H), 5.00 (m, 1H), 4.55 (m, 1H), 3.68 (m, 1H), 3.09 (s, 3H), 2.52 (t, 4H, J = 7.2 Hz), 2.44 (t, 1H, J = 7.2 Hz), 2.24 (m, 1H), 1.92 (m, 2H), 1.03 (m, 1H); 31P NMR (D2O) δ 0.70 (s), 0.31 (s); HRMS (ESI MS m/z) calculated for C19H24N5O10P2− (M–H)−, 544.1004; found, 544.0998; HPLC RT 5.0 min (98%) in solvent System A, 15.4 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[Hydroxycarbonyl]-1-hexynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (10)
Yield: 2.6 mg (60%). 1H NMR (D2O) δ 8.56 (s, 1H), 5.23 (m, 1H), 6.00 (m, 1H), 4.56 (m, 1H), 3.65 (m, 1H), 3.13 (s, 3H), 2.53 (t, 4H, J = 6.2 Hz), 2.27 (t, 1H, J = 6.6 Hz), 2.03 (m, 1H), 1.89 (m, 2H), 1.70 (m, 1H), 1.21 (m, 1H), 0.99 (m, 1H); 31P NMR (D2O) δ 1.94 (s), 1.29 (s); HRMS (ESI MS m/z) calculated for C2OH26N5O10P2− (M–H)−, 558.1160; found, 558.1155; HPLC RT 5.2 min (98%) in solvent System A, 16.0 min (99%) in System B.
General procedure for 2-(aminoalkylamino-carbonylalkynyl) derivatives 29–34
A solution of the appropriate phosphate-protected ester derivative 26–28 (0.0135 mmol) in methanol (12 μ L) was treated with ethylenediamine (0.22 mL, 3.2 mmol) or its homologue. The reaction mixture was stirred overnight at room temperature. The residue was purified by flash chromatography EtOAc:MeOH (5:1) to obtain the desired compounds as solids.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(2-aminoethylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (29)
Yield from 26 and ethylenediamine: 7.4 mg (69%). 1H NMR (CDC13) δ 8.24 (s, 1H), 5.47 (dd, 1H, J = 15.0, 8.1 Hz), 5.08 (d, 1H, J = 7.2 Hz), 4.58 (dd, 1H, J = 11.4, 5.1 Hz), 4.02 (dd, 1H, J = 11.1, 6.3 Hz), 3.14 (br s, 3H), 2.80 (t. 2H, J = 7.2 Hz), 2.6080 (t. 2H, J = 7.2 Hz), 2.40 (dd, 1H, J = 15.3, 8.4 Hz), 2.22 (m, 1H), 1.90 (m, 1H), 1.41, 1.37, 1.36 (3s, 36H), 1.23 (m, 1H), 1.13 (m, 1H); HRMS (ESI MS m/z) calculated for C36H62N7O9P2+ (M+H)+, 798.4079; found, 798.4035.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(2-aminoethylaminocarbonyl)-1-pentynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (30)
Yield from 27 and ethylenediamine: 7.4 mg (68%). 1H NMR (CDC13) δ 8.07 (s, 1H), 6.66 (br s, 1H), 5.83 (m, 1H), 5.43 (dd, 1H, J = 14.1, 7.5 Hz), 5.11 (d, 1H, J = 6.9 Hz), 4.69 (dd, 1H, J = 10.8, 5.0 Hz), 3.97 (dd, 1H, J = 10.8, 6.3 Hz), 3.33 (c, 2H, J = 5.7 Hz), 3.25 (m, 3H), 2.84 (t, 2H, J = 6.0 Hz), 2.54 (t, 2H, J = 6.6 Hz), 2.47 (t, 2H, J = 7.2 Hz), 2.31 (dd, 1H, J = 15.6, 8.4 Hz), 2.11 (c, 1H, J = 7.5 Hz), 2.01 (t, 1H, J = 7.2 Hz), 1.43, 1.40, 1.35 (3s, 36H), 1.08 (m, 1H), 1.00 (m, 1H); HRMS (ESI MS m/z) calculated for C37H64N7O9P2+ (M+H)+, 812.4235; found, 812.4221.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(2-aminoethylaminocarbonyl)-1-hexynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (31)
Yield from 28 and ethylenediamine: 8.1 mg (73%). 1H NMR (CDC13) δ 8.04 (s, 1H), 5.29 (dd, 1H, J = 14.4, 7.5 Hz), 5.08 (d, 1H, J = 7.2 Hz), 4.59 (dd, 1H, J = 11.1, 4.5 Hz), 3.80 (dd, 1H, J = 10.9, 6.5 Hz), 3.25 (c, 3H, J = 5.5 Hz), 3.17 (s, 1H), 2.77 (t, 2H, J = 6.0 Hz),2.48 (t, 2H, J = 6.7 Hz), 2.24 (t, 2H, J = 7.3 Hz), 2.22 (m, 1H), 2.01 (m, 1H), 1.40, 1.32, 1.33 (3s, 36H), 1.04 (m, 1H), 0.90 (m, 1H); HRMS (ESI MS m/z) calculated for C38H66N7O9P2+ (M+H)+, 826.4392; found, 826.4434.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(3-aminopropylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (32)
Yield from 26 and 1,3-diaminopropane: 10.4 mg (95%). 1H NMR (CDC13) δ 8.09 (s, 1H), 5.39 (dd, 1H, J = 14.0, 7.2 Hz), 5.09 (d, 1H, J = 7.2 Hz), 4.66 (dd, 1H, J = 10.8, 4.2 Hz), 3.93 (dd, 1H, J = 10.5, 6.0 Hz), 3.43 (m, 2H), 3.19 (br s, 3H), 3.11 (m, 1H), 3.05 (m, 1H), 2.97 (m, 2H), 2.85 (t. 2H, J = 7.2 Hz), 2.60 (m, 2H), 2.13 (m, 1H), 1.88 (m, 2H), 1.76 (m, 1H), 1.41, 1.39, 1.37 (3s, 36H), 1.28 (m, 1H), 1.22 (m, 1H); HRMS (ESI MS m/z) calculated for C37H64N7O9P2+ (M+H)+, 812.4235; found, 812.4225.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxy-phosphoryloxymethyl)-4-{2-[(4-aminobutylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (33)
Yield from 26 and 1,4-diaminobutane: 7.8 mg (70%). 1H NMR (CDC13) δ 8.17 (s, 1H), 5.39 (m, 1H), 5.11 (t, 1H, J = 6.3 Hz), 4.94 (m, 1H), 4.67 (dd, 1H, J = 11.4, 5.4 Hz), 3.97 (m, 1H), 3.03 (m, 2H), 3.22 (m, 2H), 3.10 (s, 1H), 2.77 (t, 2H, J = 7.2 Hz), 2.57 (t, 2H, J = 6.9 Hz), 2.38 (m, 2H), 2.31 (c, 2H, J = 7.5 Hz), 2.15 (m, 1H), 2.05 (m, 1H), 1.42, 1.39, 1.36 (3s, 36H), 1.84-1.54 (m, 2H), 1.10 (m, 1H), 0.94 (m, 1H); HRMS (ESI MS m/z) calculated for C38H66N7O9P2+ (M+H)+, 826.4392; found, 826.4452.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxy-phosphoryloxymethyl)-4-{2-[(6-aminohexylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (34)
Yield from 26 and 1,6-diaminohexane: 7.8 mg (68%). 1H NMR (CDC13) δ 8.20 (s, 1H), 5.36 (m, 1H), 5.13 (d, 1H, J = 6.9 Hz), 4.68 (dd, 1H, J = 15.9, 6.9 Hz), 3.88 (dd, 1H, J = 11.1, 6.3 Hz), 3.31 (m, 2H), 2.99 (m, 1H), 2.76 (t, 2H, J = 8.0 Hz), 2.57 (t, 2H, J = 8.0 Hz), 2.32 (m, 2H), 2.18 (m, 1H), 1.75 (m, 2H), 1.49, 1.478, 1.26 (3s, 36H), 1.10 (m, 1H), 0.94 (m, 1H); HRMS (ESI MS m/z) calculated for C40H70N7O9P2+ (M+H)+, 854.4705; found, 854.4731.
General procedure for 2-(aminoalkylamino-carbonylalkynyl) derivatives 11–16
A mixture of the appropriate phosphate-protected derivative 29–34 (0.0045 mmol) in CH2C12 (0.5 mL) was treated with TFA (11 μL) and the reaction mixture was stirred at room temperature for 3 h. After removal of the solvent, the crude product was purified by HPLC with System E and isolated in the triethylammonium salt form to obtain the desired compounds as solids.
(1'R,2'S,4'S,5'S)-4-{2-[(2-Aminoethylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (11)
Yield from 29: 3.1 mg (71%). 1H NMR (D2O) δ 8.63 (s, 1H), 5.09 (dd, 1H, J = 16.2, 9.0 Hz), 4.92 (d, 1H, J = 6.3 Hz), 4.50 (dd, 1H, J = 11.4, 4.2 Hz), 3.63 (m, 1H), 3.58 (t. 3H, J = 6.3 Hz), 2.82 (t. 2H, J = 6.7 Hz), 2.67 (t. 2H, J = 6.6 Hz), 2.25 (dd, 1H, J = 14.7, 7.6 Hz), 1.95 (m, 1H), 1.18 (m, 1H), 1.00 (m, 1H); 31P NMR (D2O) δ 3.19 (s), 2.60 (s); HRMS (ESI MS m/z) calculated for C20H28N7O9P2− (M-H)−, 572.1429; found, 572.1412. HPLC RT 5.3 min (98%) in solvent System A, 5.6 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(2-Aminoethylaminocarbonyl)-1-pentynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (12)
Yield from 30: 3.0 mg (68%). 1H NMR (D2O) δ 8.54 (s, 1H), 5.15 (dd, 1H, J = 15.9, 8.6 Hz), 4.94 (d, 1H, J = 6.6 Hz), 4.55 (dd, 1H, J = 11.5, 5.6 Hz), 3.7 (dd, 1H, J = 11.4, 4.5 Hz), 5.7 (t, 2H, J = 5.7 Hz), 3.17 (t, 2H, J = 6.0 Hz), 3.09 (br s, 3H), 2.55 (m, 2H), 2.24 (dd, 1H, J = 14.5, 7.6 Hz), 1.97 (m, 3H), 1.21 (m, 1H), 1.00 (m, 1H); 31P NMR (D2O) δ 2.02 (s), 1.58 (s); HRMS (ESI MS m/z) calculated for calculated for C21H30N7O9P2− (M-H)−, 586.1580; found, 586.1592. HPLC RT 5.9 min (98%) in solvent System A, 5.8 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(2-Aminoethylaminocarbonyl)-1-hexynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (13)
Yield from 31: 3.3 mg (73%). 1H NMR (D2O) δ 8.55 (s, 1H), 5.18 (dd, 1H, J = 11., 7.2 Hz), 5.00 (d, 1H, J = 6.3 Hz), 4.57 (dd, 1H, J = 11.4, 5.7 Hz), 3.66 (dd, 1H, J = 11.1, 4.8 Hz), 3.52 (t, 2H, J = 6.0 Hz), 3.16 (t, 2H, J = 6.0 Hz), 3.11 (s, 3H), 2.55 (t, 2H, J = 6.9 Hz),2.40 (t, 2H, J = 7.5 Hz), 2.26 (m, 1H), 2.00 (m, 1H), 1.94 (m, 1H), 1.84 (m, 2H),1.69 (m, 2H), 1.21 (m, 1H), 1.00 (m, 1H); 31P NMR (D2O) δ 1.92 (s), 1.43 (s); HRMS (ESI MS m/z) calculated for C22H32N7O9P2− (M-H)−, 600.1737; found, 600.1735. HPLC RT 6.4 min (98%) in solvent System A, 6.3 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(3-Aminopropylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (14)
Yield from 32: 2.2 mg (50%). 1H NMR (CDC13) δ 8.75 (s, 1H), 5.05 (m, 1H), 5.03 (m, 1H), 4.96 (m, 1H), 4.47 (m, 1H), 3.64 (m, 1H), 3.35 (t, 2H, J = 5.1 Hz), 3.11 (s, 2H), 2.81 (t, 2H, J = 6.3 Hz), 2.61 (t, 2H, J = 6.3 Hz), 2.24 (m, 1H), 1.93 (m, 2H), 1.85 (m, 2H), 1.65 (m, 1H), 1.16 (m, 1H), 0.99(m, 1H); HRMS (ESI MS m/z) calculated for C21H30N7O9P2− (M-H)−, 586.1580; found, 586.1594. 31P NMR (D2O) δ 1.90 (s), 1.19 (s); HPLC RT 5.5 min (99%) in solvent System A, 5.0 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(4-Aminobutylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane(15)
Yield from 33: 2.2 mg (49%). 1H NMR (CDC13) δ 8.55 (s, 1H), 5.11 (dd, 1H, J = 10.5, 6.9 Hz), 4.94 (d, 1H, J = 7.2 Hz), 4.53 (dd, 1H, J = 12.0, 6.0 Hz), 3.59 (dd, 1H, J = 13.2, 6.0 Hz), 3.07 (m, 2H), 2.78 (t, 2H, J = 6.3 Hz), 2.56 (m, 4H), 2.18 (m, 1H), 2.00 (m, 4H), 1.91 (m, 2H), 1.54, 1.44 (m, 4H), 1.17(m, 1H), 0.96 (m, 1H); 31P NMR (D2O) δ 1.99 (s), 1.63 (s); HRMS (ESI MS m/z) calculated for C22H32N7O9P2− (M-H)−, 600.1737; found, 600.1763. HPLC RT 5.8 min (98%) in solvent System A, 5.4 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(6-Aminohexylaminocarbonyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (16)
Yield from 34: 2.9 mg (63%). 1H NMR (CDC13) δ 8.55 (s, 1H), 6.03 (bs, 1H), 5.15 (m, 1H), 5.03 (m, 1H), 4.48 (m, 1H), 3.71 (m, 1H), 3.65 (m, 2H), 3.44 (m, 2H), 3.15 (s, 2H), 2.99 (t, 2H, J = 7.8 Hz), 2.66 (m, 1H), 2.29 (m, 1H), 1.98 (m, 1H), 1.88 (m, 1H), 1.69 (m, 1H), 1.43 (m, 1H), 1.15 (m, 1H), 0.97 (m, 1H); 31P NMR (D2O) δ 3.09 (s), 2.48 (s); HRMS (ESI MS m/z) calculated for C24H36N7O9P2+ (M+H)+, 628.2055; found, 628.2070. HPLC RT 7.4 min (99%) in solvent System A, 6.6 min (98%) in System B.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxy-phosphoryloxymethyl)-4-{2-[(6-(6-oxo-6-(2-(5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)ethylamino)hex-1-ynyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (35)
HATU (2.8 mg, 0.0074 mmol, 1.2 eq) was added to a solution of 30 (5 mg, 0.0062 mmol), biotin (1.67 mg, 0.00682 mmol, 1.1 eq), and DIEA (1.4 μL, 0.008 mmol, 1.3 eq) in DMF (0.5 mL), and the resulting mixture was stirred overnight at room temperature. The residue was purified by flash chromatography EtOAc:MeOH (10:1 to 3:1) to give 3.9 mg of 35 (61%) as a solid. 1H NMR (CDC13) δ 8.07 (s, 1H), 5.73 (m, 2H), 5.02 (m, 4H,), 4.69 (m, 1H), 4.51 (m, 1H), 4.25 (m.,4H), 3.80 (m, 1H),3.44 (m, 4H), 3.30 (m, 3H), 3.20 (m, 2H), 2.82 (m, 1H), 2.52 (m, 2H), 1.62 (m, 6H), 1.43, 1.40, 1.35 (3s, 36H), 1.08 (m, 1H); HRMS (ESI MS m/z) calculated for C47H78N9O11P2S+ (M+H)+, 1038.5011; found, 1038.5016.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxy-phosphoryloxymethyl)-4-{2-[6-(6-oxo-6-(2-(6-(5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)hexanamido)ethylamino)hex-1-ynyl)]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (36)
A solution of 30 (5 mg, 0.0062 mmol) and biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinimide ester sodium salt (5.18 mg, 0.0093 mmol, 1.5 eq) in DMF (1 mL) was treated with bicarbonate buffer (0.002 M Na2CO3, 0.048 M NaHCO3, 0.15 M NaCl) until pH 8.5. The mixture was stirred overnight at room temperature. The residue was purified by flash chromatography using EtOAc:MeOH (10:1 to 3:1) to give 5.6 mg of 36 (78%) as a solid. 1H NMR (CDC13) δ 8.08 (s, 1H), 5.44 (dd, 1H, J = 14.4, 7.8 Hz), 5.08 (d, 1H, J = 6.9 Hz), 4.69 (dd, 1H, J = 11.1, 5.1 Hz), 4.51 (m, 2H), 4.36 (m, 2H), 4.14 (t, 1H, J = 7.2 Hz), 4.01 (dd, 1H, J = 11.1, 6.0 Hz), 3.38 (m, 3H), 3.32 (m, 2H), 2.92 (m, 2H), 2.53 (t, 2H, J = 6.6 Hz), 2.46 (t, 2H, J = 7.5 Hz), 2.22 (m, 1H), 1.92 (m, 6H), 1.63, 1.49, 1. 47 1.46 (3s, 36H), 1.27 (m, 1H), 1.12 (m, 1H); HRMS (ESI MS m/z) calculated for C53H89N10O12P2S+ (M+H)+, 1151.5952; found, 1151.5901.
(1'R,2'S,4'S,5'S)-4-{2-[(6-(6-Oxo-6-(2-(5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)ethylamino)hex-1-ynyl)-1-propynyl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (17)
A solution of 35 (3 mg, 0.0029 mmol) in CH2C12 (0.3 mL) was treated with TFA (9 μL), and the reaction mixture was stirred at room temperature for 3 h. After removal of the solvent, the crude product was purified by HPLC with System E to give 0.6 mg of 17 (20%, isolated in the triethylammonium salt form) as a solid. 1H NMR (CDC13) δ 8.68 (s, 1H), 5.00 (m, 4H,), 4.88 (m, 1H), 4.48 (m, 1H),4.23 (m, 4H), 3.83 (m, 1H), 3.59 (m, 4H), 3.33 (m, 3H), 3.12 (m, 2H), 3.01(m, 1H), 2.82 (m, 1H), 2.52 (c, 2H, J = 6.7 Hz), 2.23 (t, 2H, J = 7.8 Hz), 1.97 (m, 2H), 1.58 (m, 1H), 0.44 (m, 1H); HRMS (ESI MS m/z) calculated for C31H44N9O11P2S− (M-H)−, 812.2362; found, 812.2358. RT 6.7 min (98%) in solvent System A, 12.4 min (98%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[6-(6-Oxo-6-(2-(6-(5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)hexanamido)ethylamino)hex-1-ynyl)]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (18)
A solution of 36 (5 mg, 0.0043 mmol) in CH2C12 (0.3 mL) was treated with TFA (10 μL), and the reaction mixture was stirred at room temperature for 3 h. After removal of the solvent, the crude product was purified by HPLC with System E to give 1.58 mg of 18 (36%, isolated in the triethylammonium salt form) as a solid. 1H NMR (D2O) 8.65 (s, 1H), 5.24 (dd, 1H, J = 14.4, 7.8 Hz), 5.04 (d, 1H, J = 6.0 Hz), 4.60 (m, 1H), 4.48 (dd, 2H, J = 8.1, 4.3 Hz), 4.39 (m, 2H), 3.66 (m, 3H), 3.16 (m, 2H), 3.04 (m, 2H), 2.57 (t, 2H, J = 6.9 Hz), 2.52 (t, 2H, J = 7.5 Hz), 2.30 (m, 1H), 1.59 (m, 6H), 1.45 (m, 1H); 31P NMR (D2O) δ 2.78 (s), 2.13 (s); HRMS (ESI MS m/z) calculated for C37H55N10O12P2S− (M-H)−, 925.3102; found, 925.3126. HPLC RT 7.24 min (99%) in solvent System A, 12.3 min (99%) in System B.
(1'R,2'S,4'S,5'S)-4-{2-[(6-(6-(4-(Fluorosulfonyl)phenethylamino)-6-oxohex-1-ynyl)]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (19)
HATU (2.8 mg, 0.0074 mmol, 1.2 eq) was added to a solution of 9 (2 mg, 0.0021 mmol), 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride 37 (1.67 mg, 0.00682 mmol, 1.1 eq) and DIEA (1.4 μL, 0.008 mmol, 1.3 eq) in DMF (0.5 mL), and the resulting mixture was stirred overnight at room temperature. The crude product was purified by HPLC with System E to give 0.6 mg of 19 (25%, isolated in the triethylammonium salt form) as a solid. 1H NMR (D2O) δ 8.53 (s, 1H), 8.17 (s, 1H), 8.12 (d, 2H, J = 8.3 Hz), 7.73 (d, 2H, J = 8.3 Hz), 5.21 (m, 1H), 4.92 (m, 1H), 3.48 (m, 1H), 3.25 (s, 3H), 2.97 (m, 4H), 2.31 (m, 1H), 2.28 (m, 1H), 2.17 (m, 2H), 1.31 (m, 1H); HRMS (ESI MS m/z) calculated for C27H33N6O11P2S− (M-F)−, 711.1403; found, 711.1498; HPLC RT 10.9 min (96%) in solvent System A, 17.6 min (96%) in System B.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-(1,5-hexadiyn-1-yl)-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (38)
To a solution of 25 (10 mg, 0.013 mmol) in DMF (0.2 mL), 1,5-hexadiyne (50% in pentane, 12 mL, 0.13 mmol), tetrakis(triphenylphosphine)palladium(0) (2.9 mg, 0.0025 mmol), copper(I) iodide (1.0 mg, 0.0051 mmol) and triethylamine (3.5 mL, 0.025 mmol) were added. The mixture was stirred overnight at room temperature. The residue was concentrated and purified by flash chromatography using CHCl3:EtOAc (1:1) to CHCl3:EtOAc:MeOH(5:5:1) and purified again by HPLC with System D to give 5.0 mg of 38 (53%) as a pale yellow oil. 1H NMR (CDC13) δ 8.16 (s, 1H), 5.90-5.77 (br, 1H), 5.33 (dd, 1H, J = 14.4, 7.8 Hz), 5.19 (d, 1H, J = 6.9 Hz), 4.68 (dd, 1H, J = 11.1, 4.8 Hz), 3.83 (dd, 1H, J = 11.1, 6.3 Hz), 3.23 (brs, 3H), 2.72 (t, 2H, J = 7.5 Hz), 2.61-2.53 (m, 2H), 2.29 (dd, 1H, J = 15.0, 8.1 Hz), 2.15-2.03 (m, 2H), 1.50, 1.49, 1.47 (3s, 36H), 1.13 (dd, 1H, J = 5.7, 4.5 Hz), 0.96 (dd, 1H, J = 8.4, 6.3 Hz); HRMS (ESI MS m/z) calculated for C35H56N5O8P2+ (M+H)+, 736.3604; found, 736.3604.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-(1,6-heptadiyn-1-yl)-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (39)
To a solution of 25 (10 mg, 0.013 mmol) in DMF (0.2 mL), 1,6-heptadiyne (15 mL, 0.13 mmol), tetrakis(triphenylphosphine)palladium(0) (2.9 mg, 0.0025 mmol), Copper(I) iodide (1.0 mg, 0.0051 mmol) and triethylamine (3.5 mL, 0.025 mmol) were added. The mixture was stirred overnight at room temperature. The residue was concentrated and purified by flash chromatography using CHCl3:EtOAc (1:1) to CHCl3:EtOAc:MeOH(5:5:1) and purified again by HPLC with System D to give 4.8 mg of 39 (50%) as pale yellow oil. 1H NMR (CDC13) δ 8.15 (s, 1H), 5.89-5.78 (br, 1H), 5.35 (dd, 1H, J = 14.4, 7.8 Hz), 5.20 (d, 1H, J = 6.9 Hz), 4.67 (dd, 1H, J = 11.1, 4.8 Hz), 3.83 (dd, 1H, J = 11.1, 6.3 Hz), 3.24 (brs, 3H), 2.60 (t, 2H, J = 7.2 Hz), 2.39 (dt, 2H, J = 7.2, 2.7 Hz), 2.30 (dd, 1H, J = 15.3, 8.1 Hz), 2.17-2.04 (m, 1H), 1.98 (t, 1H, J = 2.7 Hz), 1.91 (dt, 2H, J = 7.2, 7.2 Hz), 1.50, 1.49, 1.47, 1.46 (4s, 36H), 1.09 (dd, 1H, J = 8.4, 6.9 Hz), 0.96 (dd, 1H, J = 8.1, 6.6 Hz); HRMS (ESI MS m/z) calculated for C36H58N5O8P2+ (M+H)+, 750.3761; found, 750.3761.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-(1,7-octadiyn-1-yl)-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (40)
To a solution of 25 (10 mg, 0.013 mmol) in DMF (0.2 mL), 1,7-octadiyne (17 mL, 0.13 mmol), tetrakis(triphenylphosphine)palladium(0) (2.9 mg, 0.0025 mmol), Copper(I) iodide (1.0 mg, 0.0051 mmol) and triethylamine (3.5 mL, 0.025 mmol) were added. The mixture was stirred overnight at room temperature. The residue was concentrated and purified by flash chromatography using CHCl3:EtOAc (1:1) to CHCl3:EtOAc:MeOH(5:5:1) and purified again by HPLC with System D to give 3.7 mg of 40 (38%) as pale yellow oil. 1H NMR (CDC13) δ 8.15 (s, 1H), 5.86-5.73 (br, 1H), 5.33 (dd, 1H, J = 15.0, 8.4 Hz), 5.21 (d, 1H, J = 6.9 Hz), 4.68 (dd, 1H, J = 10.8, 4.5 Hz), 3.83 (dd, 1H, J = 11.1, 6.3 Hz), 3.24 (brs, 3H), 2.50 (t, 2H, J = 6.9 Hz), 2.30-2.22 (m, 2H), 2.10 (dd, 1H, J = 15.0, 7.8 Hz), 1.96 (t, 1H, J = 2.7 Hz), 1.82-1.65 (m, 4H), 1.50, 1.49, 1.47, 1.46 (4s, 36H), 1.13 (dd, 1H, J = 6.0, 4.2 Hz), 0.97 (dd, 1H, J = 7.8, 4.2 Hz); HRMS (ESI MS m/z) calculated for C37H60N5O8P2+ (M+H)+, 764.3917; found, 764.3917.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-(1,8-nonadiyn-1-yl)-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (41)
To a solution of 25 (10 mg, 0.013 mmol) in DMF (0.2 mL), 1,8-nonadiyne (19 mL, 0.13 mmol), tetrakis(triphenylphosphine)palladium(0) (2.9 mg, 0.0025 mmol), copper(I) iodide (1.0 mg, 0.0051 mmol) and triethylamine (3.5 mL, 0.025 mmol) were added. The mixture was stirred overnight at room temperature. The residue was concentrated and purified by flash chromatography using CHCl3:EtOAc (1:1) to CHCl3:EtOAc:MeOH(5:5:1) and purified again by HPLC with System D to give 5.4 mg of 41 (55%) as pale yellow oil. 1H NMR (CDC13) δ 8.16 (s, 1H), 5.90-5.77 (br, 1H), 5.33 (dd, 1H, J = 14.4, 7.8 Hz), 5.19 (d, 1H, J = 6.9 Hz), 4.68 (dd, 1H, J = 11.1, 4.8 Hz), 3.83 (dd, 1H, J = 11.1, 6.3 Hz), 3.23 (brs, 3H), 2.72 (t, 2H, J = 7.5 Hz), 2.61-2.53 (m, 2H), 2.29 (dd, 1H, J = 15.0, 8.1 Hz), 2.15-2.03 (m, 2H), 1.50, 1.49, 1.47 (3s, 36H), 1.13 (dd, 1H, J = 5.7, 4.5 Hz), 0.96 (t, 1H, J = 7.5 Hz); HRMS (ESI MS m/z) calculated for C38H62N5O8P2+ (M+H)+, 778.4074; found, 778.4074.
General procedure for tert-Bu-protected triazole derivatives 42–45
Freshly prepared aqueous sodium ascorbate (1 M, 1 mmol equivalent) was added to a mixture of the appropriate tert-Bu-protected dialkyne derivative (compound 38 – 41, 1.66 mmol equivalent) and N-(2-azidoethyl)acetamide (24) (3.4 mmol equivalent) in a mixture of THF (46 mL per mmol equivalent) and water (46 mL per mmol equivalent), followed by addition of 7.5% aqueous copper(II) sulfate pentahydrate (3.2 μL per mmol equivalent). The reaction mixture was stirred at room temperature overnight, and the reaction mixture was concentrated. The crude product was used in the next reaction without purification.
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(4-(2-acetamidoethyl)-1H-1,2,3-triazol-4-yl)-but-1-yn-1-yl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (42)
Prepared according to the general procedure from compound 38 (5.0 mg, 6.8 mmol).
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(5-(2-acetamidoethyl)-1H-1,2,3-triazol-4-yl)-pent-1-yn-1-yl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (43)
Prepared according to the general procedure from compound 39 (4.8 mg, 6.4 mmol).
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(6-(2-acetamidoethyl)-1H-1,2,3-triazol-4-yl)-hex-1-yn-1-yl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (44)
Prepared according to the general procedure from compound 40 (2.1 mg, 2.7 mmol).
(1'R,2'S,4'S,5'S)-Phosphoric acid di-tert-butylester 1-(di-tert-butoxyphosphoryloxymethyl)-4-{2-[(7-(2-acetamidoethyl)-1H-1,2,3-triazol-4-yl)-hept-1-yn-1-yl]-6-methylaminopurin-9-yl}-bicyclo[3.1.0]hex-2-yl ester (45)
Prepared according to the general procedure from compound 41 (5.4 mg, 6.9 mmol).
General procedure for phosphate-deprotected triazole derivatives 20–23
A solution of a tert-Bu-protected triazole derivative prepared above (compound 42 – 45, crude) in CH2C12 (0.21 mL) was treated with TFA (0.07 mL), and the reaction mixture was stirred at room temperature for 3 h. After removal of the solvent, the crude product was purified by HPLC with a Luna 5 l RP-C18(2) semipreparative column (250 × 10.0 mm; Phenomenex, Torrance, CA) and using the following conditions: flow rate of 2 mL/min; 10 mM triethylammonium acetate (TEAA)-CH3CN from 100:0 to 70:30 in 20 min to give the desired products 20 – 23 as white amorphous solids (triethylammonium salt form).
(1'R,2'S,4'S,5'S)-4-{2-[(5-(2-Acetamidoethyl)-1H-1,2,3-triazol-4-yl)-pent-1-yn-1-yl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (21)
Prepared according to the general procedure from compound 43 to give 3.6 mg of 21 (65% in 2 steps, isolated as triethylammonium salt form). 1H NMR (D2O) δ 8.70-8.55 (br, 1H), 7.89 (s, 1H), 5.33-5.18 (br, 1H), 5.08-5.01 (m, 1H), 4.70-4.55 (br, 1H), 4.48 (t, 2H, J = 5.7 Hz), 3.73-3.65 (m, 1H), 3.59 (t, 2H, J = 5.7 Hz), 3.13 (brs, 3H), 2.95 (t, 2H, J = 6.6 Hz), 2.52 (t, 2H, J = 6.6 Hz), 2.37-2.21 (m, 1H), 2.10-1.97 (m, 3H), 1.93 (s, 3H), 1.26-1. 19 (m, 1H), 1.06-0.97 (m, 1H); 31P NMR (D2O) δ 4.0 – −1.0 (br); HRMS (ESI MS m/z) calculated for C24H32N9O9P2− (M-H)−, 652.1804; found, 638.1798. RT 7.9 min (98%) in solvent System C.
(1'R,2'S,4'S,5'S)-4-{2-[(6-(2-Acetamidoethyl)-1H-1,2,3-triazol-4-yl)-hex-1-yn-1-yl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (22)
Prepared according to the general procedure from compound 44 to give 1.1 mg of 22 (44% in 2 steps, isolated as triethylammonium salt form). 1H NMR (D2O) δ 8.63-8.50 (br, 1H), 7.83 (s, 1H), 5.31-5.18 (br, 1H), 5.06-5.02 (m, 1H), 4.67-4.50 (m, 1H), 4.51 (t, 1H, J = 6.0 Hz), 3.71-3.63 (m, 1H), 3.62 (t, 1H, J = 6.0 Hz), 3.12 (brs, 3H), 2.81 (t, 2H, J = 6.6 Hz), 2.54 (t, 2H, J = 6.6 Hz), 2.36-2.23 (m, 1H), 2.08-1.93 (m, 1H), 1.93-1.82 (m, 3H), 1.89 (s, 3H), 1.75-1.62 (m, 2H), 1.26-1. 21 (m, 1H), 1.05-0.97 (m, 1H); 31P NMR (D2O) δ 2.2 – 0.2 (br); HRMS (ESI MS m/z) calculated for C25H34N9O9P2− (M-H)−,666.1960; found, 666.1955. RT 8.2 min (98%) in solvent System C.
(1'R,2'S,4'S,5'S)-4-{2-[(7-(2-Acetamidoethyl)-1H-1,2,3-triazol-4-yl)-hept-1-yn-1-yl]-6-methylaminopurin-9-yl}-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane (23)
Prepared according to the general procedure from compound 45 to give 3.6 mg of 23 (59% in 2 steps, isolated as triethylammonium salt form). 1H NMR (D2O) δ 8.65-8.50 (br, 1H), 7.78 (s, 1H), 5.34-5.16 (br, 1H), 5.07-5.00 (m, 1H), 4.69-4.51 (m, 1H), 4.36 (t, 2H, J = 6.3 Hz), 3.75-3.68 (m, 1H), 3.49 (t, 2H, J = 6.0 Hz), 3.11 (brs, 3H), 2.78 (t, 2H, J = 6.0 Hz), 2.51 (t, 2H, J = 6.6 Hz), 2.36-2.21 (m, 1H), 2.10-1.96 (m, 1H), 1.96-1.88 (m, 1H), 1.89 (s, 3H), 1.80-1.63 (m, 4H), 1.58-1.48 (2H, m), 1.27-1. 21 (m, 1H), 1.07-0.98 (m, 1H); 31P NMR (D2O) δ 2.0 – −1.0 (br); HRMS (ESI MS m/z) calculated for C26H36N9O9P2− (M-H)−, 680.2117; found, 680.2111. RT 8.5 min (98%) in solvent System C.
PAMAM Dendrimer derivatives
Dialysis was performed using Spectra/Por® Dialysis membrane (Molecular weight cutoff of 3500) purchased from Spectrum Laboratories, Inc. (Rancho Dominguez, CA, USA). NMR spectra were recorded on a Bruker DRX-600 spectrometer at 25.0 °C under an optimized parameter setting for each sample, unless otherwise mentioned. 1H NMR chemical shifts were measured relative to the residual solvent peak at 2.50 ppm in DMSO-d6 and at 4.80 ppm in D2O. Complete NMR peak assignments were made possible with 2D COSY and NOESY experiments. Each PAMAM dendrimer conjugate was not unimolecular, and thus, the characterization by NMR represented the average value from its polymeric distribution. The integration values were reported only for the peaks clearly resolved (i.e., with a relatively good baseline-separation) in the 1H NMR spectra and to two decimal places.
G3 PAMAM, conjugated with 4-pentynoic acid (47)
HATU (20.6 mg, 55 μmol, 38 eq) was added to a mixture of 46 (10 mg, 1.44 μmol), 4-pentynoic acid (5 mg, 50 μmol, 35 eq), and DIEA (10 μL, 60 μmol, 42 eq) in DMF (0.5 mL). The reaction mixture was stirred at room temperature overnight, and the product was purified by dialysis in water. The mixture was lyophilized to give compound 47 (7.6 mg, 70%) as a white foamy solid, which was determined to contain 8 moieties of 4-pentynoic acid per G3 dendrimer. MALDI-MS: calcd. 7545; found 7536.
G4 PAMAM, conjugated with tert-Bu-protected P2Y1 antagonist 38 (52)
Freshly prepared aqueous sodium ascorbate (1 M, 10 μL, 10 μmol) was added to a mixture of compound 51 (1.0 mg, 0.06 μmol) and compound 38 (0.2 mg, 0.3 μmol) in a mixture of t-butanol (0.1 mL) and water (0.1 mL), followed by addition of 7.5% aqueous cupric sulfate (17 μL, 5.0 μmol). The reaction mixture was stirred at room temperature overnight, and the product was purified by dialysis in water. The mixture was lyophilized to give compound 52 (0.57 mg, 43%) as a white amorphous. 1H NMR (D2O, 400 MHz) δ 8.30 (brs), 5.30-5.25 (br), 5.17-5.13 (br), 4.36-4.30 (br), 4.03-3.90 (m), 3.87-3.03 (m), 2.28-2.20 (br), 2.09-2.00 (br), 1.47-1.36 (br), 1.30-1.00 (m), 1.18 (s), 1.16 (s), 0.80-0.67 (m). MALDI-MS: calcd. 21,029.
G4 PAMAM, conjugated with P2Y1 antagonist (53)
TFA (10 μL) was added to a solution of compound 52 (0.21 mg, 0.01 μmol) in CH2Cl2 (10 μL). The reaction mixture was stirred at room temperature overnight, and concentrated. The product was purified by dialysis in water. The mixture was lyophilized to give compound 53 (0.15 mg, 78%) as a white amorphous solid. 1H NMR (D2O, 400 MHz) δ 4.00-1.70 (br). MALDI-MS: calcd. 19,458; found 19,601.
Pharmacological testing
2-MeSADP was purchased from Sigma (St. Louis, MO). The affinities of bisphosphate analogues for the hP2Y1 receptor were directly determined by using [125I]MRS2500 2 in a radioligand binding assay, as we recently described in detail (25,26). Binding and functional parameters were estimated using GraphPAD Prism software (GraphPAD, San Diego, CA).
Binding of [125I]MRS2500, 2
P2Y1 receptor expression in Sf9 insect cells
Sf9 insect cell membranes expressing recombinant hP2Y1 receptors were prepared as described in detail previously (25). Briefly, suspension cultures of Sf9 insect cells grown to a density of 1.4 × 106 cells/ml were infected with empty vector or recombinant baculovirus encoding the hP2Y1 receptor. Cells were harvested after two days by low speed centrifugation, and the cells were lysed using a Parr cavitation apparatus. A membrane fraction was isolated by high speed centrifugation, and the membranes were resuspended at a concentration of ~ 3.5 mg/ml in 20 mM Tris, pH 8.0, 250 mM sucrose, 2 mM 2-mercaptoethanol, 100 μg/ml phenylmethanesulphonylfluoride, 100 μM benzamidine, and 50 μg/ml tosylphenylalanyl chloromethyl ketone. Membranes were stored at −80°C and diluted approximately 50-fold for binding assays as described below.
Binding assay at the hP2Y1 receptor
Assays with membranes isolated from P2Y1 receptor-expressing Sf9 insect cells typically were with 0.2–0.5 nM [125I]MRS2500 (26) and 100–300 ng of membranes in assay buffer (20 mM HEPES, 145 mM NaCl, 5 mM MgCl2, pH 7.5) in a total volume of 25 μl. All incubations were carried out in 12 × 75 mm conical tubes at 4° C and were terminated after 30 min by the addition of 3.5 ml of ice cold assay buffer followed by vacuum filtration over Whatman GF/A glass microfiber filters. The filters were washed with 7 ml of ice-cold assay buffer and radioactivity on each filter was quantified in a gamma counter. Specific binding was defined as total [125I]MRS2500 bound minus binding occurring in the presence of 30 μM 2-MeSADP or 10 μM MRS2179. Radioactivity was determined in a Beckman 5500B γ-counter.
Statistical Analysis
Binding and functional parameters were calculated using Prism 4.0 software (GraphPAD, San Diego, CA). IC50 values obtained from competition curves were converted to Ki values using the Cheng-Prusoff equation (27). Data were expressed as the mean ± standard error. Statistical analysis was performed using Analysis of Variance (ANOVA) with post hoc test or Student's test where appropriate with p values less than 0.05 being considered significant.
Studies of human washed platelets
Preparation
Human washed platelets were prepared as previously described (28). Briefly, fresh blood obtained from healthy donors was centrifuged at 175×g for 15 min at 37 °C. Then, platelet rich plasma was removed, and centrifuged at 1570×g for 15 min at 37 °C. The platelet pellet was washed twice in Tyrode's buffer (137 mM NaCl, 2 mM KCl, 12 mM NaHCO3, 0.3 mM NaH2PO4, 1 mM MgCl2, 2 mM CaCl2, 5.5 mM glucose, 5 mM Hepes, pH 7.3) containing 0.35% human serum albumin. The final resuspension was at a density of 3×105 platelets/μL in Tyrode's buffer containing 0.02 U/mL of apyrase (adenosine 5'-triphosphate diphosphohydrolase, EC 3.6.1.5), a concentration sufficient to prevent desensitization of platelet P2Y receptors during storage. Platelets were kept at 37 °C throughout all experiments.
Platelet aggregation studies
Aggregation was measured at 37 °C by a turbidimetric method in a dual-channel Payton aggregometer (Payton Associates, Scarborough, Ontario, Canada). The stock solutions for platelet aggregation studies were 1.5 mM solutions of dendrimer 53 or MRS2500 2 in water (HPLC grade, Aldrich), to give final concentrations of 1 or 4 μM 53 and 1 μM 2. An aliquot of platelet suspension (450 μL) was stirred at 1100 rpm and activated by addition of ADP (5 mM stock solution to give a final concentration of 5 μM), in the presence or absence of a monomeric or dendrimeric P2Y1 antagonist and in the presence of human fibrinogen (0.8 mg/mL), in a final volume of 500 μL. The extent of aggregation was estimated quantitatively by measuring the maximum curve height above the baseline level.
Molecular modeling
The homology model of the P2Y1 receptor has been constructed in MOE,1 using a two templates strategy. In particular, our established rhodopsin-based P2Y1 homology model (1) has been for the construction of almost the entirety of the receptor, while the A2A adenosine receptor (PDB ID: 3EML) (29) has been used as the template for the construction of the second extracellular loop (EL2) – alignment shown in Figure S1 of the Supporting Information, page S19. In this way, we obtained a model closely resembling our previous one (18), but with a more solvent-exposed EL2. In particular, the A2A receptor was used as the template to model only the portion of EL2 downstream of Cys 202. The remaining portion of the loop, for which the published crystal structures of GPCRs suggest a greater variability among the receptors, was instead modeled without the use of a template. The Charmm27 force field and the Born model for implicit solvation were employed. The level of energy minimizations for the intermediate and final models was set to medium. This setting employed a MOE internal protocol, which allows only for restrained energy minimizations that do not prevent the model from drifting far away from the template.
Molecular docking
The remainder of the modeling was performed with the Schrödinger package through the Maestro interface.2 The structure of 2 was taken from the previously published model (18). Compound 20 was sketched in Maestro from 2, and its susbtituent at the 2-position was then subjected to 500 steps of energy minimization with the MacroModel engine,4 using the MMFF force field and the GBSA implicit model for solvation. The homology model of the receptor was subjected to the Protein Preparation Wizard workflow. This added hydrogens, which were subsequently minimized, and also optimized the protonation state of His residues and the orientation of hydroxyl groups, Asn residues, and Gln residues. The molecular docking was performed with Glide,3 using the XP protocol and the extended sampling. A docking grid with a side of 30 Ang was centered on the centroid of Arg 128, Lys 280, and Arg 310, which are known to be involved in nucleotide binding. Additionally, the docking of the phosphate moieties was directed within a sphere of 3.4 Ang radius centered on the centroids of Arg 128, Lys 280, and Arg 310. No scaling factors were used for the van der Waals (vdW) radii of the P2Y1 receptor atoms, while a scaling factor of 0.5 was applied to the vdW radii of the ligand atoms with partial charge lower than 0.15. All the hydroxyl groups within the binding pocket were considered as rotatable. A maximum of 50 poses per ligand were generated, all of which were subjected to a post-docking energy minimization. The highest scoring complexes were then subjected to a 500 steps of energy minimization with the MacroModel engine,4 using the MMFF force field and the GBSA implicit model for solvation.
Systematic torsional sampling
Conformational analyses were performed with the Systematic Pseudo-Monte Carlo (SPMC) method, as implemented in MacroModel,4 using the MMFF force field and the GBSA implicit model for solvation. The torsion samping level was set to extended, the number of search steps to 5000, and the number of minimization steps to 500 per search step.
RESULTS
Chemical synthesis
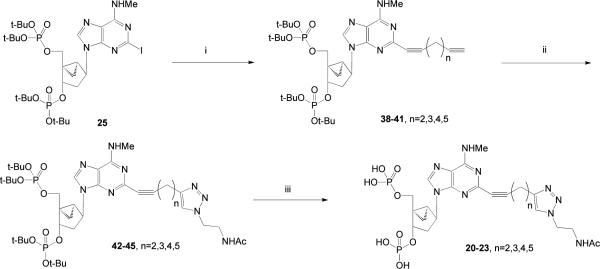
Novel (N)-methanocarba P2Y1 receptor antagonist derivatives (Table 1) containing C2 chains with terminal ester 5 – 7, carboxylic acid 8 – 10, and amino 11 – 16 groups were synthesized by the routes shown in Scheme 1 (15,16,30). A functionalized chain at the 2 position of the adenine ring was formed by a Sonogashira reaction (31) of the protected 2-iodo derivative 25 with (PPh3)2PdCl2, CuI, and the appropriate acetylene. The deprotection of t-butyl protecting groups of the phosphates in derivatives 26 – 28 with TFA provided the ester derivatives 5 – 7. Each of the terminal ester groups was then hydrolyzed with aqueous KOH to yield the corresponding carboxylic acid 8 – 10. Alternatively, the ester group was directly aminolyzed using 1,2-diaminoethylene, 1,3-diaminopropane, or 1,4-diaminobutane to provide, after deprotection with TFA, amine congeners 11 – 16.
Table 1.
Pharmacological data for inhibition of radioligand binding at the hP2Y1 receptor by the novel nucleotide derivatives.
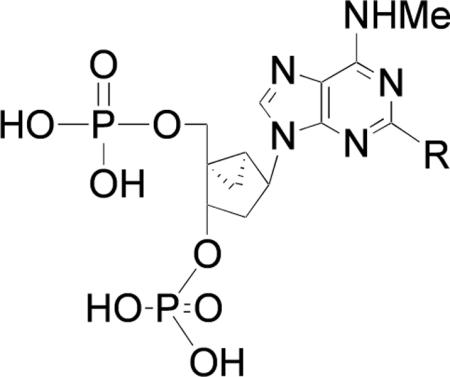
| Compound | R | Binding, Ki, nMa,b |
|---|---|---|
| 2 | I | 0.78 ± 0.08 (18) |
| 3 | C≡CH | 95 ± 39 (18) |
| 4 | C≡C(CH2)3CH3 | 430 ± 200 (18) |
| 5 | C≡C(CH2)2COOCH3 | 363 ± 18 |
| 6 | C≡C(CH2)3COOCH3 | 2200 ± 770 |
| 7 | C≡C(CH2)4COOCH3 | 649 ± 57 |
| 8 | C≡C(CH2)2COOH | 23 ± 3 |
| 9 | C≡C(CH2)3COOH | 376 ± 101 |
| 10 | C≡C(CH2)4COOH | 350 ± 139 |
| 11 | C≡C(CH2)2CONH(CH2)2NH2 | 1440 ± 30 |
| 12 | C≡C(CH2)3CONH(CH2)2NH2 | 1150 ± 100 |
| 13 | C≡C(CH2)4CONH(CH2)2NH2 | 3960 ± 960 |
| 14 | C≡C(CH2)2CONH(CH2)3NH2 | 3580 ± 140 |
| 15 | C≡C(CH2)2CONH(CH2)4NH2 | 132 ± 17 |
| 16 | C≡C(CH2)2CONH(CH2)6NH2 | 2410 ± 460 |
| 17 | C≡C(CH2)3CONH(CH2)2NH-biotin | >10,000 |
| 18 | C≡C(CH2)3CONH(CH2)2NHCO(CH2)5NH-biotin | 1210 ± 230 |
| 19 | C≡C(CH2)3CONH(CH2)2-(4-SO2F)-Ph | >10,000 |
| 20 |
|
300 ± 200 |
| 21 |
|
2700 ± 600 |
| 22 |
|
8300 ± 1500 |
| 23 |
|
6300 ± 2000 |
Affinities determined by using [125I]MRS2500 2 in a radioligand binding assay, except for 2 – 4, which were determined using [3H]MRS2279. The hP2Y1 receptor was expressed to high levels in Sf9 insect cells with a recombinant baculovirus. Membranes prepared from these cells were incubated for 30 min at 4°C in the presence of ~1 nM [125I]MRS2500 2.
Mean ± standard error given for three separate determinations.
2, MRS2500; 8, MRS2816; 15, MRS2900; 20, MRS2939.
Scheme 1.
Synthesis of C2 derivatives of P2Y1 receptor antagonists containing terminal ester 5–7, carboxylic acid 8–10, and amino 11–16 groups. Reagents and conditions: (i) CuI, (Ph3P)2PdCl2,TEA, DMF, rt; (ii) TFA, CH2Cl2, rt; (iii) ethylenediamine, MeOH, rt; (iv) KOH, H2O, rt; (v) TFA, CH2Cl2, rt.
Ligands intended for irreversible receptor binding and avidin complexation were sythesized by attaching an appropriate prosthetic group to the functionalized chains at the C2 position. Biotin conjugates of varying length 17 and 18 were synthesized by amide coupling of the phosphate-protected intermediate amine 30 (Scheme 2), which was the precursor of amine congener 12. Amine derivative 12 was found to be the most potent of the aminoethyl congeners in the series 11 – 13 (see below). The biotin conjugate 17 was prepared by a HATU coupling with biotin via protected intermediate 35, and the extended chain analogue 18 was prepared from the corresponding biotin-ε-aminocaproyl active ester via protected intermediate 36. In the final step, the phosphate groups were cleanly deprotected by reaction with TFA to provide 17 and 18.
Scheme 2.
Preparation of biotin derivatives of a P2Y1 receptor antagonist for avidin complexation. Reagents and conditions: (i) biotin, DIEA, HATU, DMF, rt; (ii) biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinimide ester sodium salt, bicarbonate buffer, DMF, rt; (iii) TFA, CH2Cl2.
Sulfonyl fluoride derivatives have been used as covalently binding affinity labels of GPCRs (32,33). To prepare the sulfonyl fluoride derivative 19, we tried to use a phosphate-protected free carboxylic acid derivative as starting material. Unfortunately, the reaction to deprotect ester derivative 27 with base, using either KOH at room temperature or LiOH at 4 °C, failed to afford the desired protected phosphate compound, and a mixture containing the deprotected phosphate ester and deprotected carboxylic acid was recovered in a very low yield. Finally, the condensation of phosphate-unprotected carboxylic acid 9 with the corresponding amine 37 (containing the preformed arylsulfonyl fluoride) in DMF in presence of HATU and DIEA afforded the amine derivative 19 (Scheme 3). The presence of the free phosphate groups in 9 did not interfere in this coupling reaction.
Scheme 3.
Preparation of a sulfonyl fluoride derivative 19 of a P2Y1 receptor antagonist. Reagents and conditions: (i) DIEA, HATU, DMF, rt.
The functionalized congeners 5 – 16 are designed for amide coupling schemes. Alternately, we introduced dialkyne groups on an extended adenine C2 substituent, as synthetic intermediates leading to selective P2Y1 antagonists. The proximal alkyne was intended to promote receptor recognition, and the distal alkynes reacted with azides to form triazole derivatives through click chemistry (22). The synthesis of the phosphate-protected dialkynyl intermediates 38 – 41 was performed using a Sonogashira coupling (31) on the corresponding 2-iodo intermediate 25 (Scheme 4). We then used Cu(I)-catalyzed [3 + 2] cycloaddition (click reaction) of N-(2-azidoethyl)acetamide (24) and the terminal acetylene group of dialkynyl nucleotide intermediates 38 – 41 to provide protected triazole derivatives 42 – 45. The reactions are generally selective for the terminal and least hindered alkyne when more than one alkyne is present in a reactant (34), which was confirmed in this series. Subsequent deprotection of the 3′ and 5′-phosphate groups with TFA provided biologically active triazole-containing nucleotides 20 – 23.
Scheme 4.
Use of click chemistry to couple P2Y1 antagonists to small azido-bearing molecules AcNH(CH2)2 present in the final products 20–23 is a group to serve as a model for the attachment to a PAMAM dendrimer. Reagents and conditions: (i) diyne, CuI, (Ph3P)2PdCl2,TEA, DMF, rt; (ii) AcNHCH2CH2N3, CuSO4 aq., sodium ascorbate; (iii) TFA.
The optimal chain length for preservation of receptor recognition was found to be two methylene groups between the alkynyl groups, i.e. the product of intermediate 38. This dialkyne was used in subsequent reactions to maintain the P2Y1 receptor binding affinity.
Three approaches were attempted to couple functionalized P2Y1 receptor antagonists to PAMAM dendrimers. In Scheme 5A, a Sonogashira reaction was used in an attempt to couple compound 25 to an alkyne-derivatized PAMAM dendrimer 47. The isolated material was insoluble in DMSO and likely was a cross-linked polymeric side product. We also attempted to condense the short-chain amine congener 11 (in its phosphate-protected form 29) with a G2.5 (32 terminal carboxylic acid groups) PAMAM dendrimer 49 using DIEA, HATU, in DMF at room temperature overnight, but the desired protected conjugate 50 was not obtained (Scheme 5B).
Scheme 5.
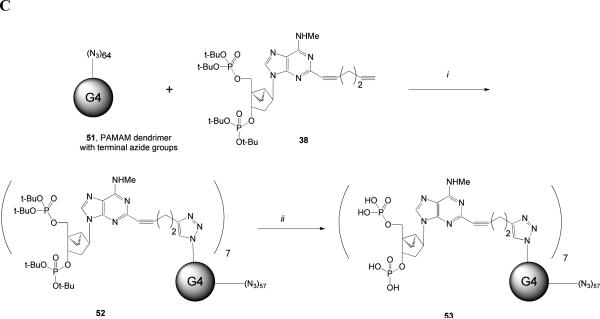
Coupling of P2Y1 receptor antagonists to PAMAM dendrimers in a manner that preserves the interaction with the receptor. (A) Attempted use of the Sonogashira reaction to couple a 2-iodoadenine derivative to a terminal alkyne G3 dendrimer. Reagents and conditions: (i) HATU, 4-pentynoic acid, DIEA, DMF (ii) CuI, (Ph3P)2PdCl2,TEA, DMF, rt. (B) Attempted coupling of a 2-amine derivatized nucleotide to a terminal carboxylic acid G2.5 dendrimer. Reagents: (i) Compound 30, HATU, DIEA, DMSO. (C) Successful use of click chemistry to couple an alkyne-functionalized, protected nucleotide derivative to a terminal azido-bearing G4 dendrimer to yield dendrimer conjugate 53 (MRS2966). Reagents: (i) CuSO4 aq., sodium ascorbate; (ii) TFA.
A third synthetic approach (Scheme 5C) consisted of a click reaction on a PAMAM dendrimer of Generation 4 (G4) containing terminal azido groups (23) that were suitable for the conjugation of P2Y1 receptor antagonists. The azido groups of 51 were first coupled to the nucleotide intermediate 38, which provided the biologically optimal chain length of the C2 chain. The bisphosphate-protected dendrimer conjugate 52 was then deprotected using TFA to provide a dendrimeric nucleoside bisphosphate derivative 53 that was expected to act as a multivalent P2Y1 receptor antagonist. This conjugate contained an average of seven nucleotide moieties on each dendrimer structure out of 64 theoretical terminal groups, as determined by mass spectrometry.
Pharmacological evaluation
The novel bisphosphate monomeric nucleotide derivatives 4 – 23 were evaluated in radioligand binding experiments using the new radiotracer [125I]MRS2500 2 (Table 1) (26). The three terminal ester derivatives 5 – 7 displayed micromolar affinity at the hP2Y1 receptor. The shortest analogue 5, having two methylenes in the alkynyl chain, was slightly more potent than the longer analogues. The carboxylic acid derivatives 8 – 10 were more potent than the corresponding ester derivatives. There was a clearly favored analogue among the carboxylic acid derivatives 8 – 10, i.e., compound 8 with a Ki of 23 nM. A comparison of the hexynyl derivative 4 with the corresponding carboxylic acid derivative 8 indicates a gain of approximately 40-fold in affinity due to the presence of the terminal carboxylate group.
The Ki values of the corresponding ethylenediamine derivatives 11 – 13, derived from the three homologous carboxylic derivatives, were all slightly higher than 1.0 μM. Thus, affinity enhancement was observed by addition of a negatively charged group to the chain, but not a positively charged terminal ethylamino group.
Therefore, we extended the alkylamino moiety of compound 11 to empirically locate an optimal chain length in the terminal amino series. Compounds 14 – 16 were all prepared as extended amine derivatives of the carboxylic acid 8. The highest affinity (Ki = 132 nM) among the amine derivatives was observed for compound 15. The isomeric compound 13, which differed from 15 only in the position of the intermediate amide group within the chain, displayed a Ki of 3.96 μM. Biotin conjugate 18 containing an extended ε-aminocaproyl spacer chain was more potent in binding than the shorter analogue 17. The sulfonyl fluoride derivative 19 was not appreciably potent in binding to the P2Y1 receptor.
Receptor binding affinities of terminal triazolo derivatives 20 – 23 in the P2Y1 receptor-antagonist series prepared as model compounds for linking to carriers by click chemistry showed a great dependence on the chain length. The shortest homologue 20 having two methylenes in the spacer group was the most potent with a Ki value of 300 nM, and the two longest homologues, 22 and 23, were 20 – 30-fold weaker in binding. Therefore, the precursor compound 38 was used in the dendrimer conjugation reactions to achieve the biologically optimal linkage.
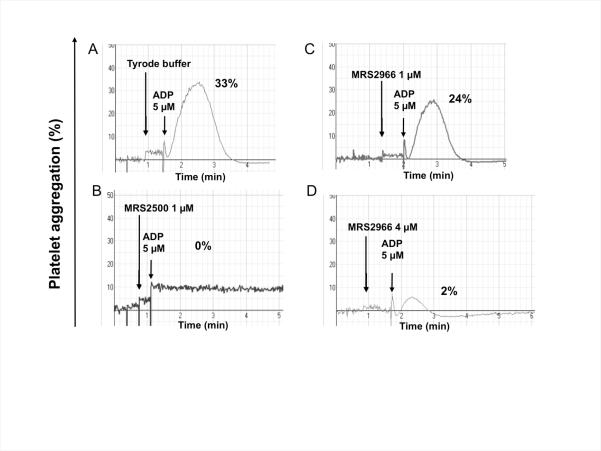
We tested the dendrimer derivative 53 on aggregation of human platelets induced by ADP (Figure 1). Compound 53 (MRS2966, 1 and 4 μM) inhibited platelet aggregation induced by ADP (5 μM) in a concentration-dependent fashion. However, the inhibition by 4 μM 53 was not total (2% residual aggregation compared to 33% in the ADP control), and the dendrimer conjugate 53 was less potent than MRS2500, which completely inhibited aggregation at 1 μM.
Figure 1.
Aggregation of human platelets induced by ADP (A) is inhibited by P2Y1 receptor antagonists: (B) the potent, small molecule antagonist MRS2500 2 and (C,D) dendrimer derivative 53 (MRS2966). The concentration of the dendrimer-ligand complex 53 was measured as the concentration of the dendrimer, not the nucleotide moiety. The results are representative of two separate determinations.
Molecular modeling
We have successfully used rhodopsin-based homology modeling to explain the structure-activity relationships (SAR) of the P2Y receptors and design mutagenesis experiments (1,18,35). However, the solution of the crystal structure of the β-adrenergic (36–38) and the adenosine A2A receptors (29), as well as controlled homology modeling experiments (39,40), revealed that the second extracellular loop (EL2) of rhodopsin is not a good template for modeling the corresponding regions of other GPCRs because it hovers singularly low over the binding pocket, occluding its entrance from the extracellular space. For this reason, here we modified our established rhosopsin-based P2Y1 model by substituting the EL2 with a model based on the A2A adenosine receptor (29). More specifically, we modeled on the basis of the A2A receptor only the portion of EL2 downstream of the conserved Cys involved in the disulfide bridge with the third transmembrane domain (TM3), while the remaining residues where modeled without the use of a template, due to the large structural variability revealed by the published crystal structures for this region. Clearly, the lack of a template is detrimental for the accuracy of this portion of the model; however, since this region appears to be further away from the putative ligand binding pocket it is not likely to affect the docking results.
The molecular docking of 2 into the binding pocket of our revised P2Y1 model confirmed the well-established binding mode that fits well with the experimental data (Figure 2, panels A and B) (1,35). Notably, we have shown that this binding mode can explain in a quantitative manner the SAR of P2Y1 antagonists based on the general scaffold of adenosine-3′,5′-bisphosphate (A3P5P) (18). Briefly, the phosphate moieties dock between TM3, TM6 and TM7, and are coordinated by Arg 128, Lys 280 and Arg 310, three cationic residues that have been experimentally found to be a key component of ligand recognition. Around the glycosidic bond, the nucleobase was arranged in an anti conformation with respect to the pseudosugar moiety, resulting in the substituent at the 2 position of the purine ring pointing towards the space between TM1 and TM7, and oriented towards the core of the receptor. The exocyclic amino group was engaged in a hydrogen bond with Gln 307 in TM7, another key interaction supported by the experiments. In agreement with this docked conformation, in 80% of the crystal structures found in the PDB, A3P5P is found in an anti conformation (see Table S1 of Supporting Information, page S20).
Figure 2.

Molecular models of the hP2Y receptor in complex with compound 2 (panels A and B) and compound 20 (panels C and D), as obtained through molecular docking at the homology model of the receptor. Panels A and C show the receptor-ligand complexes from the plane of the membrane, while panels B and D offer a more detailed view of the ligand binding site, with the intermolecular hydrogen bonds indicated by yellow dotted lines. The backbone structure of the receptor is represented as a ribbon with the colors of the rainbow – TM1: orange; TM2: orange/yellow; TM3: yellow; TM4: green; TM5: green/blue; TM6: cyan; TM7: purple. The carbon atoms are colored in gray for the receptor, in green for compound 2, and in yellow for compound 20. The residue numbering according to a standard GPCR indexing system (47) for the residues shown is: L54(1.35), F112(2.65), D116(EL1), W117(EL1), R128(3.29), F131(3.32), H132(3.33), C202(EL2), Y203(EL2), D204(EL2), T205(EL2), F276(6.51), H277(6.52), N283(6.58), A294(EL3), Y303(7.32), Q307(7.36), G311(7.40), S314(7.43). For both ligands, a schematic representation of the interactions with the receptor is given in the supporting information (Figure S2, pages S22 – S23).
However, for the triazole model compound 20, the results of our molecular docking experiments suggested docking of the nucleoside with the nucleobase oriented in a syn conformation with respect to the sugar moiety, because of the large substituent at the 2 position (Figure 2, panels C and D). This allowed for the maintaining of all the key interactions, including not only those of the phosphate moieties with the three key cationic residues, but also that of the exocyclic amino group with the Gln in TM7, thanks to a concomitant 180° rotation of the latter. As is evident from Figure 2, the syn conformation of the nucleobase allowed the substituent to point towards the extracellular space, thus explaining the possibility of coupling the structural equivalent of 20 to PAMAM dendrimers, while maintaining antagonistic activity. Notably, the terminal amido group of the substituent established a favorable interaction with Asp 116 in the first extracellular loop (EL1). Although the modeling of the loops generally yields less accurate results than that of the transmembrane domains (40), the short nature of EL1 conferred a relatively high confidence on the accuracy of this model.
We subjected to systematic torsional sampling six nucleotides (2, 5, 8, 11, 17, 20) bearing different substituents at the 2 position and having different receptor affinities. For all of these nucleotides, including compound 20, the syn conformation was slightly disfavored over anti, with an energy content that was 0.20 – 0.25% higher (see Table S2 of the Supporting Information, page S21). This suggests that the putative ability of some of the compounds with large appendages at the 2 position to bind to the P2Y1 receptor in the syn conformation is not due to the conformational preferences of the compounds in solution, but rather to steric fit and the establishment of favorable interactions with the receptor. The absence of good templates for the modeling of the loops prevented us from accurately establishing the nature of these interactions. However, as mentioned, our model suggested a hydrogen bond between compound 20 and Asp 116 in EL1.
DISCUSSION
MRS2500 (2) contains a bicyclo[3.1.0]hexane ring, i.e. methanocarba ring system constrained in the (N)-conformation (2′-exo), which is preferred in the receptor binding site (15). This high affinity antagonist has been radiolabeled, and its application for quantification of the P2Y1 receptor underscores the utility of this chemical series as molecular probes of this important signaling protein. MRS2500 is also sufficiently stable for in vivo use, and its antithrombotic properties have been demonstrated against thrombus formation induced by laser or ferric chloride and in systemic thromboembolism. The presence of the methanocarba ring in MRS2500 impedes the action of 5′-nucleotidase to cleave the 5′-monophosphate and other ectonucleotidases that otherwise would greatly limit the half-life in vivo (5). Thus, this series of P2Y1 receptor antagonists has potential for use as research tools in vitro and in vivo or perhaps as therapeutic agents.
Molecular modeling, based on homology of the receptor to the high resolution structure of rhodopsin, and ligand docking have played a role in studies of the SAR (structure activity relationship) at the P2Y1 receptor (1,35). The recognition of the phosphate group(s) in the putative binding site is associated with three conserved cationic residues, which serve as a ligand anchor in the modeling. Another key interaction is established between Gln 307 in TM7 and the exocyclic amino group. Combining docking and QSAR we generated a consensus model correlating antagonist structures with their apparent binding affinities (18). A QSAR model of the P2Y1 receptor antagonists based on alignment of 45 docked nucleotide antagonists was constructed using Comparative Molecular Field Analysis (CoMFA), Comparative Molecular Similarity Indices Analysis (CoMSIA), and molecular descriptors. Consensus scoring of free binding energy based on multiple models predicted accurately the potency of 75% of a test set. This experimentally supported model, in which the adenine ring is oriented in the anti conformation with respect to the pseudosugar moiety, suggests the tolerance for only small substituents at the 2 position. However, the results obtained in the present study suggest that, when the pseudo-nucleotides bear large substituents at the 2 position, the adenine ring adopts the syn conformation upon binding to the receptor. Thus, the 2 position of the adenine ring is oriented towards extracellular space, allowing the presence of a large appendage. Large substituents in some (e.g., 20) but not all (e.g., 17 and 19) of the dervatives presented here are tolerated in receptor binding. The preservation of receptor affinity in many of the functionalized congeners and the P2Y1 receptor antagonistic activity of the macromolecular conjugate 53 are consistent with this proposed new binding mode.
A series of small molecule congeners of N6-methyl-(N)-methanocarba-2′-deoxyadenosine-3′,5′-bisphosphates containing extended 2-alkynyl chains was synthesized and tested in radioligand binding at the hP2Y1 receptor. The chains of these functionalized congeners contained hydrophilic moieties, a reactive substituent, or biotin, linked via an amide. Although the functionalized congeners are thought to be chain functionalized in a region that is insensitive in the receptor binding, only intermediate P2Y1 receptor affinity was achieved in these compounds. Variation of the chain length and position of an intermediate amide group revealed high affinity of both a carboxylic congener 8 (Ki 23 nM) and an extended amine congener 15 (Ki 132 nM), both of which contained a 2-(1-pentynoyl) group. The terminal carboxylic acid or amino groups of the model compounds allowed incorporation of hydrophilic or chemically reactive substituents. These were intended as general sites for conjugation to groups such as fluorescent reporter groups. Thus, a consistent pattern of SAR depending on chain length and terminal functionality was observed. Although 17 and 19 were weak in binding to the receptor, their activities illustrate that the distal regions of the chain can have a major effect on the receptor interaction. Not all chain-extended derivatives anchored on the purine core with a 2-alkynyl group are suitable probes of the P2Y1 receptor.
PAMAM dendrimers are tree-like polymers that have wide application in drug delivery in vivo (41,42). Their advantages as peptide-like biocompatible drug carriers are the ability to control the size, functionality and degree of substitution. We recently reported the first example of using a PAMAM dendrimer scaffolding for the polyvalent, nanoscale presentation of nucleosides to selectively activate adenosine receptors to induce intracellular signal transduction (43–45). The effective inhibition of ADP-induced platelet aggregation was previously demonstrated using PAMAM dendrimers with multiple copies of an A2A adenosine receptor agonist (43,46).
Different approaches to synthesize dendrimer conjugates were compared here. Conjugation of P2Y1 receptor antagonists to dendrimers by amide formation or palladium-catalyzed reaction between an alkyne on the dendrimer and an iodoaryl-derivatized nucleotide was unsuccessful. Only click coupling to an azide-containing PAMAM dendrimer was successful. The resulting multivalent nucleotide conjugate, which had a chain length that in model compounds was favored in receptor binding, inhibited human platelet aggregation. Thus, click chemistry was an efficient synthetic approach to attaching a purinergic antagonist to a PAMAM dendrimer, and this linkage containing a triazole moiety maintained the interaction with the receptor.
In conclusion, this study is the first example of attaching multiple ligands to PAMAM dendrimers that target the P2Y receptors to produce a desired biological effect, i.e. antithrombotic action of P2Y1 antagonists. Although the potency of 53 in inhibition of ADP-induced platelet aggregation was less than the high affinity antagonist 2, at 4 μM it inhibited aggregation by >90%. Thus, this multivalent nucleotide dendrimer conjugate provides an initial approach to novel antithrombotic agents. The preservation of receptor interaction of the functionalized congeners was also consistent with new P2Y1 receptor modeling and ligand docking. The model proposed a conformation change in the angle of the nucleobase to explain the availability of the 2-alkynyl group for conjugation to carriers. In the future, we will explore structural modification of the spacer group to enhance potency. The multivalent conjugates may now be examined in in vivo testing to determine bioavailability and pharmacokinetic characteristics.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases and a grant (GM38213) from the National Institute of General Medical Sciences. We thank Dr. John Lloyd and Dr. Noel Whittaker (NIDDK) for the mass spectral determinations. Research conducted at the Center for Nanophase Materials Sciences at Oak Ridge National Laboratory is sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy. S. de Castro thanks Ministerio de Educación y Ciencia (Spain) for financial support. H. Maruoka thanks Asubio Pharmaceuticals for financial support.
Abbreviations
- A3P5P
adenosine-3′,5′-bisphosphate
- cAMP
adenosine 3′,5′-cyclic phosphate
- CHO
Chinese hamster ovary
- EDC
N-ethyl-N′-dimethylaminopropylcarbodiimide
- DCM
dichloromethane
- DIEA
diisopropylethylamine
- DMEM
Dulbecco's modified Eagle's medium
- DMF
N,N-dimethylformamide
- EDTA
ethylenediaminetetraacetic acid
- EtOAc
ethyl acetate
- Gi
GTP-binding protein containing αi subunit
- Gq
GTP-binding protein containing αq subunit
- GPCR
G protein-coupled receptor
- HATU
2-(1H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyl uronium hexafluorophosphate methanaminium
- HEK
human embryonic kidney
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HLPC RT
high performance liquid chromatography retention time
- HRMS
high resolution mass spectroscopy
- MeOH
methanol
- MRS2179
N6-methyl-2′-deoxyadenosine-3′,5′-bisphosphate
- MRS2500
((1′R,2′S,4′S,5′S)-4-(2-iodo-6-methylamino-purin-9-yl)-1-[(phosphato)-methyl]-2-(phosphato)-bicyclo[3.1.0]hexane)
- PAMAM
polyamidoamine
- PBS
phosphate-buffered saline
- PDB
Protein Data Bank
- PKC
protein kinase C
- TEA
triethylamine
- TEAA
triethylammonium acetate
- TFA
trifluoroacetic acid
- TLC
thin layer chromatography
Footnotes
MOE. [2008.10]. 2008. Chemical Computing Group Inc.
Maestro. [9.0]. 2009. Schrödinger, LLC.
Glide. [5.5]. 2009. Schrödinger, LLC.
MacroModel. [9.7]. 2009. Schrödinger, LLC.
Supporting Information Available. HPLC traces of monomeric compounds submitted for biological testing, sequence alignment for P2Y receptors, documentation of the anti conformation in crystal structures, and schematic representations of the receptor-ligand interactions. This material is available free of charge via the Internet at http://pubs.acs.org/BC.
REFERENCES
- (1).Costanzi S, Mamedova L, Gao ZG, Jacobson KA. Architecture of P2Y nucleotide receptors: structural comparison based on sequence analysis, mutagenesis, and homology modeling. J. Med. Chem. 2004;47:5393–5404. doi: 10.1021/jm049914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Knight GE, Fumagalli M, Gachet C, Jacobson KA, Weisman GA. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol. Rev. 2006;58:281–341. doi: 10.1124/pr.58.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gachet C. Regulation of platelet functions by P2 receptors. Ann. Rev. Pharmacol. Toxicol. 2006;46:277–300. doi: 10.1146/annurev.pharmtox.46.120604.141207. [DOI] [PubMed] [Google Scholar]
- (4).Jin J, Kunapuli SP. Coactivation of two different G protein-coupled receptors is essential for ADP-induced platelet aggregation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8070–8074. doi: 10.1073/pnas.95.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hechler B, Nonne C, Roh EJ, Cattaneo M, Cazenave JP, Lanza F, Jacobson KA, Gachet C. MRS2500 [2-iodo-N6-methyl-(N)-methanocarba-2'-deoxyadenosine-3',5'-bisphosphate], a potent, selective, and stable antagonist of the platelet P2Y1 receptor with strong antithrombotic activity in mice. J. Pharmacol. Exp. Ther. 2006;316:556–563. doi: 10.1124/jpet.105.094037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Paredes-Gamero EJ, Leon CMMP, Borojevic R, Oshiro MEM, Ferreira AT. Changes in intracellular Ca2+ levels induced by cytokines and P2 agonists differentially modulate proliferation or commitment with macrophage differentiation in murine hematopoietic cells. J. Biol. Chem. 2008;283:31909–31919. doi: 10.1074/jbc.M801990200. [DOI] [PubMed] [Google Scholar]
- (7).Hechler B, Freund M, Ravanat C, Magnenat S, Cazenave JP, Gachet C. Reduced atherosclerotic lesions in P2Y1/apolipoprotein E double-knockout mice: the contribution of non-hematopoietic-derived P2Y1 receptors. Circulation. 2008;118:754–763. doi: 10.1161/CIRCULATIONAHA.108.788927. [DOI] [PubMed] [Google Scholar]
- (8).Sellers LA, Simon J, Lundahl TS, Cousens DJ, Humphrey PP, Barnard EA. Adenosine nucleotides acting at the human P2Y1 receptor stimulate mitogen-activated protein kinases and induce apoptosis. J. Biol. Chem. 2001;276:16379–16390. doi: 10.1074/jbc.M006617200. [DOI] [PubMed] [Google Scholar]
- (9).Amadio S, Vacca F, Martorana A, Sancesario G, Volonté C. P2Y1 receptor switches to neurons from glia in juvenile versus neonatal rat cerebellar cortex. BMC Developmental Biol. 2007;7:1–17. doi: 10.1186/1471-213X-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tonazzini I, Trincavelli ML, Montali M, Martini C. Regulation of A1 adenosine receptor functioning induced by P2Y1 purinergic receptor activation in human astroglial cells. J. Neurosci. Res. 2008;86:2857–2866. doi: 10.1002/jnr.21727. [DOI] [PubMed] [Google Scholar]
- (11).Fujita T, Tozaki-Saitoh H, Inoue K. P2Y1 receptor signaling enhances neuroprotection by astrocytes against oxidative stress via IL-6 release in hippocampal cultures. Glia. 2009;57:244–257. doi: 10.1002/glia.20749. [DOI] [PubMed] [Google Scholar]
- (12).Massé K, Bhamra S, Eason R, Dale N, Jones EA. Purine-mediated signalling triggers eye development. Nature. 2007;449:1058–1062. doi: 10.1038/nature06189. [DOI] [PubMed] [Google Scholar]
- (13).Shalito I, Kopyleva O, Serebruany V. Novel antiplatelet agents in development: Prasugrel, ticagrelor, and cangrelor and beyond. Am. J. Ther. 2009 Feb 28; doi: 10.1097/MJT.0b013e318187de4f. PMID: 19262362. [DOI] [PubMed] [Google Scholar]
- (14).Boyer JL, Mohanram A, Camaioni E, Jacobson KA, Harden TK. Competitive and selective antagonism of P2Y1 receptors by N6-methyl 2′-deoxyadenosine 3′,5′-bisphosphate. Brit. J. Pharmacol. 1998;124:1–3. doi: 10.1038/sj.bjp.0701837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kim HS, Ohno M, Xu B, Kim HO, Choi Y, Ji XD, Maddileti S, Marquez VE, Harden TK, Jacobson KA. 2-Substitution of adenine nucleotide analogues containing a bicyclo[3.1.0]hexane ring system locked in a northern conformation: enhanced potency as P2Y1 receptor antagonists. J. Med. Chem. 2003;46:4974–4987. doi: 10.1021/jm030127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cattaneo M, Lecchi A, Ohno M, Joshi BV, Besada P, Tchilibon S, Lombardi R, Bischofberger N, Harden TK, Jacobson KA. Antiaggregatory activity in human platelets of potent antagonists of the P2Y1 receptor. Biochem. Pharmacol. 2004;68:1995–2002. doi: 10.1016/j.bcp.2004.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Houston D, Ohno M, Nicholas RA, Jacobson KA, Harden TK. [32P]2-iodo-N6-methyl-(N)-methanocarba-2'-deoxyadenosine-3',5'-bisphosphate ([32P]MRS2500), a novel radioligand for quantification of native P2Y1 receptors. Br. J. Pharmacol. 2006;147:459–467. doi: 10.1038/sj.bjp.0706453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Costanzi S, Tikhonova IG, Ohno M, Roh EJ, Joshi BV, Colson A-O, Houston D, Maddileti S, Harden TK, Jacobson KA. P2Y1 antagonists: combining receptor-based modeling and QSAR for a quantitative prediction of the biological activity based on consensus scoring. J. Med. Chem. 2007;50:3229–3241. doi: 10.1021/jm0700971. [DOI] [PubMed] [Google Scholar]
- (19).Léon C, Hechler B, Freund M, Eckly A, Vial C, Ohlmann P, Dierich A, LeMeur M, Cazenave JP, Gachet C. Defective platelet aggregation and increased resistance to thrombosis in purinergic P2Y1 receptor-null mice. J. Clin. Invest. 1999;104:1731–1737. doi: 10.1172/JCI8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Léon C, Freund M, Ravanat C, Baurand A, Cazenave JP, Gachet C. Key role of the P2Y1 receptor in tissue factor-induced thrombin-dependent acute thromboembolism: studies in P2Y1-knockout mice and mice treated with a P2Y1 antagonist. Circulation. 2001;103:718–723. doi: 10.1161/01.cir.103.5.718. [DOI] [PubMed] [Google Scholar]
- (21).Jacobson KA. Functionalized congener approach to the design of ligands for G protein–coupled receptors (GPCRs) Bioconjugate Chem. 2009;20:1816–1835. doi: 10.1021/bc9000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov. Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- (23).Tosh DK, Yoo LS, Chinn M, Hong K, Kilbey SM, Barrett MO, Fricks IP, Harden TK, Gao ZG, Jacobson KA. Polyamidoamine (PAMAM) dendrimer conjugates of “clickable” agonists of the A3 adenosine receptor and coactivation of the P2Y14 receptor by a tethered nucleotide. Bioconjugate Chem. 2010;21:372–384. doi: 10.1021/bc900473v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Saito S, Tamai H, Usui Y, Inaba M, Moriwake T. Efficient synthesis of secondary carboxamides with ω-substituted ethyl and propyl groups on nitrogen atom by nucleophilic ring opening of cyclic imidates. Chem. Lett. 1984;7:1243–1246. [Google Scholar]
- (25).Waldo GL, Corbitt J, Boyer JL, Ravi G, Kim HS, Ji XD, Lacy J, Jacobson KA, Harden TK. Quantitation of the P2Y1 receptor with a high affinity radiolabeled antagonist. Mol. Pharmacol. 2002;62:1249–1257. doi: 10.1124/mol.62.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ohlmann P, de Castro S, Gachet C, Jacobson KA, Harden TK. Pharmacol. Res. Quantification of recombinant and platelet P2Y1 receptors utilizing a [125I]-labeled high affinity antagonist 2-iodo-N6-methyl-(N)-methanocarba-2′-deoxyadenosine-3′,5′-bisphosphate ([125I]MRS2500) in press, doi:10.1016/j.phrs.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- (28).Cazenave JP, Ohlmann P, Cassel D, Eckly A, Hechler B, Gachet C. Preparation of washed platelet suspensions from human and rodent blood. Methods Mol. Biol. 2004;272:13–28. doi: 10.1385/1-59259-782-3:013. [DOI] [PubMed] [Google Scholar]
- (29).Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1207. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yoshimura Y, Moon HR, Choi Y, Marquez VE. Enantioselective synthesis of bicyclo[3.1.0]hexane carbocyclic nucleosides via a lipase-catalyzed asymmetric acetylation. Characterization of an unusual acetal byproduct. J. Org. Chem. 2002;67:5938–5945. doi: 10.1021/jo020249u. [DOI] [PubMed] [Google Scholar]
- (31).Chincshilla R, Nájera C. The Sonogashira reaction: A booming methodology in synthetic organic chemistry. Chem. Rev. 2007;107:874–922. doi: 10.1021/cr050992x. [DOI] [PubMed] [Google Scholar]
- (32).Shryock JC, Snowdy S, Baraldi PG, Cacciari B, Spalluto G, Monopoli A, Ongini E, Baker SP, Belardinelli L. A2A-Adenosine receptor reserve for coronary vasodilation. Circulation. 1998;98:711–718. doi: 10.1161/01.cir.98.7.711. [DOI] [PubMed] [Google Scholar]
- (33).Scammells PJ, Baker SP, Belardinelli L, Olsson RA. Substituted 1,3-dipropylxanthines as irreversible antagonists of A1 adenosine receptors. J. Med. Chem. 1994;37:2704–2712. doi: 10.1021/jm00043a010. [DOI] [PubMed] [Google Scholar]
- (34).Bock VD, Hiemstra H, van Maarseveen JH. CuI-Catalyzed alkyneazide click cycloadditions from a mechanistic and synthetic perspective. Eur. J. Org. Chem. 2006:51–68. [Google Scholar]
- (35).Ohno M, Costanzi S, Kim HS, Kempeneers V, Vastmans K, Herdewijn P, Maddileti S, Gao ZG, Harden TK, Jacobson KA. Nucleotide analogues containing 2-oxa-bicyclo[2.2.1]heptane and l-alpha-threofuranosyl ring systems: interactions with P2Y receptors. Bioorg. Med. Chem. 2004;12:5619–5630. doi: 10.1016/j.bmc.2004.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- (38).Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Costanzi S. On the applicability of GPCR homology models to computer-aided drug discovery: a comparison between in silico and crystal structures of the beta2-adrenergic receptor. J. Med. Chem. 2008;51:2907–2914. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Michino M, Abola E, GPCR Dock 2008 participants. Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat. Rev. Drug. Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nwe K, Brechbiel MW. Growing applications of “click chemistry” for bioconjugation in contemporary biomedical research. Cancer Biother. Radiopharm. 2009;24:289–302. doi: 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Bioch. Soc. Transact. 2007;35:61–67. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]
- (43).Kim Y, Hechler B, Klutz A, Gachet C, Jacobson KA. Toward multivalent signaling across G protein-coupled receptors from poly(amidoamine) dendrimers. Bioconjugate Chem. 2008;19:406–411. doi: 10.1021/bc700327u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Klutz AM, Gao ZG, Lloyd J, Shainberg A, Jacobson KA. Enhanced A3 adenosine receptor selectivity of multivalent nucleoside-dendrimer conjugates. J. Nanobiotechnol. 2008;6:12. doi: 10.1186/1477-3155-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Das A, Zhou Y, Ivanov AA, Carter RL, Harden TK, Jacobson KA. Enhanced potency of nucleotide-dendrimer conjugates as agonists of the P2Y14 receptor: Multivalent effect in G protein-coupled receptor recognition. Bioconjugate Chem. 2009;20:1650–1659. doi: 10.1021/bc900206g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kim Y, Hechler B, Gao ZG, Gachet C, Jacobson KA. PEGylated dendritic unimolecular micelles as versatile carriers for ligands of G protein-coupled receptors. Bioconjugate Chem. 2009;20:1888–1898. doi: 10.1021/bc9001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.