Abstract
Epstein-Barr virus (EBV) is an oncogenic γ-herpes virus associated with malignancies that develop in both lymphoid and epithelial cells including nasopharyngeal carcinoma (NPC). The EBV protein latent membrane protein 2A (LMP2A) is expressed in NPC and can modulate epithelial proliferation, transformation, and differentiation, and as such, may promote malignancy. A key regulator of epithelial cell differentiation is the transcription factor p63, a member of the p53 family. This study examines the potential contribution of p63 to LMP2A-mediated inhibition of epithelial differentiation. Stable expression of LMP2A increased the protein level and stability of the ΔNp63α isoform, and in two epithelial cell lines, LMP2A interacted with ΔNp63α under stable and transient expression systems. LMP2A and ΔNp63α were localized to the cytoplasm and nuclear membrane and co-immunoprecipitated in the same fractions. Following induction of epithelial cell differentiation by calcium, expression of differentiation markers was impaired in both ΔNp63α and LMP2 expressing cells. Induction of p63α, association of p63α with LMP2A, and impairment of differentiation required the PY and ITAM signaling motif of LMP2A. By associating with and being regulated by LMP2A,ΔNp63α may function as a unique regulator of LMP2A effects on epithelial differentiation and contribute to EBV-associated epithelial cancers.
Keywords: Epstein-Barr virus, nasopharyngeal carcinoma, differentiation, p63, latent membrane protein-2A
Introduction
Epstein-Barr virus (EBV) is an oncogenic γ-herpes virus associated with several malignancies including Burkitt’s lymphoma, Hodgkin’s lymphoma and nasopharyngeal carcinoma (NPC) (Brooks et al., 1992; Busson et al., 1992; Raab-Traub, 1992a; Raab-Traub, 1992b; Raab-Traub, 2002; Young & Rowe, 1992). Most individuals are infected early in life and develop life-long latency, with some primary infections inducing infectious mononucleosis. Although primarily B-cell tropic, EBV infects other cell types, such as epithelial cells, and is considered a major factor in the development of nasopharyngeal carcinoma (NPC) (Brooks et al., 1992; Busson et al., 1992; Raab-Traub, 1992a; Raab-Traub, 1992b). There are at least 3 different gene expression patterns in latently infected cells, Latency I, II, and III. Many of the EBV-associated cancers including NPC and Hodgkin’s lymphoma have the EBV latency pattern II, characterized by expression of the nuclear protein EBNA-1 and the membrane proteins latent membrane protein 1 and 2 (LMP1 and LMP2) (Brooks et al., 1992; Busson et al., 1992; Raab-Traub, 1992a; Raab-Traub, 1992b). LMP2A mimics B-cell receptor signaling to promote cell survival and proliferation of B-cells (Merchant et al., 2001). In epithelial cells, LMP2A can induce transformation, promote motility, and inhibit differentiation, all functions that can promote malignant cell growth (Bultema et al., 2009; Fukuda & Longnecker, 2007; Scholle et al., 2000). One of the key regulators of epithelial cell development and differentiation is the transcription factor p63.
p63 is member of the p53 family of transcription factors and is preferentially expressed in epithelial cells (Candi et al., 2008; Candi et al., 2007; Medawar et al., 2008; Murray-Zmijewski et al., 2006; Ogawa et al., 2008; Truong & Khavari, 2007). p63 exists as 6 distinct isoforms generated by two different promoters, to produce TAp63 and ΔNp63, and C-terminal alternative splicing, to produce TAp63α, ΔNp63α, TAp63β, ΔNp63β, TAp63γ, and ΔNp63γ. The TAp63 isoforms contain an N-terminal transactivating domain missing in ΔNp63α, consequently TAp63 has traditionally been considered an activating transcription factor (Candi et al., 2008; Candi et al., 2007; Medawar et al., 2008; Murray-Zmijewski et al., 2006; Ogawa et al., 2008; Truong & Khavari, 2007). The isoforms can be distinguished by their molecular weights (MW) such that all of the TA isoforms are approximately 10 kd larger than the corresponding ΔN isoform. Recently, ΔNp63α has been demonstrated to be the main p63 isoform expressed in keratinocytes and has been shown to be essential for epithelial development and differentiation (Candi et al., 2008; Koster et al., 2004). ΔNp63α is highly expressed in basal, proliferating keratinocytes and is associated with inhibition of terminal differentiation marker expression as cells progress through the differentiation pathway (Bamberger et al., 2002; Candi et al., 2008; Ogawa et al., 2008). In addition, ΔNp63 expression has been detected in human malignancies including NPC and may participate in disease pathogenesis by promoting cell proliferation and inhibiting differentiation (Candi et al., 2007; Guo et al., 2006; Okuyama et al., 2007; Truong & Khavari, 2007; Yip & Tsao, 2008).
To evaluate the potential role of p63 in LMP2A-mediated inhibition of differentiation, p63 expression was determined in epithelial cells expressing LMP2A and in cells induced to differentiate. The data indicate that LMP2A increased the expression levels and stability of ΔNp63α, and physically associated with ΔNp63α at the nuclear membrane. Both p63 and LMP2A- impaired expression of the differentiation markers involucrin, keratin 1, and keratin 10. The effects on p63 and inhibition of differentiation by LMP2A required the PY and ITAM signaling motifs.
Results
LMP2A Induces Expression and Stability of p63α in Epithelial Cells
Several signaling motifs have been identified in LMP2A including a potential src kinase binding site (YEEA), an immunoreceptor tyrosine-based activation motif (ITAM) that binds the Syk tyrosine kinase, and two PY motifs that bind WW domain-containing ubiquitin ligases. LMP2A has previously been shown to inhibit differentiation in epithelial cells and the PY motif is required for the inhibition of involucrin expression upon differentiation in human foreskin keratinocytes (Morrison & Raab-Traub, 2005; Scholle et al., 2000). The ubiquitin ligase Itch interacts with the LMP2A PY motifs and regulates degradation of LMP2A and LMP2A-associated signaling mediators (Ikeda et al., 2003; Ikeda et al., 2000; Longnecker et al., 2000; Rossi et al., 2006a; Rossi et al., 2006b). One target of Itch is p63, a transcription factor belonging to the p53 family, that has been identified as a key regulator of epithelial cell growth and differentiation (Candi et al., 2008; Candi et al., 2007; Medawar et al., 2008; Murray-Zmijewski et al., 2006; Ogawa et al., 2008; Truong & Khavari, 2007). ΔNp63α is associated with impaired epithelial cell differentation and with nasopharyngeal carcinoma, and as such may provide a key target for LMP2A signaling. To determine whether LMP2A affects ΔNp63α, its expression and stability were studied in HaCaT cells stably expressing LMP2A.
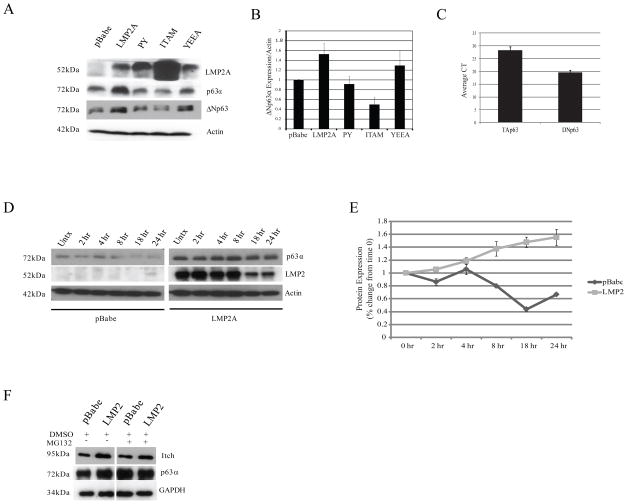
Using antibody specific for total p63α, immunoblotting of whole cell lysates revealed that LMP2A increased the protein levels of an approximately 72 kd protein, compared to pBabe, consistent with the size of ΔNp63α (Figure 1A). Antibody specific for ΔNp63 indicated that the 72 kd protein was ΔNp63α and quantitation relative to actin indicated that LMP2 increased the levels approximately 50% (Figure 1A, B). The LMP2A-induced increase in ΔNp63α expression was impaired by mutation of the LMP2A PY motifs or by deletion of the ITAM motif (Figure 1A, B). In contrast, the YEAA motif did not impair the increased ΔNp63α expression. The ITAM mutant that also lacks one PY domain was consistently expressed at higher levels perhaps reflecting its impaired activity and interaction with ubiquitin ligases. These findings suggest that the LMP2A-induced increase in ΔNp63α levels required PY and ITAM. Quantitative RT-PCR that distinguished the two distinct amino terminal forms revealed that the ΔNp63 isoform was predominant in HaCaT cells with essentially no expression of TAp63 (Figure 1C), further implicating ΔNp63α as the isoform targeted by LMP2A. Importantly, LMP2A did not increase the relative transcription of either form (data not shown), suggesting that ΔNp63α regulation by LMP2A was post-transcriptional.
Figure 1. LMP2A Increases Expression and Stability of p63α.
A LMP2A, Itch, p63α, and ΔNp63 expression in HaCaT cells stably expressing full-length LMP2A and LMP2A mutants PY, ITAM, and YEEA were determined by western blot. Actin expression was measured as the loading control. B ΔNp63α protein expression in HaCaT cells was determined by western blot and is expressed graphically. Equal loading and normalization was measured using an actin-specific antibody. C Endogenous expression of the p63 isoforms TAp63 and ΔNp63 in HaCaT cells was determined by SYBR-green quantitative RT-PCR. Expression is shown graphically by CT (cycle threshold) indicating that ΔNp63 is the predominant isoform in HaCaT cells. D Stability of LMP2A and p63α protein levels were determined by inhibiting de novo protein synthesis with 20mM cycloheximide for 0–24 hours in HaCaT cells stably expressing pBabe or LMP2A. Cell lysates were collected and protein levels of p63α and actin were determined by western blot. E p63α protein levels were normalized to actin and expressed relative to time 0 to determine turnover rate in HaCaT cells stably expressing pBabe and LMP2A. Time (hrs) indicates duration of treatment with 20mM cycloheximide. F pBabe and LMP2A expressing HaCaT cells were treated with DMSO or 20μM of the proteasome inhibitor MG132 for 4 hours. p63α expression was determined by western blot in whole cell lysates.
To determine the effect of LMP2A on p63α protein stability, HaCaT cells expressing pBabe or LMP2A were treated with cycloheximide to inhibit protein synthesis, and p63α expression was determined by western blot. pBabe cells had a time-dependent decrease in p63α expression while in LMP2A cells, p63α expression did not decrease in the presence of a protein synthesis inhibitor, suggesting LMP2A increased the expression p63α through effects on its stability (Figure 1D, E). A half-life for p63α in LMP2A cells could not be calculated because a slight increase in p63α expression relative to actin from time 0 to 24 hours gave a line of best fit with a positive slope, indicating no degradation of p63α (Figure 1E). LMP2A expression decreased in a time-dependent manner following cycloheximide treatment indicating that LMP2A did not impair the protein degradation machinery in HaCaT cells (Figure 1D). The increased stability of ΔNp63α has also been described in NPC cell lines (Guo et al., 2006)
To determine if LMP2A-induced p63α protein stability was caused by impaired proteasome-mediated degradation, pBabe and LMP2A expressing cells were treated with the proteasome inhibitor MG132. In pBabe cells, treatment with MG132 increased p63α expression compared to treatment with the DMSO vehicle control to levels equivalent to those in LMP2A expressing cells indicating that p63α is regulated by proteasome-mediated degradation (Figure 1F). In contrast, p63α levels in LMP2A cells were unchanged when treated with MG132 compared with DMSO and were similar to p63α levels in pBabe cells treated with MG132 (Figure 1F). These findings suggest that LMP2A impairs proteasome-mediated degradation of p63α in pBabe cells (Figure 1F). The Itch ubiquitin ligase that is known to regulate p63 was increased by LMP2A and inhibition of the proteasome did not affect Itch levels (Figure 1F).
LMP2A and p63α Interact in Cytoplasmic and Nuclear Membrane Fractions in Epithelial Cells
To assess the effects of LMP2A on the subcellular localization of Itch and p63α, HaCaT cells stably expressing LMP2A, LMP2A mutants, and ΔNp63α were fractionated into cytoplasm, nucleoplasm, and nuclear membrane. Protein expression was determined by western blot (Figure 2A) and the levels of the specific proteins in each fraction are presented as % of the total individual protein with standard error calculated from 3 or more experiments (Figure 2B, C, D). The purity of the fractions was determined using the endoplasmic reticulum marker GRP78 and the nuclear membrane marker emerin (data not shown). In the pBabe vector control cell, Itch was predominantly detected in the cytoplasm and nucleoplasm fractions with low expression detected in the nuclear membrane (Figure 2A, B). Stable expression of LMP2A, PY, YEEA, or ΔNp63α did not affect Itch localization or levels, however loss of the ITAM motif decreased cytoplasmic and increased nuclear expression of Itch (Figure 2A, B).
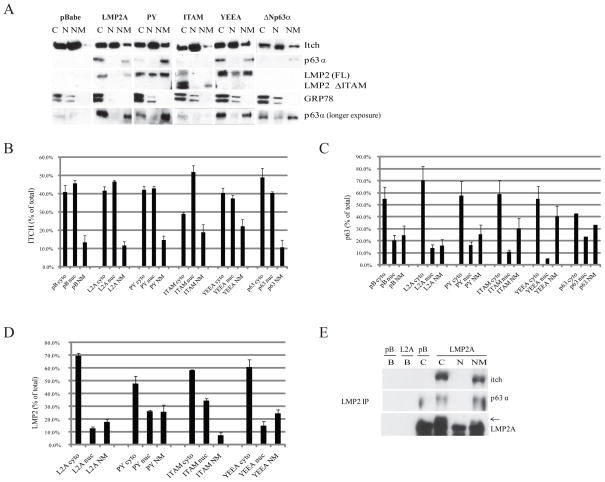
Figure 2. LMP2A Immunoprecipitates with p63α in Cytoplasmic and Nuclear Membrane Fractions.
A Lysates from HaCaT cells stably expressing pBabe, LMP2A, PY, ITAM, YEEA, and ΔNp63α were fractionated into cytoplasmic (cyto - C), nuclear (nuc – N), and nuclear membrane (NM) fractions. Protein expression in each fraction was determined by western blot. Data is also expressed graphically as % of the total amount of the protein of interest detected in each fraction. B Itch (n=4). C p63α (n=3). D LMP2A (n=3). E Localization of LMP2A-Itch-p63α complexes in HaCaT cells was determined by immunoprecipitation of LMP2A in fractionated lysates followed by western blot for p63α and Itch. LMP2A was immunoprecipitated from pBabe (pB) cytoplasmic, and from LMP2A cytoplasmic (C), nucleoplasmic (N), and nuclear membrane (NM) fractions. For beads-only controls (B), protein G beads were added to pBabe and LMP2A cytoplasmic fractions, without an antibody IP, to assess non-specific protein binding.
In the vector control cells, p63α localized primarily to the cytoplasm and nuclear membrane fractions (Figure 2A, C). Stable expression of LMP2A, PY, ITAM, YEEA, or ΔNp63α did not significantly affect p63 localization (Figure 2A, C). However, the low levels of expression of p63α in pBabe, ITAM, and ΔNp63α cells compared with LMP2A-induced expression required longer exposures to detect bands for quantitation (Figure 2A, C). The lower overall expression of p63α in pBabe and ITAM cells was consistent with the lower expression detected in whole cell lysates (Figure 1A). Full length LMP2A and all LMP2A mutants were predominately cytoplasmic with some expression in the nuclear membrane fraction (Figure 2A, D).
To determine the potential interaction of LMP2A and p63α, LMP2A was immunoprecipitated using cytoplasm from the pBabe control cells and from the cytoplasm, nucleoplasm, and nuclear membranes from LMP2A expressing HaCaT cells. Itch and p63α were detected with LMP2A precipitated from the cytoplasm and nuclear membrane (Figure 2E). Neither Itch nor p63α were detected in the immunoprecipitated material from the pBabe lysates with the LMP2 antibody or by using beads alone in the absence of antibody indicating that their precipitation required LMP2 and did not reflect a non-specific interaction of Itch or p63α with the LMP2 antibody. These findings indicate that LMP2A interacts with endogenous p63 and confirm the interaction of LMP2A and Itch that has been previously described (Ikeda et al., 2003; Ikeda et al., 2000; Longnecker et al., 2000).
The ability of LMP2A to interact with the ΔNp63α isoform was evaluated in HEK293 cells, that do not endogenously express detectable levels of p63α (data not shown), transfected with an expression construct for ΔNp63α. Endogenous Itch was detected in the cytoplasm, nucleoplasm, and nuclear membrane of the vector control HEK293 cells (Figure 3A, B). Transiently expressed LMP2A was predominantly detected in the nuclear membrane compared to the stably expressing LMP2A HaCaT cells. Localization of transiently expressed ΔNp63α was also localized primarily to the nuclear membrane with equivalent levels of detection in the cytoplasm and nucleoplasm (Figure 3A, C). The different localization of ΔNp63α and LMP2A in transient and stable expression systems suggests that initial protein expression is detected at the nuclear membrane, whereas stably expressed or endogenous proteins associate with cytoplasmic components. Surprisingly, the relative levels of nuclear ΔNp63α decreased in the presence of LMP2A.
Figure 3. LMP2A Associates with p63α in Cytoplasm and Nuclear Membrane and Requires the PY and ITAM Signaling Motifs.
A Whole cell lysates from HEK293 cells transiently expressing pBabe+pcDNA3, ΔNp63α, LMP2A, or LMP2A+ΔNp63α were fractionated into cytoplasmic (cyto - C), nuclear (nuc – N), and nuclear membrane (NM) fractions. Protein expression in each fraction was determined by western blot. Data is also expressed graphically as % of the total amount of the protein of interest that was detected in each fraction. B Itch (Standard deviations were calculated based on n=2). C p63α (Standard deviations were calculated based on n=2). D LMP2A (n=1). E Localization of LMP2A-p63α-Itch complexes was determined by immunoprecipitation of p63α from fractionated lysates (cytoplasmic – C, nucleoplasmic – N, nuclear membrane – NM) followed by western blot for LMP2A and Itch. For beads-only controls, protein G beads were added to ΔNp63α and ΔNp63α+ LMP2A cytoplasmic fractions, without an antibody IP, to determine non-specific protein binding. F Requirement of LMP2A signaling motifs for association of LMP2 with p63α was determined by immunoprecipitation of LMP2A from HEK293 cells transiently expressing pBabe, LMP2A or the PY, ITAM, or YEEA mutants of LMP2A, followed by western blot for Itch and p63α. For beads-only controls, protein G beads were added to LMP2A lysates, without an antibody IP, to determine non-specific protein binding.
The interaction of ΔNp63α with Itch and LMP2 was evaluated in the cytoplasm, nucleoplasm, and nuclear membrane fractions of the transiently transfected HEK293 cells. Itch interacted withΔNp63α in the nuclear membrane where ΔNp63α was predominantly expressed (Figure 3E). Similarly, LMP2A also interacted with ΔNp63α at the nuclear membrane. While protein localization differed in transient and stable expression systems, LMP2A and ΔNp63α interacted under both conditions.
To determine which signaling motifs were required for the interaction of LMP2A with p63α, LMP2A was immunoprecipitated from HEK293 cells transiently expressing full-length or mutated LMP2A. ΔNp63α was detected at equivalent levels with immunoprecipitated LMP2A or LMP2A with the mutated YEEA motif but was significantly decreased with the PY and ITAM mutations (Figure 3F). Similarly, the interaction of Itch with LMP2A also required the PY and ITAM motifs (Figure 3F). The decreased immunoprecipitation of both ΔNp63α and Itch with these LMP2 mutants confirmed that Itch and ΔNp63α do not nonspecifically precipitate with the LMP2A antisera. The low level of Itch detected with LMP2A likely reflects that only a subset of the total Itch is present in the nuclear membrane where LMP2A is predominantly detected. The requirement of PY and ITAM for both interactions suggests that p63α, Itch, and LMP2A may exist as a complex.
LMP2A Co-Localizes with p63α
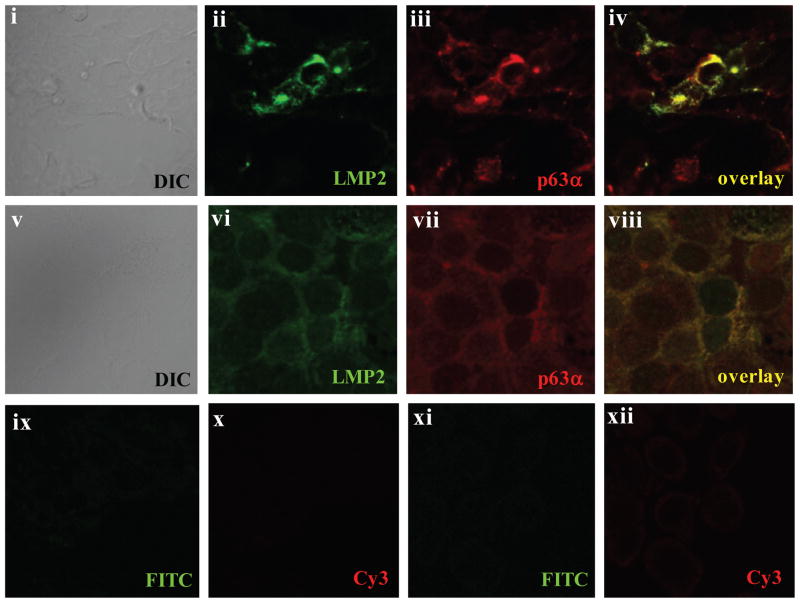
The potential interaction of LMP2A and p63α was also evaluated in HEK293 cells transiently expressing LMP2A and ΔNp63α, and in HaCaT cells stably expressing LMP2A, using immunofluorescent staining to identify colocalization. In HEK293 cells (Figure 4, panels ii through iv), LMP2A and p63α colocalized with striking perinuclear staining confirming the cell fractionation and co-immunoprecipitation analyses (Figure 3). Similarly, LMP2A and endogenous p63α primarily had perinuclear co-localization in HaCaT cells, suggesting they interact at the nuclear membrane, with some interaction evident in the cytoplasm (Figure 4 panels vi-viii). The less intense staining of both LMP2A and p63α in HaCaT cells is likely a consequence of more uniform, stable, and less abundant expression of these proteins, whereas both proteins were transiently expressed in HEK293 cells and stained 48 hours following transfection. Background staining with FITC-anti-rat or Cy3-anti-mouse antibodies was not detected in either HEK293 cells or HaCaT cells (Figure 4 panels ix–xii).
Figure 4. LMP2A Expression and Association with p63α is Localized to the Nuclear Membrane.
Localization and expression of LMP2A and p63α was determined by immunofluorescence in LMP2A and ΔNp63α expressing HEK293 cells (i–iv) and in LMP2A-expressing HaCaT cells (v–viii). Co-localization of LMP2A and p63α is indicated in panels iv (yellow – HEK293 cells) and viii (yellow – HaCaT cells). Samples were stained with secondary antibodies only to determine background staining in HEK293 cells (ix–x) and HaCaT cells (xi–xii) and images were acquired with identical exposure times and conditions as in panels i–viii. All images were obtained with a 60X oil objective using an Olympus FV500 confocal laser scanning microscope.
LMP2A Impairs Epithelial Cell Differentiation Induced by Calcium
To determine the effects of LMP2A and ΔNp63α on epithelial cell differentiation, HaCaT cells were grown for 3 weeks in calcium-free DMEM to decrease differentiation. In the original calcium-containing growth media, HaCaT cells expressed high levels of the early differentiation marker keratin 10 and the intermediate differentiation marker involucrin (Figure 5A, day –5). Following treatment for 3 weeks without calcium, HaCaT cells with or without LMP2A acquired an undifferentiated morphology (data not shown; (Deyrieux & Wilson, 2007)) with reduced levels of keratin 10 and involucrin (Figure 5A). The relative levels of keratin 10 and involucrin relative to GAPDH confirmed this decrease and indicated that LMP2A did not significantly affect the expression of these markers in de-differentiated cells (Figure 5B).
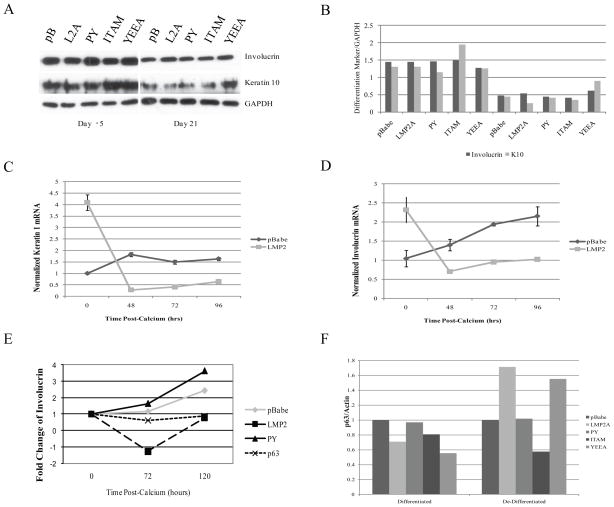
Figure 5. LMP2A is Associated with Inhibition of Differentiation Marker Expression.
HaCaT cells were induced to de-differentiate in calcium-free culture conditions for 3 weeks. A Expression of the differentiation markers involucrin and keratin 10 were determined by western blot before and after de-differentiation in calcium-free media (day -5 and day 21, respectively). B Expression levels of involucrin and keratin 10 were normalized to GAPDH and are represented graphically in the right panel. C,D To evaluate induction of differentiation by CaCl2, de-differentiated cells were induced to differentiate in media containing 2.8mM CaCl2. Cells were collected and lysed 0, 5, 24, 48, 72, and 96 hours following addition of CaCl2. mRNA expression of keratin 1 (C) and involucrin (D) was determined by quantitative real-time PCR, was normalized to GAPDH, and is expressed relative to time 0. Standard errors were calculated from 3 experiments each run in triplicate. E Protein expression of involucrin in HaCaT cells stably expressing pBabe, LMP2A, PY, and ΔNp63α induced to differentiate with 2.8mM CaCl2 for 0–120 hours was determined by western blot, normalized to actin, and expressed graphically relative to time 0. For each time point, involucrin levels were also normalized to involucrin from HaCaT cells allowed to proliferate without calcium, in order to determine specific calcium-induced involucrin expression. D To determine if LMP2A-induced p63α was associated with epithelial cell differentiation, p63α expression was determined by western blot from HaCaT cells before (differentiated) and after (de-differentiated) de-differentiation in Ca2+-free media. p63α levels were normalized to actin, and are expressed graphically.
It is known that differentiation can be induced in proliferating keratinocytes by exposure to calcium. The effects on expression of keratin 10 and involucrin mRNA in response to the addition of 2.8mM CaCl2 to the growth media of the de-differentiated HaCaT cells were determined. HaCaT pBabe cells had a slight time dependent increase in calcium-induced mRNA expression of the early differentiation marker keratin 1 (Figure 5C) and a clear induction of mRNA for the intermediate differentiation marker involucrin (Figure 5D). Whereas expression of the early marker keratin 1 peaked at 48 hours, expression of the intermediate marker involucrin continued to increase for 96 hours in pBabe cells. In contrast, the mRNA for keratin 1 and involucrin in HaCaT LMP2A cells decreased in response to calcium and remained low over the duration of the experiment (Figure 5C, D). Interestingly, LMP2A expressing cells had higher mRNA levels for both markers at time zero, in particular for the early marker keratin 1, although the difference in involucrin was not at the protein level (Figure 5A, day 21). Involucrin protein expression increased in response to calcium over time in the pBabe control cells, however this induction of involucrin was impaired in cells stably expressing LMP2A confirming the decreased expression of involucrin mRNA (Figure 5E). The impaired induction of the differentiation markers keratin 1 and involucrin in the presence of LMP2A confirms that LMP2 inhibits differentiation (Figure 5E). Similarly, involucrin was not induced in HaCaT cells that stably expressed ΔNp63α indicating that ΔNp63α expression can mimic the effect of LMP2A on calcium-induced differentiation. Calcium-induced involucrin expression in the PY mutant was similar to that observed in pBabe cells, confirming that the PY motif is required for the inhibition of differentiation by LMP2A (Figure 5E) (Morrison and Raab-Traub, 2005).
To determine whether LMP2A-induced ΔNp63α was associated with the state of differentiation of epithelial cells, p63α protein expression was determined by western blot in HaCaT cells, normalized to actin, prior to de-differentiation and following de-differentiation (Fig. 5F). The p63α protein levels were slightly lower in LMP2A compared to pBabe cells prior to de-differentiation (Figure 5F), suggesting that LMP2A does not induce p63α or promote a de-differentiated phenotype when it is expressed in differentiated cells. However, LMP2A clearly induced ΔNp63α in the de-differentiated cells and both LMP2A and ΔNp63α inhibited expression of differentiation markers after exposure to calcium. The ability of LMP2A to induce expression of ΔNp63α was impaired with mutation of the PY or ITAM signaling motifs (Figure 5F). This data supports LMP2A-induced p63α as a mediator for inhibiting keratinocyte progress through the calcium-induced differentiation pathway.
Discussion
The findings in this study reveal that in epithelial cells, expression of EBV latent membrane protein 2A (LMP2A) increased the protein levels and stability of p63α and physically associated with p63α in the cytoplasm and at the nuclear membrane. Both LMP2A and ΔNp63α impaired cellular differentiation induced by calcium and the effects of LMP2A on p63α and intermediate differentiation marker expression required the PY motifs. These findings confirm previous findings that LMP2A-induced inhibition of involucrin in epithelial cells was dependent on its PY motif (Morrison & Raab-Traub, 2005; Scholle et al., 2000). This is the first study to identify effects of LMP2A on p63α and to identify the requirement for specific LMP2A signaling motifs in regulating ΔNp63α.
It is known that p63α protein levels can be regulated through ubiquitination mediated by the ubiquitin ligase Itch that physically associates with p63α (Melino et al., 2006). Itch also associates with other target proteins, including LMP2A, through interaction of the Itch WW domain with the target protein PY domain (Ikeda et al., 2003; Ikeda et al., 2000; Ikeda et al., 2001; Longnecker et al., 2000; Winberg et al., 2000). The data presented here suggest that LMP2A can modulate the effects of Itch on ΔNp63α resulting in increased protein levels. LMP2A co-immunoprecipitated with Itch and this complex was detected only in the cytoplasm and nuclear membrane. Similarly, LMP2A-co-immunoprecipitated with ΔNp63α in the cytoplasm and nuclear membrane, suggesting the possibility that LMP2A, ΔNp63α, and Itch form a complex.
The interaction of LMP2A with p63α required both the PY and the ITAM signaling domains of LMP2A as either mutant reduced complex formation. As expected, Itch was unable to interact with LMP2A in the presence of the PY mutation. The ITAM was also required for the interaction of Itch and LMP2A, although this may reflect that the ITAM deletion mutant is also missing a PY domain. It is unknown if p63α directly binds to the PY or ITAM motifs of LMP2. p63α contains PY domains that can bind Itch and a sterile α motif (SAM) that could interact with the SH2 domains of Syk, leading to the possibility that p63α interacts with LMP2A via Itch and/or Syk as signaling mediators or as scaffold proteins. A recent study revealed that after induction of DNA damage, the stability of ΔNp63 was decreased due to its interaction with RACK1 and that the RACK1 effects were blocked by the interaction of ΔNp63 with Stxbp4 (Li et al., 2009). LMP2A may affect the expression of additional proteins that regulate p63 stability such as Stxbp4 or RACK1.
Both LMP2A and ΔNp63α inhibited expression of the intermediate epithelial differentiation marker involucrin suggesting that ΔNp63 may, in part, mediate the effect of LMP2A on differentiation. LMP2A is expressed in the epithelial cancer nasopharyngeal carcinoma (Busson et al., 1992) and by promoting transformation, motility, and inhibiting differentiation, LMP2A has been implicated in promoting epithelial malignancy (Fukuda & Longnecker, 2007; Scholle et al., 2000). ΔNp63α is expressed at high levels in NPC suggesting that its stabilization by LMP2A may be a contributing factor to the undifferentiated state of NPC. ΔNp63α has also been shown to affect the transcription of several target genes including those encoding cell cycle proteins (Candi et al., 2007; Testoni & Mantovani, 2006; Wu et al., 2003). It is presently unknown if ΔNp63α is active as a transcription factor in the presence of LMP2A. ΔNp63α was barely detected in the nucleoplasm in fractionated cells but nuclear ΔNp63α was detectable by immunostaining. However, in an initial microarray analysis several previously identified transcriptional targets of ΔNp63α were upregulated in LMP2A-expressing cells including matrilin-2, Dlx3, and IL-8 (data not shown). This may indicate that ΔNp63α is transcriptionally active in the presence of LMPA and could potentially regulate expression of genes involved in the regulation of differentiation. It is known that ΔNp63α inhibits p21 expression, a gene that is required for keratinocyte terminal differentiation (Candi et al., 2007; Westfall et al., 2003).
It is intriguing that LMP2A regulates a protein so integral to epithelial function and differentiation. Although ΔNp63α inhibits expression of terminal differentiation markers like involucrin, it is also essential for the initiation of differentiation and of early differentiation markers like keratin 1 (Ogawa et al., 2008). This recent finding may explain why LMP2A cells had approximately 4-fold higher mRNA levels of keratin 1 compared to pBabe vector control cells. In addition, pBabe and LMP2A cells had similar morphological changes in response to calcium (data not shown), suggesting that the initiation of the differentiation program was not impaired by LMP2A. These findings are consistent with the effects of LMP2A on ΔNp63α contributing to the effects of LMP2A on epithelial differentiation.
Both LMP2A and ΔNp63α have been suggested to have oncogenic potential and are associated with epithelial cell malignancies. The effects of LMP2A on ΔNp63α expression and stability may be an important contributing factor to the development of EBV associated epithelial malignancies including NPC.
Materials and Methods
Differentiation of HaCaT Cells
The human keratinocyte HaCaT cell line was maintained in calcium-free DMEM supplemented with 10% heat-inactivated FBS, from which calcium was removed, and 1% antibiotic/antimycotic. To remove calcium, Chelex 100 resin (BioRad Laboratories, Hercules, CA) was incubated with FBS on a shaker at 4°C for 1 hr (2.5g/50ml) and was removed by two sequential vacuum filtrations with a 0.22μm filter. To induce differentiation, HaCaT cells were supplemented with 2.8mM CaCl2 for 0 to 120 hours. This protocol was adapted from a study investigating HaCaT cells as an in vitro model of epithelial cell differentiation (Deyrieux & Wilson, 2007).
Plasmids and LMP2A-Expressing Cell Lines
Stable HaCaT cell lines expressing the pBabe vector or the pBabe vector subcloned with HA-tagged LMP2A, LMP2A Δ PY, LMP2A Δ ITAM, LMP2A Δ YEEA, or with ΔNp63α were generated by transduction with recombinant retroviruses expressing each vector and selection with 0.5μg/ml puromycin as described previously (Morrison & Raab-Traub, 2005; Scholle et al., 2000). The N-terminus of LMP2A contains two PY motifs (aa56-60 and aa96-100), one ITAM motif (aa74-88) and one YEEA motif (aa112-115). The LMP2A Δ PY mutant contains four proline-alanine mutations at aa57, aa58, aa98, and aa99 to disrupt both PY motifs. The LMP2AΔ ITAM mutant contains a tyrosine-phenylalanine mutation at aa74, a deletion from aa75-111 and is consequently missing both the ITAM motif and the second PY motif. The LMP2A Δ YEEA mutant contains a tyrosine-phenylalanine mutation at aa112. The LMP2A mutants were a generous gift from Dr. R. Longnecker.
To stably express ΔNp63α in HaCaT cells, ΔNp63α was subcloned into pBabe from pcDNA3-ΔNp63α graciously provided by Dr. M. Oren. HaCaT cells stably over-expressing pBabe-ΔNp63α were generated by retroviral transduction and selection with puromycin (0.5μg/ml).
HEK293 cells were maintained in DMEM containing 10% FBS and 1% antibiotic/antimycotic and were transiently transfected for 48 hours using FuGENE with the pBabe and pcDNA3 vectors alone, pBabe-LMP-2A, pcDNA3-ΔNp63α (a generous gift from Dr. M. Oren), and pBabe-LMP-2A + pcDNA3-ΔNp63α, as per manufacturer’s protocol.
Western Blot and Immunoprecipitation
Whole-cell lysates were generated by lysis of cell pellets with RIPA buffer (50mM Tris-HCl pH 7.4, 150mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1mM EDTA) and were subjected to western blot. For whole cell lysate immunoprecipitation, cell pellets were lysed with NETN buffer (0.5% NP-40, 50mM Tris-HCl pH 8.0, 1mM EDTA, 120mM NaCl) and were incubated at 4°C overnight with 2–10μg primary antibody (LMP2 or p63α). 50μl protein G bead slurry was added for 1 hour, beads were washed 5 times, and bound proteins were eluted with sample buffer heated at 95°C for 10 minutes.
RIPA/NETN buffer whole cell lysates and IP lysates were subjected to SDS PAGE, transferred to nitrocellulose membranes, blocked for 1 hour in 5% milk/TBS-Tween, and incubated overnight with primary antibody diluted in 5% milk/TBS-Tween (mouse anti-Itch 1:250 BD Biosciences, mouse anti-involucrin 1:1000 Sigma-Aldrich, anti-GRP78 1:200 Santa Cruz, rabbit anti-GAPDH 1:500 Santa Cruz, goat anti-actin 1:500 Santa Cruz, rabbit anti-ΔNp63 1:250 BioLegend, mouse anti-p63α 1:200 Santa Cruz, rat anti-LMP2 clone 14B7 1:2000 Abcam Inc, mouse anti-keratin 10 1:100 NeoMarkers). Membranes were washed and incubated for 1 hour with horseradish peroxidase conjugated secondary antibodies (Amersham Biosciences and Dako) diluted 1:1000 in 5% milk/TBS-Tween. Western blots were washed and developed with Pierce Supersignal West Pico System. Band volumes were measured with ImageJ64 software (National Institutes of Health).
Cell Fractionations
Cell pellets were isolated and resuspended in 200–300 μl of hypotonic lysis buffer (10 mM Hepes (pH 7.6)/10 mM NaCl/1.5 mM MgCl2/0.1% Nonidet P-40/10% glycerol, 0.5 mM DDT/0.4 mM PMSF/1 mM NaF/0.1 mM NaVO4/Complete Mini protease inhibitors (Roche)). Lysates were incubated on ice for 15 min and centrifuged for 5 min at 1,000 x g (4°C). Supernatants were reserved and cell pellets were washed once with 100 μl of hypotonic lysis buffer and centrifuged for 5 min at 1,000 x g (4°C). Supernatants were combined with the previous supernatant to yield the cytoplasmic fraction.
The nuclear pellets were manually homogenized using a dounce homogenizer with 1mM EDTA containing protease inhibitors. Nuclear lysates were centrifuged at 300 x g for 10 min at 4°C. The clear supernatant was collected and centrifuged at 20,000 x g for 20 min at 4°C to separate the particulate fraction. Supernatants were collected and reserved (nucleoplasm) and pellets were resuspended in homogenization buffer (nuclear membranes).
Real-Time Polymerase Chain Reaction
RNA lysis buffer was added to freshly isolated cell pellets and lysates were stored at −80°C until extraction. Total RNA was purified fusing the Qiagen RNeasy Plus kit according to the manufacturer’s instructions. mRNA expression levels of involucrin, keratin 1, TAp63, ΔNp63, and actin were determined using the Qiagen QuantiTech SYBR-green real-time PCR kit according to the manufacturer’s instructions and SYBR-green labeled PCR products were detected using an Applied Biosystems 7900HT real-time PCR system.
Immunofluorescence
Cells were plated into 8 well permanox-coated chamber slides and were allowed to attach overnight. Slides were gently washed with PBS and fixed for 15 minutes with ice-cold acetone:methanol (1:1) and blocked for 15 minutes at room temperature with 3% bovine serum albumin in PBS. Primary antibodies were prepared in PBS (rat anti-LMP2 clone 4E11 1:2000, mouse anti-p63α) and incubated for 1 hour at room temperature in a humidified chamber. Slides were washed and incubated in a humidified chamber for 1 hour at room temperature with secondary antibodies prepared in PBS (anti-rat FITC 1:100, anti-mouse Cy3 1:100). Slides were then washed and mounted with glass coverslips using anti-fade mounting media. Samples were visualized using an Olympus FV500 confocal laser scanning microscope at the Microscopy Services Laboratory at the University of North Carolina, Chapel Hill.
Acknowledgments
Work supported by the National Institutes of Health grants CA19014, CA32979, and CA103634.
The authors gratefully acknowledge Dr. M. Oren for providing the ΔNp63α plasmid and Dr. R. Longnecker for providing the LMP2A PY mutant. Funding for this study was provided by the National Institutes of Health grants CA19014, CA32979, and CA103634.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Bamberger C, Pollet D, Schmale H. J Invest Dermatol. 2002;118:133–8. doi: 10.1046/j.0022-202x.2001.01649.x. [DOI] [PubMed] [Google Scholar]
- Brooks L, Yao QY, Rickinson AB, Young LS. J Virol. 1992;66:2689–97. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema R, Longnecker R, Swanson-Mungerson M. Oncogene. 2009;28:1471–6. doi: 10.1038/onc.2008.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. J Virol. 1992;66:3257–62. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R. Cell Mol Life Sci. 2008;65:3126–33. doi: 10.1007/s00018-008-8119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, Krammer PH, Melino G. Cell Cycle. 2007;6:274–85. doi: 10.4161/cc.6.3.3797. [DOI] [PubMed] [Google Scholar]
- Deyrieux AF, Wilson VG. Cytotechnology. 2007;54:77–83. doi: 10.1007/s10616-007-9076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Longnecker R. J Virol. 2007;81:9299–306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Pan ZG, Li DJ, Yun JP, Zheng MZ, Hu ZY, Cheng LZ, Zeng YX. J Transl Med. 2006;4:23. doi: 10.1186/1479-5876-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Caldwell RG, Longnecker R, Ikeda M. J Virol. 2003;77:5529–34. doi: 10.1128/JVI.77.9.5529-5534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ikeda A, Longan LC, Longnecker R. Virology. 2000;268:178–91. doi: 10.1006/viro.1999.0166. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ikeda A, Longnecker R. J Virol. 2001;75:5711–8. doi: 10.1128/JVI.75.12.5711-5718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. Genes Dev. 2004;18:126–31. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Peart MJ, Prives C. Mol Cell Biol. 2009;29:3953–63. doi: 10.1128/MCB.00449-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Merchant M, Brown ME, Fruehling S, Bickford JO, Ikeda M, Harty RN. Exp Cell Res. 2000;257:332–40. doi: 10.1006/excr.2000.4900. [DOI] [PubMed] [Google Scholar]
- Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, Aberdam D. PLoS ONE. 2008;3:e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G, Knight RA, Cesareni G. Cell Cycle. 2006;5:1735–9. doi: 10.4161/cc.5.16.3260. [DOI] [PubMed] [Google Scholar]
- Merchant M, Swart R, Katzman RB, Ikeda M, Ikeda A, Longnecker R, Dykstra ML, Pierce SK. Int Rev Immunol. 2001;20:805–35. doi: 10.3109/08830180109045591. [DOI] [PubMed] [Google Scholar]
- Morrison JA, Raab-Traub N. J Virol. 2005;79:2375–82. doi: 10.1128/JVI.79.4.2375-2382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. Cell Death Differ. 2006;13:962–72. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Okuyama R, Egawa T, Nagoshi H, Obinata M, Tagami H, Ikawa S, Aiba S. J Biol Chem. 2008;283:34241–9. doi: 10.1074/jbc.M804101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama R, Ogawa E, Nagoshi H, Yabuki M, Kurihara A, Terui T, Aiba S, Obinata M, Tagami H, Ikawa S. Oncogene. 2007;26:4478–88. doi: 10.1038/sj.onc.1210235. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N. Semin Cancer Biol. 1992a;3:297–307. [PubMed] [Google Scholar]
- Raab-Traub N. Infect Agents Dis. 1992b;1:173–84. [PubMed] [Google Scholar]
- Raab-Traub N. Semin Cancer Biol. 2002;12:431–41. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. Proc Natl Acad Sci U S A. 2006a;103:12753–8. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, De Simone M, Pollice A, Santoro R, La Mantia G, Guerrini L, Calabro V. Cell Cycle. 2006b;5:1816–22. doi: 10.4161/cc.5.16.2861. [DOI] [PubMed] [Google Scholar]
- Scholle F, Bendt KM, Raab-Traub N. J Virol. 2000;74:10681–9. doi: 10.1128/jvi.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testoni B, Mantovani R. Nucleic Acids Res. 2006;34:928–38. doi: 10.1093/nar/gkj477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Khavari PA. Cell Cycle. 2007;6:295–9. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. Mol Cell Biol. 2003;23:2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg G, Matskova L, Chen F, Plant P, Rotin D, Gish G, Ingham R, Ernberg I, Pawson T. Mol Cell Biol. 2000;20:8526–35. doi: 10.1128/mcb.20.22.8526-8535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Nomoto S, Hoque MO, Dracheva T, Osada M, Lee CC, Dong SM, Guo Z, Benoit N, Cohen Y, Rechthand P, Califano J, Moon CS, Ratovitski E, Jen J, Sidransky D, Trink B. Cancer Res. 2003;63:2351–7. [PubMed] [Google Scholar]
- Yip YL, Tsao SW. Int J Oncol. 2008;33:713–24. doi: 10.3892/ijo_00000057. [DOI] [PubMed] [Google Scholar]
- Young LS, Rowe M. Semin Cancer Biol. 1992;3:273–84. [PubMed] [Google Scholar]