Abstract
Replication protein A (RPA) is a single-stranded DNA-binding complex that is essential for DNA replication, repair and recombination in eukaryotic cells. In addition to this canonical complex, we have recently characterized an alternative Replication Protein A complex (aRPA) that is unique to primates. aRPA is composed of three subunits: RPA1 and RPA3, also present in canonical RPA, and a primate-specific subunit RPA4, homologous to canonical RPA2. aRPA has biochemical properties similar to the canonical RPA complex but does not support DNA replication. We describe studies to identify what properties of aRPA prevent it from functioning in DNA replication. We show aRPA has reduced interaction with DNA polymerase α (pol α) and that aRPA is not able to efficiently stimulate DNA synthesis by pol α on aRPA-coated DNA. Additionally, we show that aRPA is unable to support de novo priming by pol α. Because pol α activity is essential for both initiation and for Okazaki strand synthesis, we conclude that the inability of aRPA to support pol α loading causes aRPA to be defective in DNA replication. We also show that aRPA stimulates synthesis by DNA polymerase α in the presence of PCNA and RFC. This indicates that aRPA can support extension of DNA strands by DNA polymerase α. This finding along with the previous observation that aRPA supports early steps of nucleotide excision repair and recombination, indicate that aRPA can support DNA repair synthesis that requires polymerase δ, PCNA and RFC and support a role for aRPA in DNA repair.
Keywords: DNA replication, DNA repair, DNA polymerase alpha, DNA polymerase delta
Replication Protein A (RPA) is a single-stranded DNA-binding protein that functions in many aspects of DNA metabolism (1–3). Since its discovery as an essential factor for simian virus 40 (SV40) DNA replication, RPA has been shown to play a crucial role in multiple processes of DNA metabolism. In addition to being essential in DNA replication, RPA is required for DNA recombination, DNA repair and the response to DNA damage (1–3). RPA functions by binding and protecting exposed single stranded DNA (ssDNA) and interacting with a multitude of proteins involved in DNA metabolism (1–3).
The canonical RPA is a heterotrimeric protein composed of 70 kDa (RPA1), 32 kDa (RPA2) and 14 kDa (RPA3) subunits that are conserved among eukaryotes (1, 3, 4). In addition to the three subunits that make up the canonical RPA complex, a fourth subunit termed RPA4 has been identified in primates (5, 6). RPA4 is 63% homologous to RPA2 and based on sequence comparison has a similar domain structure to RPA2: an N-terminal putative phosphorylation domain, a central DNA binding domain, DBD G, and a C-terminal winged helix domain (5, 6). It has been shown that RPA4 can replace RPA2 in the trimeric complex creating a complex containing RPA1, RPA3 and RPA4, termed alternative RPA (aRPA) (7). This alternative complex has hydrodynamic properties indistinguishable from the canonical RPA complex but does not support SV40 or chromosomal DNA replication (6, 7). It has been hypothesized that aRPA functions in DNA repair processes. Supporting this hypothesis, in vivo studies have shown that RPA4 localizes to sites of DNA damage when cells are challenged with inhibitors of either topoisomerase I or II (6). In vitro studies have shown that aRPA can support the dual incision/excision steps of nucleotide excision repair and stimulate Rad51 dependent strand invasion during the initial steps of recombination-mediated repair (8).
The role of RPA in DNA replication has been characterized in detail using the SV40 system. SV40 initiation requires the concerted action of four proteins, SV40 large T-antigen (Tag), polymerase α/primase (pol α), topoisomerase I (topo I) and RPA (9–11). Tag assembles at the origin of replication, bi-directionally unwinds the double-stranded DNA and recruits other proteins to establish a replication fork (12). Topo I stimulates pol α by binding to Tag and releases torsional stress induced by unwinding of the parental strands (13, 14). RPA is required to stabilize the emerging ssDNA and along with Tag, recruits pol α(15, 16). Pol α is a heterotetrameric complex of p180, p68, p58 and p48 subunits that synthesizes a short RNA primer on the leading strand and at the beginning of each Okazaki fragment on the lagging strand (15, 17). After about 10-ribonucleotides are incorporated, the complex transitions to DNA synthesis for about 20 deoxynucleotides creating the initial RNA-DNA primers used to start DNA replication and each Okazaki fragment (18). It has been shown that RPA acts as an auxiliary factor for pol α by stimulating synthesis and increasing processivity during initiation of DNA replication (19). During initiation, RPA interacts with pol α to keep the polymerase at the primed site. To switch from initiation to elongation, RFC interacts with RPA disrupting the pol α – RPA interaction and causing the release of pol α (20). RFC then loads PCNA and remains at the primed site by interacting with RPA. DNA polymerase δ (pol δ) can then access the primed site via contact with RPA. pol δ is one of the replicative polymerases in eukaryotes and is the major polymerase used for lagging-strand synthesis (21). Pol δ competes with RFC for RPA, resulting in displacement of RFC from the 3′ terminus, and replacement with pol δ (20). RFC remains at the site by interacting with the PCNA ring. While in SV40 replication, pol δ can support synthesis of both leading and lagging strands (22), it is believed that generally once the elongation complex is established, pol δ extends the primers generated by pol α on the lagging strand while DNA polymerase ε continuously synthesizes DNA on the leading strand (21, 23, 24).
The current model suggests multiple roles for RPA in DNA replication. These include binding to exposed ssDNA being created by the helicase, helping recruit polymerase α/primase, and coordinating the polymerase switch from polymerase α to polymerase δ/polymerase ε. Throughout the course of replication, RPA serves as a common interaction partner for many proteins and through a protein-mediated hand-off mechanism coordinates the ordered assembly of the proteins (3). We have previously shown that aRPA does not support SV40 DNA replication at the initiation and elongation steps. However, it is not known what activity prevents aRPA from functioning in DNA replication. The present study examines the role of aRPA during the initiation and elongation reactions of DNA replication using purified recombinant proteins. In particular, we wished to understand how aRPA affects the activities of pol α and pol δ. We also show that unlike RPA, aRPA has altered interactions with pol α and does not support efficient loading of or priming by pol α. The pattern of DNA synthesis by pol α in the presence of aRPA also suggests that aRPA cannot stabilize pol α on the DNA. In contrast, we find that aRPA does support pol δ synthesis in the presence of PCNA and RFC. These findings suggest that the defect of aRPA in replication is in promoting efficient priming by pol α but can function in processive DNA synthesis by pol δ.
Experimental Procedures
Plasmids
pGBM-RFC1, pET-RFC4/2 and pCDFK-RFC5/3 were generous gifts from Dr. Yuji Masuda (25). pET-hPold1 and pCOLA-hPold234 were generous gifts from Dr. Yoshihiro Matsumoto (26). p11d-tRPA and p11d-aRPA were described previously (7). pT7-hPCNA was a generous gift from Dr. Bruce Stillman, Cold Spring Harbor Laboratory. This plasmid was used as a template for in vitro site directed mutagenesis to insert an N-terminal 6x Histidine tag. The primers used to generate pT7-His-hPCNA were 5′-CCGTTTACTTTAAGAAGGAGATATACATATGCATCACCATCATCACCACGGATCCGCTATGTTCGAGGCGCGCCTGGTCCAGGGCTCC-3′ and 5′-GGAGCCCTGGACCAGGCGCGCCTCGAACATAGCGGATCCGTGGTGATGATGGTGATGCATATGTATATCTCCTTCTTAAAGTAAACG-3′. The mutations were confirmed by DNA sequencing.
Protein Purification
Recombinant RPA and aRPA were expressed in BL21(DE3) cells and purified as previously described (7, 27, 28). Recombinant human pol α was expressed and purified as described previously (29). Recombinant human pol δ was expressed in BL21(DE3) cells harboring pRARE2 (Novagen) co-transformed with pET-hPold1 and pCOLA-hPold234 for 18h at 16°C after induction with 0.2 mM isopropyl β-D-1-thiogalactopyranoside and purified as described previously (25). Purity of pol δ (~85%) was confirmed by SDS-PAGE visualized with Coomassie Blue staining. Recombinant human RFC was expressed in BL21(DE3) cells harboring pRARE2 co-transformed with pGBM-RFC1, pET-RFC4/2 and pCDFK-RFC5/3 for 18h at 16°C after induction with 0.2 mM isopropyl β-D-1-thiogalactopyranoside and purified as described previously except the size exclusion column was omitted (25). Purity of RFC (~85%) was confirmed by SDS-PAGE visualized with Coomassie Blue staining. Recombinant human PCNA was expressed in BL21(DE3) cells transformed with pT7-His-hPCNA for 4 hr at 37°C. The cells were harvested and lysed similar to RPA and aRPA. The supernatant loaded onto a Ni-NTA Agarose (Qiagen) column equilibrated with Buffer J (30 mM HEPES (pH 7.8), 0.25% (w/v) myo-inositol, 1 mM tris(2-carboxyethyl)phosphine and 0.02% Tween-20 (v/v) supplemented with 20 mM imidazole. Following a 3 column volume wash, PCNA was eluted with 5 column volume linear gradient from 20–250 mM imidazole. The peak fractions were pooled and dialyzed for 16hr against Buffer J supplemented with 150 mM KCl to remove the imidazole. Purity of PCNA (>95%) was confirmed by SDS-PAGE visualized with Coomassie Blue staining.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA was used to examine interactions between purified proteins as described previously (28). Briefly, wells in microtiter plates were coated with 1 μg of RPA or aRPA for interactions with PCNA and pol α and 1 μg of RFC and pol δ in 50 μL of water and incubated for 1 hour. Plates were washed with phosphate buffered saline with 0.2% Tween 20 and blocked with 5% milk in phosphate buffered saline. The indicated amount of pol α, PCNA, RPA, aRPA or BSA was added to each well, incubated for 1 hour, and washed. Primary antibodies in phosphate buffered saline with 5% milk for pol α (1:100 of SJK237), RPA/aRPA (1:300 of 719A) and PCNA (1:50 of anti-human PCNA antibody was the generous gift from Dr. Thomas Kelly) were added to the plates, incubated for 30 min, and washed. Goat-α-mouse IgG-HRP (1:1000) was added and incubated for 30 min. Plates were developed using 200 μL of 0.8 mg/mL o-phenylenediamine in 0.005 M phosphate citrate buffer with 0.03% sodium perborate. OD450 was quantified after 10–60 min using a microtiter plate reader. Background was determined by using BSA as the secondary protein and all data shown have these values subtracted. In all assays, the background values were similar and close to zero.
pol α Extension Assay
pol α activity was assayed with a singly primed d24:d66-mer oligodeoxynucleotide (d24: 5′-CTCGGACAATTTGGTGTGCTAGGT-3′; d66: 5′-AGGATGTATGTCTAGTAGGTACATAACTATTCAGTAGTATAGACCTAGCACACCAAATTGTCCGAG-3′) as a template. The d24:d66-mer was prepared by labeling the 5′-end of the d24-mer primer with [γ-32P]ATP and T4 polynucleotide kinase (NEB) according to the manufacturer’s protocol. The d66-mer template oligonucleotide was then mixed with the complementary labeled d24-mer oligonucleotide in a 1:1 molar ratio in 20 mM Tris-HCl (pH 8.0) containing 20 mM KCl and 1 mM EDTA, heated for 5 min at 90 °C, and then incubated for 2 h at 65 °C and slowly cooled to room temperature. A final volume of 15 μL contained 50 mM Tris-HCl (pH 7.6), 0.25 mg/mL BSA, 1 mM dithiothreitol, 6 mM Mg Cl2, 20 nM (3′-OH ends) of the 5′ 32P-labeled d24:d66-mer DNA template, 10 μM dNTPs, 1 nM pol α, 50 nM RPA or aRPA as indicated. Reactions were assembled on ice and initiated by the addition of dNTPs and incubated for indicated time at 37°C. When order of addition was varied, reactions were pre-incubated at 37°C for 10 minutes and then initiated by the addition of indicated proteins and dNTPs. Reactions were quenched by the addition of formamide loading buffer (80% deionized formamide, 10 mM EDTA (pH 8.0), 1 mg/mL xylene cyanol, 1 mg/mL bromophenol blue), heated at 95 °C for 5 min and products were separated in a 15% polyacrylamide sequencing gel containing 8 M urea. Products were visualized with a FLA-7000 phosphorimager (Fujifilm Global) and quantified using Multi Gauge software (Fujifilm Global).
Primer RNA-DNA Synthesis Assay
These assays were carried out as described by Khopde et al. (14). Briefly, reactions (40 μL) contained 400 ng CsCl purified pSKori, 245 nM T antigen (monomer), 7 nM pol α, and either 200 nM of RPA or aRPA with or without 55 nM topo I in replication buffer (30 mM HEPES-KOH (pH 8.0), 7 mM MgCl2, 40 mM creatine phosphate, 25 μg/mL creatine phosphokinase, 0.5 mM dithiothreitol, 50 μg/mL BSA, 4 mM ATP, 0.2 mM each CTP, GTP and UTP, 10U RNasin (Sigma)). After 1 h at 37°C, newly synthesized RNA-DNA primers were pulse labeled for 1 min with 10 μCi [α-32P]dCTP, in the presence of 100 μM dATP, dTTP, and dGTP. Purified DNA was incubated with 15 μL of 95% formamide, 10 mM EDTA, 0.01% bromophenol blue and 0.01% xylene cyanol at 90°C for 3 min and subjected to electrophoresis on 12% polyacrylamide-urea sequencing gels. Products were visualized by exposure to phosphorimager screens.
pol δ Extension on Singly Primed ssM13mp18 Assay
Pol δ activity was assayed on singly primed single-stranded M13mp18. The standard reaction (10 μL) contained 20 mM HEPES-NaOH (pH 7.5), 0.2 mg/mL BSA, 1 mM dithiothreitol, 1 mM ATP, 1 mM EDTA, 50 fmol (364 pmol for nucleotides) of singly primed ssM13mp18 (5′ 32P-labeled 36-mer primer, CAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGG is complementary to 6330-6295 nt), 555 nM RPA or aRPA, 50 nM PCNA, 50 nM RFC and 20 nM pol δ. Reactions were initiated by the addition of dNTP’s and MgCl2 to a final concentration of 150 μM and 10 mM, respectively. After incubation at 37°C for indicated time, the reactions were quenched by the addition of formamide loading buffer (80% deionized formamide, 10 mM EDTA (pH 8.0), 1 mg/mL xylene cyanol, 1 mg/mL bromophenol blue), heated at 95 °C for 5 min and products were separated in a 15% polyacrylamide sequencing gel containing 8 M urea. Products were visualized with a FLA-7000 phosphorimager (Fujifilm Global) and quantified using Multi Gauge software (Fujifilm Global).
Results
Effect of aRPA on pol α
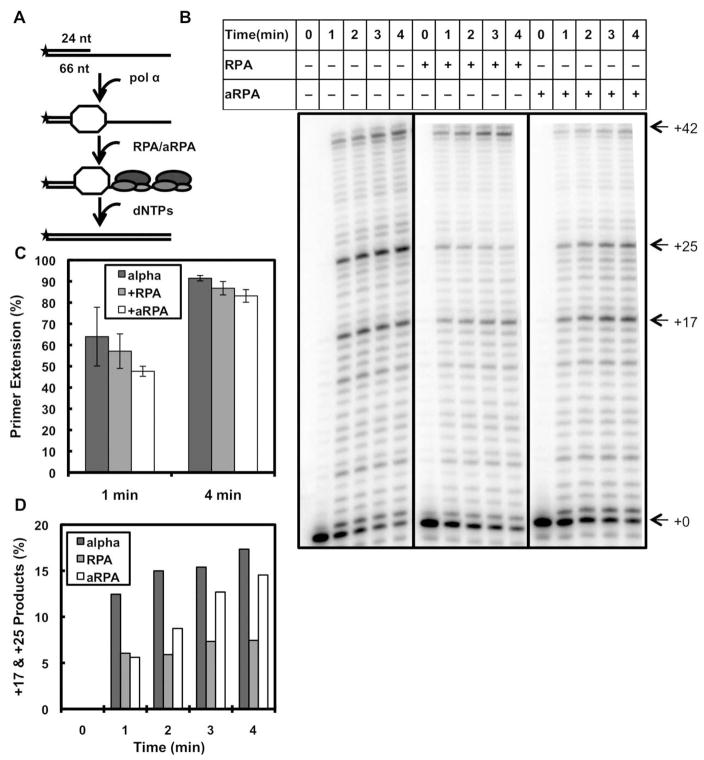
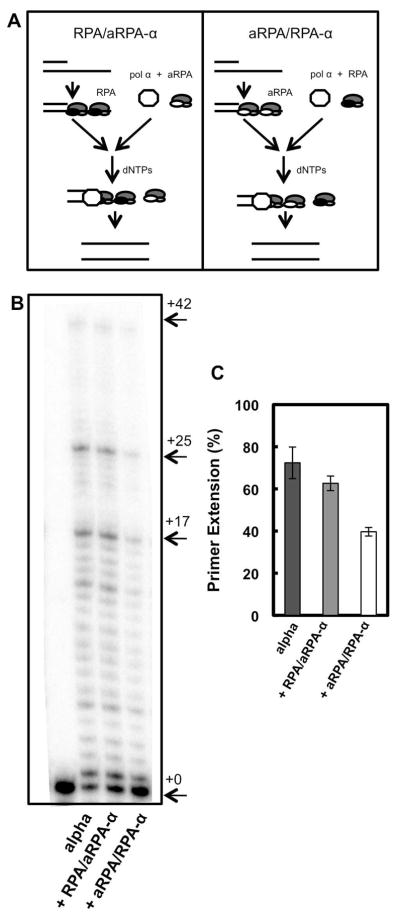
Initiation of SV40 DNA replication requires the concerted action of SV40 large T-antigen (Tag), topoisomerase I, pol α and RPA, which form an ‘initiation complex’ (10). Previously we have shown that aRPA interacts with Tag at levels similar to RPA (7). This suggests that the inability of aRPA to support DNA replication is a result of aRPA affecting one of the other proteins in initiation or at the replication fork. We initially examined the effect of aRPA on the function of pol α using a 66-mer oligonucleotide primed with a 24-mer oligonucleotide. Pol α and either RPA or aRPA were incubated with the primed template in the presence of deoxynucleotides (dNTPs) and products of the reaction were separated on a 15% denaturing polyacrylamide gel, which allowed for separation of single nucleotide incorporation events creating a laddering of products from +1 nt to +42 nt. By allowing some components to pre-bind to the DNA, we were able to examine the effect of aRPA on the polymerization of a pre-bound pol α (Figure 1A) or on the loading and subsequent polymerization of pol α (Figure 2A). When pol α was allowed to bind to the template before the addition of dNTPs, it efficiently synthesized DNA with full-length product being observed in less than one minute (Figure 1B, lanes 1–5). Intermediate length products, notably two major pause sites at +17 and +25 were observed. The major pause sites are both two purines in a row (AA and GG, respectively) and the degree of pausing is consistent with the low processivity of pol α (19, 30). Similar experiments were carried out in which pol α was allowed to bind to the primer-template junction and then either RPA or aRPA was added with the initiating dNTPs. The amount of RPA or aRPA used was enough to saturate the ssDNA region of the substrate with two RPA molecules bound per DNA substrate. In these reactions, addition of either RPA or aRPA resulted in levels of synthesis similar to that observed with pol α alone (Figure 1B–C). Addition of RPA caused a decrease in the accumulation of products at the two major pause sites by an average of 56% over the time course (Figure 1D). In contrast, there was only a slight change in the level of pausing (16%) when aRPA was added. Together these finding suggest that aRPA does not affect the polymerization of pol α that is associated with the primer-template junction.
Figure 1.
The affect of RPA and aRPA on pol α when polymerase is pre-mixed with the DNA substrate. (A) Schematic illustrating experimental setup and order of addition of proteins. Asterisk indicates the location of the 32-P label. (B) DNA pol α extension assays where pol α (1 nM) has been pre-incubated with the DNA substrate (20 nM 3′-OH ends). Following pre-incubation, either RPA (50 nM) or aRPA (50 nM) was added and the reaction initiated by the addition of dNTP’s (10 μM). Reaction products were separated by electrophoresis on a denaturing polyacrylamide sequencing gel and visualized on a Fuji FLA-7000 phosphorimager. (C) The results from three independent experiments were quantified and are presented. For each experiment the amount of DNA in each band was determined. Percent primer extension was calculated by determining the ratio of all extended products (+1 – +42 nt) to total DNA (all products plus unextended primer (+0)). Error bars indicate standard deviation. (D) The amount of products at +17 and +25 nt in the gel shown in 1(B) were quantified by dividing the sum of the amount of DNA at +17 nt and +25 nt by total DNA.
Figure 2.
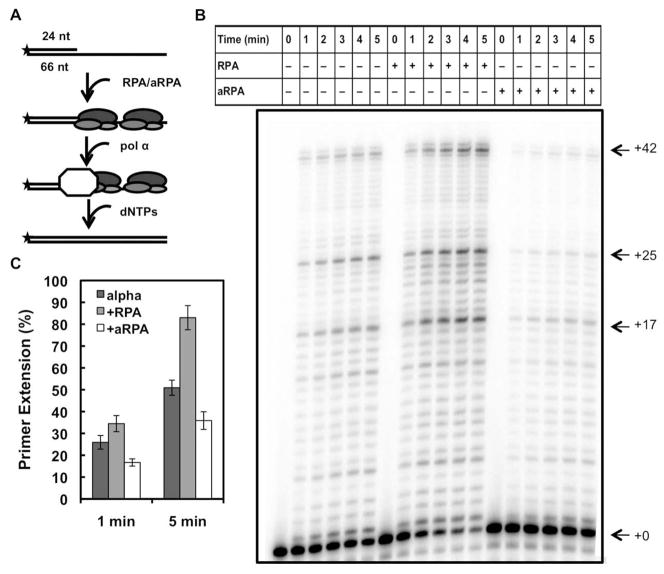
The effect on pol α when either RPA or aRPA are pre-bound to the DNA substrate. (A) Schematic illustrating experimental setup and order of addition of proteins. Asterisk indicates the location of the 32-P label. (B) DNA pol α extension assays where either RPA (50 nM) or aRPA (50 nM) was pre-incubated with the DNA substrate (20 nM 3′-OH ends). Following pre-incubation, pol α (1 nM) was added and the reaction initiated by the addition of dNTP’s (10 μM). Reaction products were separated and visualized as described in Figure 1. (C) The results from three independent experiments were quantified and are presented. Percent primer extension was determined as described in Figure 1. Error bars indicate standard deviation.
We next examined the ability of aRPA to facilitate the loading of pol α. This was done using the template described above, but the order of addition was changed: RPA or aRPA were allowed to pre-bind to exposed ssDNA on the template strand and reactions were initiated by the addition of pol α (Figure 2A). Total synthesis with pol α was reduced under these conditions (Figure 2B). This is consistent with association of the polymerase being the rate-limiting step with this type of template. When RPA is pre-bound, total DNA synthesis by pol α is similar to conditions in which pol α is pre-bound to the template (compare Figure 1B with Figure 2B). This suggests RPA promotes loading of pol α on primer template junctions. In contrast, when aRPA was pre-bound to the ssDNA of the template, there was a 64% decrease in total DNA synthesis (compared to pre-binding RPA and a decrease of 42% compared to pol α alone; Figure 2C). This suggests, that unlike RPA, aRPA does not support efficient loading of pol α and actually inhibits its association with the primer-template junction.
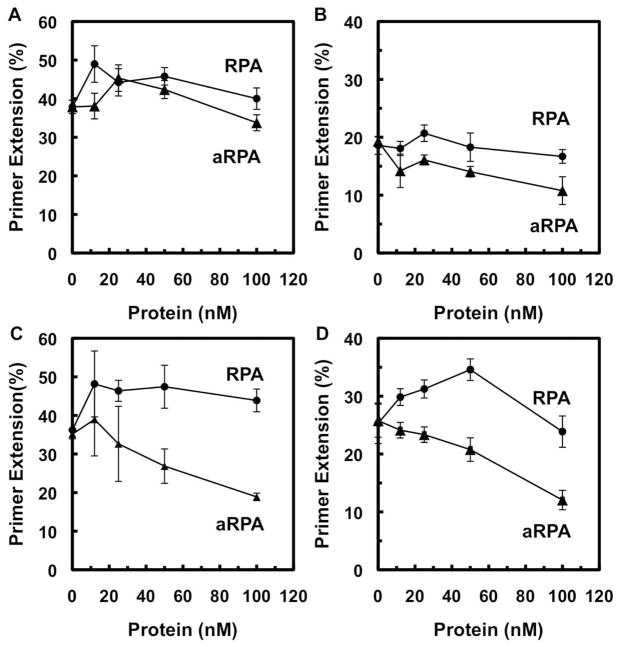
We next examined the concentration dependence of RPA and aRPA on pol α synthesis. The concentrations of RPA and aRPA were varied from 0 to 100 nM with a fixed amount of DNA substrate (20 nM). RPA and aRPA had minimal effect on pol α when the polymerase was allowed to pre-bind the DNA substrate (Figure 3A and 3B). When either RPA or aRPA was allowed to pre-bind the DNA substrate, there was minimal change in DNA synthesis at low concentrations but DNA synthesis quickly decreased when aRPA concentrations went above 30 nM (Figure 3C). However, DNA synthesis with RPA increased up to 30 nM then remained constant for multiple incorporation events (Figure 3C). We also examined incorporation of the first nucleotide to determine whether the form of RPA affected the initial polymerization reaction. Single nucleotide incorporation was examined by carrying out the reactions in the presence of only the next nucleotide in the sequence (dCTP). RPA had a minimal effect up to 50 nM but inhibited synthesis at higher concentrations (Figure 3D). In contrast, aRPA decreased the amount of single nucleotide incorporation at all concentrations examined (Figure 3B). The results suggest that over this concentration range, aRPA is inhibitory while RPA had minimal effect on synthesis by pol α.
Figure 3.
Titration of RPA and aRPA in pol α extension assay. (A) (B) Pol α was pre-incubated with the DNA substrate (20 nM 3′-OH ends) as described in Figure 1. Following pre-incubation, RPA (closed circles) or aRPA (closed triangles) was added and the reaction initiated by the addition of (A) dNTP’s (10 μM) or (B) only dCTP (10 μM). (C) (D) RPA (closed circles) or aRPA (closed triangles) was pre-incubated with the DNA substrate (20 nM 3′-OH ends) as described in Figure 2. Following pre-incubation, pol α (1 nM) was added and the reaction initiated by the addition of (C) dNTP’s (10 μM) or (D) dCTP (10 μM). Primer extension was quantified as described in Figure 1. Average of two independent experiments are shown with error bars to indicate the range of the data.
Mechanism of pol α Inhibition by aRPA
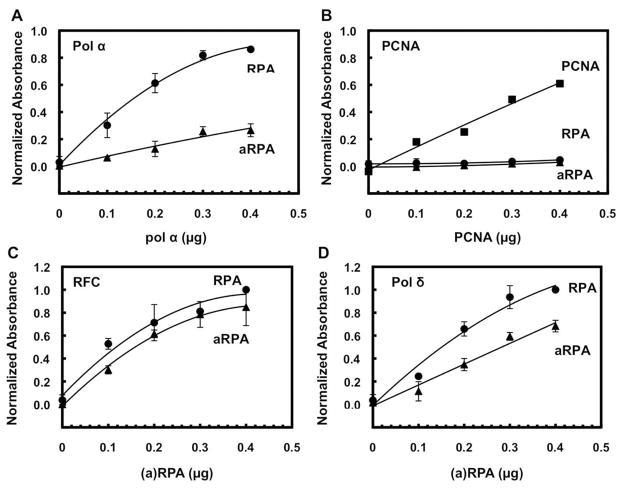
We next determined whether there were altered interactions between aRPA and pol α. Enzyme linked immunosorbant assays were carried out with purified proteins. Compared to RPA the interaction between aRPA and pol α was decreased by ~75% (Figure 4A). Both RPA1 and RPA2 interact with the pol α complex so either the pol α interaction with RPA2 is most important for the interactions monitored in these assays or the presence of RPA4 in the aRPA complex causes altered interactions of pol α with RPA1. These findings suggest that aRPA has reduced interactions with pol α and that this prevents efficient loading when aRPA is bound to the DNA. However, they do not rule out aRPA allosterically modulating the activity of pol α. To test this possibility, a series of mixing experiments were done. Either RPA or aRPA was pre-bound to the DNA substrate while at the same time DNA pol α was pre-incubated with the other form of RPA. DNA synthesis was then initiated by combining the two mixtures and primer extension monitored (Figure 5A). When RPA was pre-bound and the reaction initiated by the addition of aRPA-pol α, there was a slight decrease in DNA synthesis compared to pol α alone (Figure 5B). In contrast, when aRPA was pre-bound to ssDNA and the reaction initiated by the addition of RPA-pol α, there was a larger decrease in DNA synthesis (Figure 5B). Together these findings suggest that the majority of the inhibition of pol α by aRPA is a result of reduced protein interactions between aRPA and pol α preventing either pol α association on aRPA coated primer-template junctions or pol α from displacing aRPA from the DNA template.
Figure 4.
Enzyme linked immunosorbant assay in which interactions were measured between different forms of RPA and either (A) pol α, (B) PCNA, (C) RFC or (D) pol δ. Forms of RPA used: RPA (closed circles) and aRPA (closed triangles). The data from each experiment was normalized to the highest absorbance in each experiment, averaged and plotted. Error bars indicate the average of two or more independent replicates. BSA was used to determine nonspecific background (< 0.1) in each assay and subtracted. (B) PCNA (closed squares) was also placed directly on the place as a positive control.
Figure 5.
Mechanism of pol α inhibition. (A) Schematic illustrating experimental setup, order of addition of proteins and pre-incubation of proteins and DNA substrate. (B) DNA pol α extension assays where either RPA (50 nM) or aRPA (50 nM) was pre-incubated with the DNA substrate (20 nM 3′-OH ends) and the other form of RPA was pre-incubated with pol α (1 nM). The pre-incubated samples were mixed and the reaction initiated by the addition of dNTP’s (10 μM). Reaction products were separated by electrophoresis on a denaturing polyacrylamide sequencing gel and visualized by phosphorimaging. (C) The results from two independent experiments were quantified and are presented. Percent primer extension was determined as described in Figure 1. Error bars indicate range of data.
Effect of aRPA on the Synthesis on RNA-DNA Primers
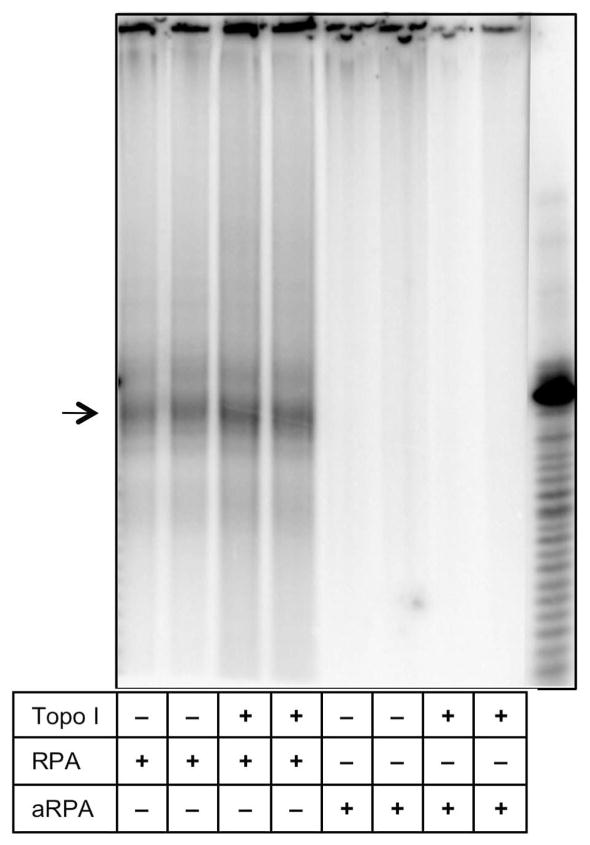
To examine the effect these altered interactions have on initiation, we examined the ability of aRPA to support pol α dependent priming using a SV40 based monopolymerase assay. Newly synthesized RNA-DNA primers were identified by labeling them in the presence of 32P-dCTP for one minute following a 1 hour incubation in the presence of rNTPs. RNA-DNA primers of approximately 36 nucleotides were synthesized and readily detected in the presence of RPA (Figure 6, lanes 1–2) or RPA and topoisomerase I (Figure 6, lanes 3–4). However, no synthesis was detected in the presence of aRPA (Figure 6, lanes 5–8). This demonstrates that aRPA is unable to support efficient initiation of DNA replication from the SV40 origin by preventing priming by pol α.
Figure 6.
Primer RNA-DNA synthesis in the presence of RPA or aRPA. Primer RNA-DNA synthesis was measured by incorporation of 32P-dCTP in a 1 min labeling reaction that contained pSKori DNA (400 ng), Tag (800 ng), pol α (100 ng) and Topo I (200 ng), RPA (900 ng) and aRPA (900 ng) as indicated. After deproteination, the DNA was denatured with formamide and subjected to electrophoresis on a 12% acrylamide sequencing gel followed by detection of the labeled DNA with a phosphorimager. Major band in lane on right represents a 42 nucleotide single stranded DNA marker. Arrow indicates predominant length primers.
Effect of aRPA on pol δ DNA synthesis
Thus far, we have shown that aRPA does not support the efficient loading of pol α onto the primer-template junction. This agrees with our earlier findings that aRPA does not support the initiation steps of SV40 DNA replication. However, we have shown that aRPA also does not support the elongation phase of SV40 DNA replication (7). One possibility is that like pol α inhibition, aRPA could also inhibit DNA synthesis by pol δ. Another possibility is that the inhibition of pol α, which is required for Okazaki fragment synthesis, is enough to uncouple leading and lagging strand synthesis thus halting DNA synthesis.
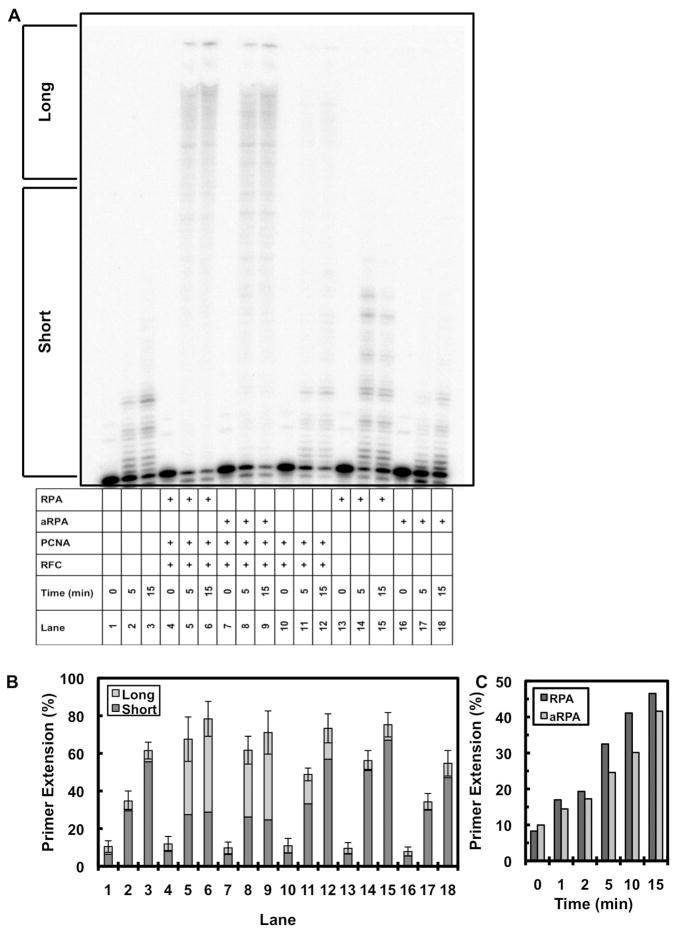
To discriminate between these two possibilities, pol δ, RFC and PCNA were purified and the effect of aRPA on pol δ DNA synthesis was examined on a singly-primed ssM13mp18 plasmid. As shown in Figure 7A lanes 2 and 3, DNA synthesis by pol δ by itself is limited to a small number of nucleotides incorporated, which is consistent with its low processivity in the absence of the accessory factors PCNA and RFC (20). When RFC and PCNA were added to the reaction (Figure 7A, lanes 11 and 12), there was an increase in the length of products formed indicating that RFC actively loaded PCNA onto the single-strand plasmid and PCNA formed a complex with pol δ. The addition of RPA to the RFC, PCNA and pol δ reaction showed a dramatic increase in the length of products formed (Figure 7A, lanes 5 and 6). Interestingly, addition of aRPA to RFC, PCNA and pol δ resulted in products that are identical to those synthesized in the presence of RPA (Figure 7A, compare lanes 8 and 9 to 5 and 6 and quantitation of the products in Figure 7B). A time course of these reactions indicated that while there was a slight lag with aRPA, overall the rate of synthesis is similar with either RPA or aRPA (Figure 7C). We conclude that aRPA does support processive DNA synthesis by pol δ in the presence of RFC and PCNA.
Figure 7.
pol δ synthesis on singly-primed single-stranded M13mp18 template. (A) pol δ activity was assayed on singly-primed single-stranded M13mp18 (50 fmol) in reaction initiated by the addition of dNTPs (150 μM). Addition of individual components is indicated by a plus sign: pol δ (20 nM), RPA (555 nM), aRPA (555 nM), PCNA (50 nM) and RFC (50 nM). The time of the reaction is indicated (0, 5, 15 min). Reaction products were separated by electrophoresis on a denaturing polyacrylamide sequencing gel and visualized by phosphorimaging. (B) The results from three independent experiments were quantified and are presented. Primer extension was quantified by dividing either short products (≤ 40 nt, dark gray) or long products (≥ 41 nt, light gray) by total DNA. Error bars indicate the standard deviation of the data for the long products. (The standard deviation for the short products is shown in supplemental figure 1.) (C) Expanded time course of complete reactions (pol δ (20 nM), PCNA (50 nM) and RFC (50 nM)) with the addition of either RPA (555 nM, dark gray) or aRPA (555 nM, light gray)
We also examined whether there were altered interactions between aRPA and RFC, PCNA and pol δ. Both RFC and pol δ interact with RPA while PCNA does not directly interact with RPA (3, 20). As shown in Figure 4C, the interaction between aRPA-RFC was the same as RPA-RFC. RPA and aRPA do not directly interact with PCNA (Figure 4B). By comparing the RPA-pol δ and the aRPA-pol δ interaction, aRPA also interacted strongly with pol δ but at a slightly reduced level (60%, Figure 4D). In the absence of the accessory proteins RFC and PCNA, RPA but not aRPA caused a modest stimulation of pol δ DNA synthesis (Figure 7A, compare lane 3 to lanes 15 and 18). This suggests that the altered interactions between aRPA and pol δ might reduce the direct stimulation of pol δ by aRPA. However, even if there is a miscommunication between aRPA and pol δ, it is readily overcome by the accessory proteins (Figure 7A).
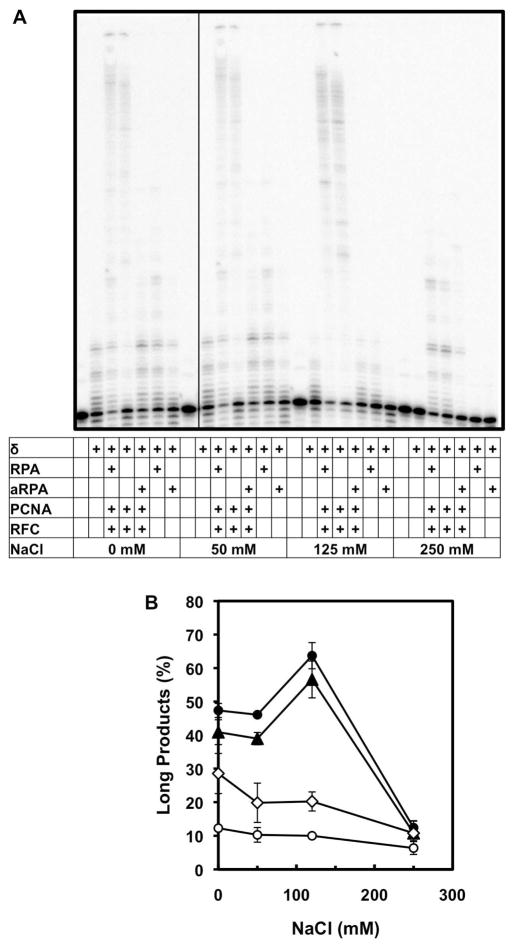
While optimizing pol δ DNA synthesis on the ssM13mp18 plasmid, we examined the salt dependence of the reaction. Pol δ is sensitive to ionic strength and most previous analyses with RFC and PCNA have been carried out under low ionic strength conditions (25, 31, 32). We find that the addition of RPA or aRPA can overcome the salt inhibition of pol δ DNA synthesis (Figure 8). Synthesis was monitored and both short (≤ 40 nt) and long (≥ 41 nt) products were quantified. At all salt conditions examined, minimal long products were observed with pol δ or with pol δ, PCNA and RFC in the absence of either form of RPA (Figure 8B-open circles and diamonds). At low salt concentrations short products were observed but the amount of synthesis decreased as salt concentration increased (Figure 8A). In contrast, when RPA or aRPA was added to RFC, PCNA and pol δ, high levels of synthesis and full-length (long) products were observed from low to near physiological ionic strength (0–125 mM NaCl; Figure 8B-closed symbols). There is inhibition of synthesis at 250 mM NaCl in the presence of RPA or aRPA; however, even at this high ionic strength, synthesis of short products was observed while there was virtually complete inhibition of pol δ in the absence of RPA. These data clearly show that aRPA can stimulate pol δ under a variety of conditions and that both RPA and aRPA stimulate pol δ activity under physiological ionic strength.
Figure 8.
Salt dependence of pol δ activity. pol δ activity was assayed on singly-primed single-stranded M13mp18 (50 fmol) in varying concentrations of NaCl initiated by the addition of dNTPs (150 μM). Reactions components: pol δ (20 nM)-open circles; pol δ (20 nM), RPA (555 nM), PCNA (50 nM) and RFC (50 nM)-closed circles; pol δ (20 nM), aRPA (555 nM), PCNA (50 nM) and RFC (50 nM)-closed triangles and pol δ (20 nM), PCNA (50 nM) and RFC (50 nM)-open diamonds. (A) Reaction products were separated by electrophoresis on a denaturing polyacrylamide sequencing gel and visualized by phosphorimaging. (B) The results from two independent experiments were quantified and are presented. Percent long products (≥ 41 nt) were calculated as described in Figure 7. Error bars indicate the range of the data.
Discussion
RPA has a central role in DNA replication, playing an essential function in both initiation and elongation (33, 34). We have previously shown that aRPA does not support DNA synthesis during the initiation and elongation phases of SV40 DNA replication or S-phase progression in human cells (6, 7). The studies presented here provide a molecular explanation for this difference in activity. We have shown that aRPA bound to ssDNA prevents the synthesis of RNA-DNA primers by pol α by preventing efficient loading of the polymerase onto the ssDNA. This effect is probably caused by altered interactions between pol α and aRPA. However, aRPA has minimal effects on the polymerization reaction by pol α once it has started synthesizing DNA. These findings indicate that aRPA is unlikely to support the association of pol α on ssDNA leading to priming and the initiation of DNA replication. This mechanism is also supported by the finding that aRPA is unable to support primer synthesis in an SV40 origin-dependent initiation reaction. This defect would also be expected to prevent priming of Okazaki fragments needed for lagging strand synthesis, which would presumably stall replication forks during elongation synthesis. Walther et al. showed that inhibition of pol α during elongation phase of SV40 replication causes a complete halt to DNA synthesis consistent with coupled synthesis of leading and lagging strands (34). This indicates that the loss of pol α loading on the lagging strand in the presence of aRPA would be expected to cause a defect in elongation. We also show that aRPA supports pol δ DNA synthesis in the presence of RFC and PCNA to the same extent as canonical RPA and that both aRPA and RPA increase the processivity of the polymerase more than PCNA and RFC alone. This suggests that aRPA can support processive DNA synthesis on primed DNA templates. This activity would have little consequence during DNA replication in the absence of priming by pol α but would further support a role for aRPA in genome maintenance. It been shown that aRPA can function in multiple aspects of DNA repair from localization to sites of damage to supporting the dual incision/excision steps of nucleotide excision repair to supporting Rad51 dependent strand invasion (8). Our findings here suggest that aRPA can complete the nucleotide excision repair process by filling the gap left when the damaged DNA is removed. This gap filling reaction is carried out by PCNA, RFC and either pol δ or pol ε, in a continuous manner using the free 3′-OH left by the removal of the damage DNA (35). Similar gap filling reactions by a high fidelity polymerase, such as pol δ and DNA polymerase ε, are common to most other forms of DNA repair (36). We suggest that aRPA, like canonical RPA, is capable of supporting gap synthesis in repair and thus help the cell maintain genome stability. Of the 14 identified human polymerases, RPA has been shown to interact with at least pol α, δ, ε, λ3 and κ (32, 37–39). Interestingly, only pol α is able to initiate strand synthesis in DNA replication. All other human DNA polymerases extend previously initiated DNA strands.
The number and role of RPA-like complexes in different processes in eukaryotic cells is diverse. Up until the last decade, it was thought that eukaryotic cells had primarily one nuclear single-stranded DNA-binding protein involved in DNA metabolism, canonical RPA. However, it is now clear that there are a number of RPA-like proteins in cells. Several domains of the tumor suppressor, BRCA2 have structural and functional similarity to the DNA-binding domains of RPA (40, 41). Other examples include the recently identified pol α accessory proteins that have homology to RPA and function in DNA metabolism (42) and the RPA-related complex, consisting of Cdc13, Stn1 and Ten1, that is involved in telomere maintenance (43–45). Mammals also have non-RPA related single-stranded DNA binding proteins that function in DNA repair (46, 47). Furthermore, a number of eukaryotes have multiple RPA complexes. Cryptosporidium parvum has two forms of RPA1 (48). Plants such as Oryza sativa and Arabidopsis thaliana have multiple copies of RPA genes that form multiple different heterotrimeric RPA complexes (49). These plant RPA complexes have non-redundant functions with respect to each other. For example, in rice the B type RPA plays a role in DNA damage repair while the C type RPA is required for DNA replication (50, 51). RPA4 and RPA2 could be functioning similarly in human cells with aRPA and canonical RPA in humans are playing the same roles as B type and C type RPA in rice, respectively.
RPA4 appears to have emerged relatively recently in evolution. RPA4-like sequences have only been identified in mammals and intact RPA4 genes have been maintained only in horse and primates (6). It has been estimated that approximately 3% of human genes are restricted to primates and these genes are termed orphan genes (52). However, very few orphan genes have been well characterized experimentally. One characterized gene, dermcidin, encodes a peptide with antimicrobial activity that is secreted in sweat glands and has been reported to be involved in neural survival and cancer (53). Another example of a primate-specific orphan gene with described function is the SPHAR, which is involved in the regulation of DNA synthesis (54). Two more genes that function in DNA metabolism pathways are FAM9B and FAM9C, expressed solely in the testis, have been suggested to play a role in mediating recombination during meiosis (55). It has also been speculated that primate specific genes are preferentially expressed in the reproductive system (56). RPA4 is expressed in reproductive tissues (placenta, ovary, prostate, testis and oocytes) but is also expressed in non-reproductive tissues (e.g. lung, esophagus, bladder) (8, 57). So while the cellular function of aRPA is not understood, it appears that the role(s) of RPA4 in the cell are not limited to reproduction.
The findings presented here, reveal the mechanism that prevents aRPA from functioning in DNA replication. They also show that aRPA can support DNA repair synthesis that depends on pol δ with its accessory proteins, RFC and PCNA. These and other recent findings on aRPA suggest that it functions in repair processes to maintain the genomic stability in non-dividing cells.
Supplementary Material
Acknowledgments
We would like to thank Cathy Hass and Troy Humpreys for useful discussion, and critical reading of this manuscript. We would also like to thank Dr. Bruce Stillman, Dr. Yuji Masuda and Dr. Yoshihiro Matsumoto for kindly providing expression plasmids for replication proteins. We thank Dr. Thomas Kelly for the antiserum to human PCNA.
Abbreviations
- RPA
human replication protein A
- aRPA
alternative RPA
- RPA1
70-kDa subunit of RPA
- RPA2
32-kDa subunit of RPA
- RPA3
14-kDa subunit of RPA
- RPA4
product of the RPA4 gene
- pol α
human DNA polymerase α/primase complex
- pol α
human DNA polymerase α
- PCNA
human proliferating cell nuclear antigen
- RFC
human replication factor C
- topo I
topoisomerase I
- SV40
simian virus 40
- Tag
SV40 large T-antigen
- nt
nucleotide
- ssDNA
single-stranded DNA
- DBD
DNA binding domain
- ssM13mp18
single-stranded M13mp18
- BSA
bovine serum albumin
- dNTPs
deoxynucleotides
Footnotes
Supporting Information Available
Supplemental materials referenced in the text may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Wold MS. Replication Protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Iftode C, Daniely Y, Borowiec JA. Replication Protein A (RPA): The eukaryotic SSB. CRC Critical Reviews in Biochemistry. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 3.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rider SD, Jr, Cai X, Sullivan WJ, Jr, Smith AT, Radke J, White MGZ. The protozoan parasite Cryptosporidium parvum possesses two functionally and evolutionarily divergent replication protein A large subunits. Journal of Bological Chemistry. 2005;280:31460–31469. doi: 10.1074/jbc.M504466200. [DOI] [PubMed] [Google Scholar]
- 5.Keshav KF, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol Cell Biol. 1995;15:3119–3128. doi: 10.1128/mcb.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haring SJ, Humphreys TD, Wold MS. A naturally occurring human RPA subunit homolog does not support DNA replication or cell-cycle progression. Nucleic Acids Res. 2010;38:846–858. doi: 10.1093/nar/gkp1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason AC, Haring SJ, Pryor JM, Staloch CA, Gan TF, Wold MS. An alternative form of replication protein A prevents viral replication in vitro. J Biol Chem. 2009;284:5324–5331. doi: 10.1074/jbc.M808963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemp MG, Mason AC, Carreira A, Reardon JT, Haring SJ, Borgstahl GE, Kowalczykowski SC, Sancar A, Wold MS. An alternative form of replication protein a expressed in normal human tissues supports DNA repair. J Biol Chem. 2010;285:4788–4797. doi: 10.1074/jbc.M109.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullock PA. The initiation of Simian Virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 10.Taneja P, Nasheuer HP, Hartmann H, Grosse F, Fanning E, Weisshart K. Timed interactions between viral and cellular replication factors during the initiation of SV40 in vitro DNA replication. Biochem J. 2007;407:313–320. doi: 10.1042/BJ20070794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanning E, Zhao K. SV40 DNA replication: from the A gene to a nanomachine. Virology. 2009;384:352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons DT. SV40 large T antigen functions in DNA replication and transformation. Advances in virus research. 2000;55:75–134. doi: 10.1016/s0065-3527(00)55002-7. [DOI] [PubMed] [Google Scholar]
- 13.Khopde S, Simmons DT. Simian virus 40 DNA replication is dependent on an interaction between topoisomerase I and the C-terminal end of T antigen. J Virol. 2008;82:1136–1145. doi: 10.1128/JVI.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khopde S, Roy R, Simmons DT. The binding of topoisomerase I to T antigen enhances the synthesis of RNA-DNA primers during simian virus 40 DNA replication. Biochemistry. 2008;47:9653–9660. doi: 10.1021/bi800825r. [DOI] [PubMed] [Google Scholar]
- 15.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 16.Weisshart K, Taneja P, Fanning E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J Virol. 1998;72:9771–9781. doi: 10.1128/jvi.72.12.9771-9781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang SG, Weisshart K, Gilbert I, Fanning E. Stoichiometry and mechanism of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase a-primase. Biochemistry. 1998;37:15345–15352. doi: 10.1021/bi9810959. [DOI] [PubMed] [Google Scholar]
- 18.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 19.Maga G, Frouin I, Spadari S, Hubscher U. Replication protein a as a “fidelity clamp” for DNA polymerase alpha. J Biol Chem. 2001;276:18235–18242. doi: 10.1074/jbc.M009599200. [DOI] [PubMed] [Google Scholar]
- 20.Yuzhakov A, Kelman Z, Hurwitz J, O’Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. Embo J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zlotkin T, Kaufmann G, Jiang YQ, Lee MYWT, Uitto L, Syväoja J, Dornreiter I, Fanning E, Nethanel T. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996;15:2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda Y, Suzuki M, Piao J, Gu Y, Tsurimoto T, Kamiya K. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res. 2007;35:6904–6916. doi: 10.1093/nar/gkm822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazlieva R, Spittle CS, Morrissey D, Hayashi H, Yan H, Matsumoto Y. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37:2854–2866. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 28.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 29.Simmons DT, Gai D, Parsons R, Debes A, Roy R. Assembly of the replication initiation complex on SV40 origin DNA. Nucleic Acids Res. 2004;32:1103–1112. doi: 10.1093/nar/gkh236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hohn KT, Grosse F. Processivity of the DNA polymerase alpha-primase complex from calf thymus. Biochemistry. 1987;26:2870–2878. doi: 10.1021/bi00384a031. [DOI] [PubMed] [Google Scholar]
- 31.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 32.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wold MS, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walther AP, Bjerke MP, Wold MS. A novel assay for examining the molecular reactions at the eukaryotic replication fork: Activities of Replication Protein A required during elongation. Nucleic Acids Res. 1999;27:656–664. doi: 10.1093/nar/27.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivji MK, Podust VN, Hubscher U, Wood RD. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 36.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 37.Dornreiter I, Erdile LF, Gilbert IU, von Winkler D, Kelly TJ, Fanning E. Interaction of DNA polymerase a-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maga G, Ramadan K, Locatelli GA, Shevelev I, Spadari S, Hubscher U. DNA elongation by the human DNA polymerase lambda polymerase and terminal transferase activities are differentially coordinated by proliferating cell nuclear antigen and replication protein A. J Biol Chem. 2005;280:1971–1981. doi: 10.1074/jbc.M411650200. [DOI] [PubMed] [Google Scholar]
- 39.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, Han M, Zdzienicka MZ, Stark JM, Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 44.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, Takeda S, Hay RT, Gautier J, West SC, Paull TT, Pandita TK, White MF, Khanna KK. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard DJ, Seidman M, Pandita TK, Khanna KK, Wang W. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem. 2009;284:23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Millership JJ, Zhu G. Heterogeneous expression and functional analysis of two distinct replication protein A large subunits from Cryptosporidium parvum. Int J Parasitol. 2002;32:1477–1485. doi: 10.1016/s0020-7519(02)00135-2. [DOI] [PubMed] [Google Scholar]
- 49.Sakaguchi K, Ishibashi T, Uchiyama Y, Iwabata K. The multi-replication protein A (RPA) system--a new perspective. Febs J. 2009;276:943–963. doi: 10.1111/j.1742-4658.2008.06841.x. [DOI] [PubMed] [Google Scholar]
- 50.Ishibashi T, Kimura S, Sakaguchi K. A Higher Plant Has Three Different Types of RPA Heterotrimeric Complex. J Biochem (Tokyo) 2006;139:99–104. doi: 10.1093/jb/mvj014. [DOI] [PubMed] [Google Scholar]
- 51.Ishibashi T, Kimura S, Furukawa T, Hatanaka M, Hashimoto J, Sakaguchi K. Two types of replication protein A 70 kDa subunit in rice, Oryza sativa: molecular cloning, characterization, and cellular & tissue distribution. Gene. 2001;272:335–343. doi: 10.1016/s0378-1119(01)00555-8. [DOI] [PubMed] [Google Scholar]
- 52.Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, Estivill X, Alba MM. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 2009;26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- 53.Schittek B, Hipfel R, Sauer B, Bauer J, Kalbacher H, Stevanovic S, Schirle M, Schroeder K, Blin N, Meier F, Rassner G, Garbe C. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat Immunol. 2001;2:1133–1137. doi: 10.1038/ni732. [DOI] [PubMed] [Google Scholar]
- 54.Digweed M, Gunthert U, Schneider R, Seyschab H, Friedl R, Sperling K. Irreversible repression of DNA synthesis in Fanconi anemia cells is alleviated by the product of a novel cyclin-related gene. Mol Cell Biol. 1995;15:305–314. doi: 10.1128/mcb.15.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Garay I, Jablonka S, Sutajova M, Steuernagel P, Gal A, Kutsche K. A new gene family (FAM9) of low-copy repeats in Xp22.3 expressed exclusively in testis: implications for recombinations in this region. Genomics. 2002;80:259–267. doi: 10.1006/geno.2002.6834. [DOI] [PubMed] [Google Scholar]
- 56.Tay SK, Blythe J, Lipovich L. Global discovery of primate-specific genes in the human genome. Proc Natl Acad Sci U S A. 2009;106:12019–12024. doi: 10.1073/pnas.0904569106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menezo Y, Jr, Russo G, Tosti E, El Mouatassim S, Benkhalifa M. Expression profile of genes coding for DNA repair in human oocytes using pangenomic microarrays, with a special focus on ROS linked decays. J Assist Reprod Genet. 2007;24:513–520. doi: 10.1007/s10815-007-9167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.