Abstract
Protein acetylation on Lys residues is recognized as a significant post-translational modification in cells, but it is often difficult to discern the direct structural and functional effects of individual acetylation events. Here we describe a new tool, methylthiocarbonyl-aziridine, to install acetyl-Lys mimics site-specifically into peptides and proteins by alkylation of Cys residues. We demonstrate that the resultant thiocarbamate modification can be recognized by the Brdt bromodomain and site-specific anti-acetyl-Lys antibodies, is resistant to histone deacetylase cleavage, and can confer activation of the histone acetyltransferase Rtt109 by simulating autoacetylation. We also use this approach to obtain functional evidence that acetylation of CK2 protein kinase on Lys102 can stimulate its catalytic activity.
Reversible acetylation of histones catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs) is recognized as a central mechanism of chromatin regulation. Beyond histones, Lys acetylation has been observed in more than 2500 mammalian proteins on more than 4000 sites and has been shown to govern many biological processes.1 Several approaches are available to explore the structural and functional consequences of protein acetylation. Traditional site-directed mutagenesis is used to replace Lys with Arg or Gln to provide indirect insights into Lys acetylation. Expressed protein ligation and unnatural amino acid mutagenesis via nonsense suppression can install acetyl-Lys (AcK) at desired protein sites but have various technical limitations.2
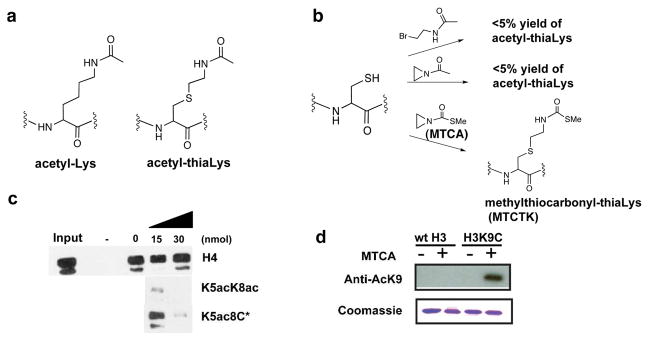
As an analog of methyl-Lys, methyl-thiaLys can be introduced at targeted protein locations via a relatively simple strategy involving Cys alkylation with N-methyl-aminoethylbromide derivatives.3 To create an acetyl-thiaLys analog (Figure 1a), we attempted to use the corresponding acetamide reagent with Cys-containing peptide; however, no significant conversion was observed. We hypothesized that the reduced ability of the amide compounds to form three-membered ring intermediates and/or intramolecular imidate formation led to diminished activity of N-acetyl-aminoethylbromide. We then explored the reaction of N-acetyl-aziridine and a Cys-containing peptide (Figure 1b). The predominant product observed by mass spectrometry was M+42 suggesting that acetyl-aziridine treatment led to an undesired acetyl transfer by nucleophilic attack at the carbonyl rather than the aziridine methylene (Figure S1).
Figure 1.
(a) Acetyl-Lys (AcK) and analog. (b) Strategies attempted to install acetyl-Lys analogs. (c) Competition assay of MTCA-modified H4 derived peptides by tetracetylated N-terminal H4 tail. Extracts from GFP-Brdt expressing cells were pre-incubated with peptides and then used in a pull-down assay with immobilized tetracetylated H4 tail or the same non-modified peptide (- lane). The GFP-Brdt retained on the beads was then revealed using an anti-GFP antibody. C* corresponds to MTCTK. (d) Western blots with site-specific anti-AcK9 Ab on MTCA-treated mutant H3.
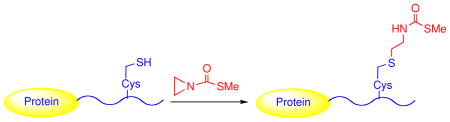
To reduce reactivity at the carbonyl, we explored methylthiocarbonyl-aziridine (MTCA) (Figure 1b).4 Cys alkylation with this reagent was proposed to provide methylthiocarbonyl-thiaLys (MTCTK), a thiocarbamate analog of AcK (Figure 1b). We hoped that this thiocarbamate could preserve key functional features of AcK but perhaps be resistant to hydrolysis by deacetylases since this could be beneficial in cell extracts. In the presence of aqueous buffer (pH 8.0) with 100 mM MTCA, mass spectrometric analysis revealed essentially complete conversion of six different Cys-containing peptides with a wide variety of residues to the desired products (Figures S2–S4). In a Cys-free peptide control, no modification was detected, indicating high selectivy of MTCA for the Cys thiol (Figure S2).
Some AcK residues mediate bromodomain interaction. To explore the potential of MTCTK to mimic this aspect of AcK function, we used a pulldown assay to assess the binding efficiency of the Brdt bromodomain with H4 acetylated tails containing MTCTK groups in place of AcK at the K5 and K8 positions.5 It was predicted and confirmed here that unacetylated or monoacetylated H4 tail peptides do not compete with the Brdt binding to a tetracetylated H4 peptide, whereas an excess of untagged di-acetylated AcK5/AcK8 H4 tail peptide can efficiently block Brdt pull-down (Figure 1c, S5). Here we show that peptides containing substitution of either AcK5 or AcK8 with the MTCTK modification can also compete with the pulldown of Brdt, although approximately 2-4-fold less efficiently than the natural AcK-containing peptides (Figure 1c, S5). These data show that Brdt’s first AcK bromodomain binding pocket can accommodate MTCTK interaction at two sites.
To explore the susceptibility of the methylthiocarbonyl linkage to deacetylase activity, we compared the reactivity of MTCTK with that of an AcK containing substrate in the context of a p53 peptide with the class I histone deacetylase HDAC8 and the class III NAD-dependent deacetylase Sir2 using an HPLC readout (Figure S6, S7).6 In contrast to the AcK p53 peptide, no thiocarbamate removal was detected under similar reaction conditions. These data suggest that this AcK mimic could be useful in the presence of deacetylase-rich cell extracts where the consequences of a “constitutive” acetylation state could be explored.
We further examined the reactivity of recombinant histone H3 mutants with MTCA. Natural Xenopus histone H3 has a single Cys (C110), and so three histone mutant proteins were prepared that replaced this site with Ala and introduced a Cys at the known AcK sites K9, K18, and K27. Alkylation reactions were monitored with site-specific anti-AcK antibodies (Abs) by western blot. MTCA treatment of C110A/K9C histone H3 led to efficient recognition by anti-AcK9 Ab whereas wt H3 displayed background detection under similar reaction conditions (Figure 1d). This experiment suggests that modification of Cys9 in C110A/K9C H3 was efficient, as confirmed by MS (Figure S8a), and further demonstrates that MTCTK is a reasonable surrogate for AcK in mediating Ab interaction. In a similar fashion, it was shown that MTCA treatment of C110A/K18C and C110A/K27C histone H3 gave rise to efficient recognition by anti-AcK18 Ab and anti-AcK27 Ab, respectively (Figure S8).
Several HATs have been suggested to undergo autoacetylation as a potential mode of regulation.1a,7 In particular, the yeast HAT Rtt109 has been shown recently to be self-acetylated on active site Lys290.1a,7 Mutation of this residue to Arg has been demonstrated to reduce catalytic activity,7 and we confirmed that here (Figure S9). We sought to examine whether introduction of the AcK mimic at this site would stimulate catalysis, and therefore we introduced a Cys at position 290. Rtt109 K290C mutant showed essentially identical HAT activity to the K290R mutant (Figure 2a and S9). Since the sequences of Rtt109 and its heterodimeric partner Vps75 have several natural Cys residues, multiple alkylations of these proteins are likely, which could complicate interpretation. To account for this, we compared directly the HAT activities of Rtt109 K290R and K290C mutants treated with MTCA under identical conditions. We observed a reproducible 4-fold relative increase in HAT activity after MTCA treatment of the Rtt109 K290C enzyme compared with the treated Rtt109 K290R enzyme (Figure 2a). It should be noted that the 4-fold activation by alkylation of Rtt109 K290C does not fully recapitulate the 20-fold rate differential between wt and K290R Rtt109 (Figure S9), and this could be related to multiple Cys alkylation in the proteins or imperfect mimicry of AcK by the MTCTK group. Nevertheless, we interpret this partial recovery of activity by introduction of the AcK analog as novel evidence that autoacetylation of Rtt109 on K290 stimulates its HAT activity.
Figure 2.
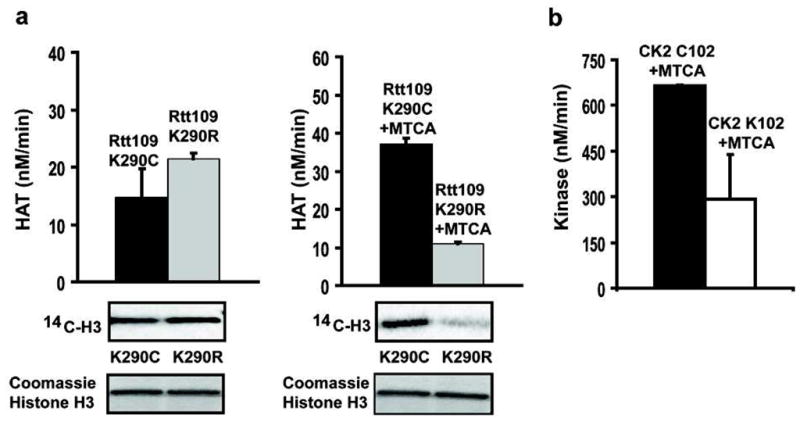
(a) HAT activity of Rtt109 K290C and RTT109 K290R HAT activity before and after MTCA treatment. (autoradiograph below). (b) Kinase activity of CK2α 3 (carrying C147A and C220A mutations) containing either Cys102 or Lys102 after MTCA treatment.
We next employed MCTA alkylation to explore the effect of the recently discovered acetylation of protein kinase CK2α on Lys102.1b,8 CK2α has two natural Cys residues (147, 220) which we mutated to Ala, and we further replaced Lys102 with Cys. Kinase assays showed that each of these mutants had essentially identical kinase activity to wild-type CK2α (Figure S10). MTCA alkylation and kinase assays revealed that MTCA-treated Cys102 showed about 2-fold higher catalytic activity compared to MTCA-treated Lys102 (Figure 2b), providing functional evidence for the acetylation-dependent activation of this important protein kinase.8
In summary, this study describes a simple approach to install AcK mimics into recombinant proteins. While the MTCTK is not identical to the AcK structure, it appears to maintain aspects of the molecular recognition by binding proteins and in enzymatic regulation. The thiocarbamate moiety appears resistant to HDAC cleavage which may provide utility in the complex environments found in transcriptional and chromatin assays.9 We submit that, along with other approaches,1,2 this Cys modification strategy offers promise in dissecting the diverse and important functions of acetylation in the proteome.
Supplementary Material
Acknowledgments
We thank S. Taverna, J. Boeke, and Cole lab members for helpful advice and discussions. We are grateful to the NIH as well as the FAMRI and Kaufman foundations for financial support. The work in SK laboratory was supported by INCa-DHOS, ANR blanche “EpiSperm” and “Empreinte” and ARC research programs.
Footnotes
Supporting Information Available. Detailed experimental procedures and Figures S1-S10. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Wang L, Tang Y, Cole PA, Marmorstein R. Curr Op Struct Biol. 2008;18:741. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Wlather TC, Olsen JV, Mann M. Science. 2009;325:834. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]; (c) Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning Z, Zeng R, Xiong Y, Guang K, Zhao S, Zhao G. Science. 2010;327:1004. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Cole PA. Nat Chem Biol. 2008;4:590. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci USA. 1998;95:6705. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311:844. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]; Neumann H, Peak-Chew SY, Chin JW. Nat Chem Biol. 2008;4:232. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 3.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. Cell. 2007;128:1003. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fishbein PL, Kohn H. J Med Chem. 1987;30:1767. doi: 10.1021/jm00393a015. [DOI] [PubMed] [Google Scholar]
- 5.Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte A, Govin J, Gaucher J, Sadoul K, Hart D, Krijgsveld J, Khochbin S, Muller CW, Petosa C. Nature. 2009;461:664. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 6.Fatkins DG, Monnot AD, Zheng W. Bioorg Med Chem Lett. 2006;16:3651. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- 7.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Nat Struct Mol Biol. 2004;11:308. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]; Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, Marmorstein R. Nat Struct Mol Biol. 2008;15:738. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stavropoulos P, Nagy V, Blobel G, Hoelz A. Proc Natl Acad Sci USA. 2008;105:12236. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinna LA, Allende JE. Cell Mol Life Sci. 2009;66:1795. doi: 10.1007/s00018-009-9148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu W, Gong D, Bar-Sagi D, Cole PA. Mol Cell. 2001;8:759. doi: 10.1016/s1097-2765(01)00369-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.