Abstract
The bacterial flagellum is a complex macromolecular machine consisting of more than 20,000 proteins, most of which must be exported from the cell via a dedicated Type III secretion apparatus. At a defined point in flagellar morphogenesis, hook completion is sensed and the apparatus switches substrate specificity type from rod and hook proteins to filament ones. How the switch works is a subject of intense interest. FliK and FlhB play central roles. In the present study two optical biosensing methods were used to characterize FliK-FlhB interactions using wild-type and two variant FlhBs from mutants with severe flagellar structural defects. Binding was found to be complex with fast and slow association and dissociation components. Surprisingly, wild-type and variant FlhBs had similar kinetic profiles and apparent affinities, which ranged between 1-10.5 μM, suggesting that the specificity switch is more complex than presently understood. Other binding experiments provided evidence for a conformational change after binding. LC-MS and NMR experiments were performed to identify a cyclic intermediate product whose existence supports the mechanism of autocatalytic cleavage at FlhB residue N269. The present results show that while autocatalytic cleavage is necessary for proper substrate specificity switching, it does not result in an altered interaction with FliK, strongly suggesting the involvement of other proteins in the mechanism.
The bacterial flagellum is a self-assembling rotary motor that drives motility and allows chemotaxis (1, 2). It is composed of three major parts: the basal body containing the motor, a hook which serves as a universal joint and a filament which acts as a propeller. Flagellar assembly is accomplished via a dedicated Type III secretion (T3S) apparatus within the basal body which consists of both membrane and soluble proteins (2, 3). Protonmotive force is used to power transport of flagellar proteins across the cytoplasmic membrane into a channel within the growing flagellum (4, 5). Export is ordered, allowing for efficient assembly of rod and hook then filament. Regulation of gene expression also plays an important role (6).
Upon hook completion, the export apparatus switches specificity of substrates from “rod/hook-type” proteins to “filament-type” ones, e.g. prior to the switch, the apparatus will not export FliC, the major component of the filament (7). The hook extends ~55 nm beyond the cytoplasmic membrane. How the apparatus detects completion at a point distant from the membrane-bound export apparatus is unknown, but several models have been proposed and debated (8-13). Hook length control also bears a remarkable similarity to needle length control in virulence T3S systems (14). Proper control of length is accordingly necessary for both flagellar motility and delivery of virulence factors (15,16).
Intergenic suppression and affinity blotting data point to an interaction between FliK and FlhB as critical to substrate specificity switching (17,18). Point mutations in flhB give rise to phenotypes of dramatically altered structures. Two mutations that result in single residue substitutions in the cytoplasmic domain of FlhB (FlhBc), N269A and P270A, cause polyhook and polyhook-filament phenotypes (7). Both variants have defects in postranslational hydrolysis at N269 with N269A being non-cleaving and P270A having dramatically slower cleavage. The affected NPTH sequence is highly conserved among FlhB homologs.
Hydrolysis at N269 is necessary for specificity switching and has been proposed to be an asparagine-mediated autocatalytic cleavage (19) as is known to occur in the degradation of aging proteins (20). A great deal of evidence supports the mechanism including enhancement of cleavage in basic conditions and an unsuccessful search for a protease (19). Crystal structures of FlhB virulence T3S orthologs EscU, SpaS and variants thereof evidenced the mechanism (21). Cleavage is unlikely to be functionally dynamic, i.e. occurring upon hook completion or otherwise effecting the switch, but is thought to be necessary for a conformational change that occurs upon binding to FliK (9).
Optical biosensing is a technique of choice for measuring biomolecular interactions. Surface plasmon resonance (SPR) is the most commonly used biosensing method, but biolayer interferometry (BLI) is an emerging method that operates on a different physical principle. Like SPR, BLI measures changes near a surface that reflect association and dissociation of biomolecules (22, 23), but is immune to refractive index changes unrelated to binding. Used in concert, BLI and SPR complement one another by alleviating concerns about artifacts specific to one method or the other such as mass transfer effects.
In this study, SPR and BLI were used to analyze interactions between FliK and FlhBc with the hypothesis that wild-type FlhBc and variants FlhBc(N269A) and FlhBc(P270A) would have altered interactions with FliK. Using immobilized FliK and FlhBc as analyte, SPR experiments revealed complex binding having fast and slow components for both association and dissociation. Remarkably, the variant FlhBcs exhibited highly similar kinetic profiles and apparent affinities. BLI results confirmed the complexity observed in SPR and were simulated to determine rate constants.
Liquid Chromatography-Mass Spectrometry (LC-MS) experiments were also conducted to identify the succinimide intermediate product whose existence supports the proposed autocatalytic mechanism. The intermediate was identified upon base treatment in the slow-cleaving variant FlhBc(P270A). While the succinimide intermediate was not observed for wild-type FlhBc, probably due to its transient existence, a two-dimensional NMR spectrum of wild-type FlhBc contained shifts consistent with the formation of iso-asparagine, a potential product of succinimide hydrolysis. These experiments provide additional, direct evidence for the asparagine-mediated cleavage, which is remarkable due to the reaction speed and its necessity for biological function. Other examples of asn-mediated cleavage are very slow and involved in protein degradation (24-26).
The present results demonstrate that while the cleavage event in FlhB is critical to morphogenesis, it does not seem to significantly alter FliK binding, suggesting that the switch mechanism involves other actors and contains additional complexity not yet reflected in the molecular ruler and other proposed models.
Experimental Procedures
Overproduction and purification of FliK and FlhBcs
His-tagged FliK and all FlhBc variants were overproduced and purified. FlhBc consists of residues 211-383; variant FlhBcs were also residues 211-383 with single substitutions N269A or P270A. Overnight cultures of E. coli BL21DE3(pLysS) cells harboring plasmids encoding His-tagged proteins were subcultured and grown at 30° C to an OD600 = 0.4. Expression was induced by addition of 0.2 mM IPTG, after which growth was continued for four hours. Cells were harvested by centrifugation and pellets were frozen at −80° C until use.
Pellets from 1 L cultures were thawed on ice and resuspended in 25 ml lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 10 mM imidazole, 0.1% Tween-20 and 200 ug/ml lysozyme). Resuspended cells were lysed by passage through a French press at 20,000 psi and then subjected to centrifugation for 20 min at 10,000 x G at 4° C. The supernatant was then centrifuged for 60 min at 100,000 x G. The resulting supernatant was transferred to a tube containing 1 ml of equilibrated Talon (BD Biosciences) immobilized metal affinity chromatography resin.
Batch binding was allowed to proceed with gentle agitation for 20 min after which the resin was pelleted by brief centrifugation and washed twice with 20 ml wash buffer (50 mM Tris pH 8.0, 500 mM NaCl, 25 mM imidazole, 0.1% Tween-20). The resin was transferred to a column and washed with an additional 10 ml. Elution was achieved by addition of elution buffer (wash buffer with 250 mM imidazole).
For SPR experiments, proteins were exchanged into binding buffer HNT (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Tween) by dialysis against 4 L twice. Protein concentration was determined by Bradford Assay using BSA as standard.
For NMR studies, N-terminally His-tagged FlhBc was overproduced as above save that cells were cultured in M9 media supplemented with 15N NH4Cl as the sole nitrogen source. Purification was performed under denaturing conditions to eliminate the C-terminal fragment (P270-E383, “FlhBcc”). SDS-PAGE confirmed that only the His-tagged “FlhBcn” (F211-N269) band was present. After elution from the affinity column, FlhBcn was dialyzed against 100 mM phosphate buffer, 10 mM Tris, 8M urea pH 8.0.
Surface plasmon resonance (SPR)
All SPR measurements were made on a Biacore X100 instrument. 500 response units (RU) of FliK were immobilized on a CM5 chip by amine crosslinking. Briefly, the surface was treated with EDC/NHS (50mM/200mM) for 7 min. FliK was then injected in 10 mM acetate, pH 5.5 for 12s. Surfaces were then blocked with 1 M ethylenediamine pH 8.0 for 7 min. Analyte injections of various concentrations in binding buffer to monitor association were made prior to changing to buffer only to monitor dissociation. Sensorgrams were analyzed with BIAEvaluation and Biacore X100 Evaluation software packages. Since rates were too fast to quantify, apparent affinity constants (KDapp) were determined by fitting response at steady state versus FlhBc concentration where response = Rmax* [FlhBc]/(KDapp + [FlhBc]).
Biolayer Interferomtery (BLI)
All BLI measurements were made on a ForteBio (Menlo Park, CA) Octet QK biosensor using streptavidin (“SA”) sensors. Assays were performed in 96-well microplates at 25° C. All volumes were 200 μl. His-FliK was biotinylated using NHS-LC-LC-biotin (succinimidyl-6-[biotinamido]-6-hexanamidohexanoate) (Pierce) at a 5:1 molar ration of biotin to protein for 30 min at 25° followed by rapid buffer exchange into PBS-T (10 mM phosphate buffer, pH 7.4, 150 mM NaCl, 0.01% Tween-20) by passage over a desalting column. FlhBc was also exchanged into PBS-T after affinity purification. Biotinylated FliK was loaded onto sensors for 900 s. After establishing a baseline in PBS-T, tethered FliK was exposed to analyte FlhBc at concentrations from 10-40 μM. Association was monitored for 900s followed by dissociation in PBS-T alone for 900 s. Raw shift data were moved to Microsoft Excel for simulation.
Simulation
Fits to raw response data were simulated using a two component model for association, Y = Yo+Afast(1-e−kobs(fast)*t)+Aslow(1-e−kobs(slow)*t), where Yo is the response at time 0, A is the amplitude of the respective component, and kobs is the observed rate constant of the respective component. Dissociations were fit to Y = Yo − Aslow(1-e−koff(slow)*t)-Afast(1-e−koff(fast)*t), where Yo is response at the beginning of dissociation, A are amplitudes and koff are dissociation rate constants for the respective components. Two component fits adequately account for the data. Addition of a third minor component did not substantially change the goodness of fit.
LC-MS
Wild-type FlhBc and FlhBc(P270A) were examined to identify the intermediate succinimide product anticipated by the proposed autocatalytic mechanism. With or without prior treatment with NaOH, purified proteins were analyzed using a NanoAcquity LC (Waters), using 0.1% v/v formic acid in water as solvent A and 0.1% v/v formic acid in acetonitrile as solvent B. 5 μl of sample were injected onto a 180 μm × 20 mm trapping column packed with 5 μm Symmetry C18 stationary phase (Waters Corporation), and trapping was carried out with 99.9% solvent B at 20 μL/min for 2 min. Sample was then separated on a 75 μm × 150 mm 1.7 μm BEH particle column, with a gradient of 5 to 90% solvent B in 15 minutes and a flow rate of 0.4 μL/min. The analytical column was heated to 55°C. The outlet of the column was coupled directly to a Waters QToF Global Ultima mass spectrometer. Mass spectra were collected in profile mode from 50 to 1990 m/z with a 1 Hz scan rate, with Masslynx 4.1 acquisition software (Waters). A lockmass scan of Glu-Fibrinopeptide B (785.8426 m/z) was collected every 30 sec during data acquisition for accurate mass correction.
MS Data Deconvolution
For protein molecular weight determination, spectra were first mass-corrected using the three lockmass scans nearest in retention time to the acquired data; lockmass scans were summed, smoothed, and centroided, and the difference between the observed monoisotopic peak for Glu-Fibrinopeptide B and the accurate mass was used to correct the experimental protein spectra. Mass spectra were summed across the elution time of the LC-MS peak, and Savitzky Golay smoothed. Molecular weight of the protein was then determined either using Maximum Entropy deconvolution at 1 Da resolution (MaxEnt 1, Waters), or by further centroiding the spectra and using the manual deconvolution tool in Masslynx 4.1. Expected mass accuracy of the deconvoluted protein masses is within 1 Da.
NMR
Wild-type FlhBcn purified after cleavage was concentrated by centrifugal filtration to 1.0 mM, pH 5.0 in 8 M urea with 5% D2O. An 1H-15N-HSQC spectrum was collected at 35 °C in a Varian INOVA 600 MHz spectrometer with a 5-mm triple resonance probe equipped with triple-axis (XYZ) pulsed magnetic field gradients. All pulse sequences were taken from the Varian BioPack user library. The spectrum was processed and analyzed using the programs nmrPipe (27) and SPARKY3 (T.D. Goddard and D.G. Kneller, University of California-San Francisco).
Results
SPR demonstrates complex FliK-FlhBc binding
Binding studies were undertaken to determine how FlhBc bound FliK and whether the previously described variants FlhBc(N269A), which causes a polyhook phenotype that results from a complete failure to switch specificity, and FlhBc(270A), which has a polyhook-filament phenotype that results from very late switching, bound FliK differently than wild-type FlhB. An SDS-PAGE gel of proteins used for SPR experiments is shown in Fig. 1. Mobilities of proteins in the gel were identical to those previously reported (7, 28). All FlhBcs had C-terminal His-tags. Wild-type FlhBc was almost completely cleaved into amino-terminal FlhBcn (residues 211-269, residue numbers relative to full-length FlhB) and carboxy-terminal FlhBcc (residues 270-383) fragments. For FlhBc(N269A) and FlhBc(270A), precursor was present along with a band previously identified as FlhBcc* pursuant to speculation that it was cleaved at an alternate site (see supplemental materials). Variants also behaved as expected relative to cleavage with FlhBc(N269A) completely uncleaved, and FlhBc(270A) mostly uncleaved but with visible FlhBcn and FlhBcc fragments.
Figure 1.
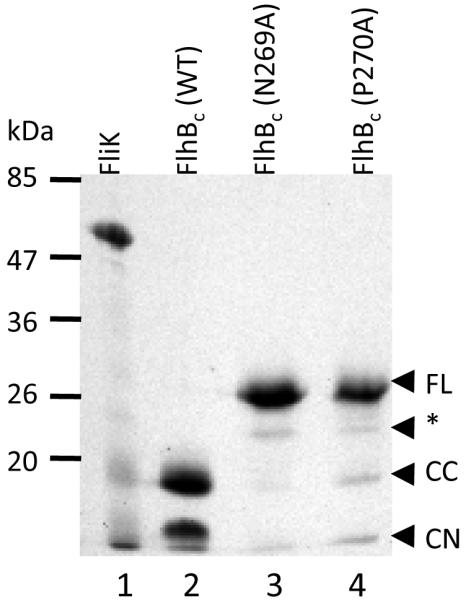
SDS-PAGE gel of purified His-tagged FliK and FlhBc proteins. Lane 1, FliK; 2, FlhBc wild type; 3, FlhBc N269A; 4, FlhBc P270A. Approximate position of molecular weight standards (kDa) are shown at left. Designation of cleavage state for FlhBcs at right are made as identified by Fraser et al (7): FL, full-length, uncleaved cytoplasmic domain; CC, FlhBcc, carboxyl terminal portion of post-cleavage FlhBc; CN, amino terminal portion of post-cleavage FlhBc; *, product of putative alternative cleavage at D237/P238 (see discussion).
FliK was tethered by amine crosslinking and allowed to bind to analyte FlhBc. Reference-subtracted sensor data from experiments with wild-type FlhBc and variants FlhBc(N269A) and FlhBc(P270A) are shown in Fig. 2. Analyte concentrations ranged from 62.5 nM-28 μM. Sensorgrams exhibited complex binding with fast association and dissociation rates beyond the sensitivity of the instrument to determine constants.
Figure 2.
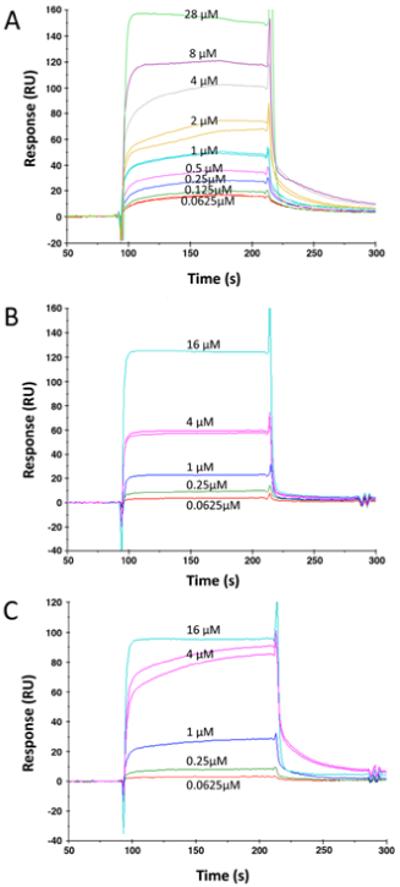
Sensorgrams for FlhBc (A), FlhBc N269A (B), and FlhBc P270A (C) binding to immobilized FliK. Concentrations each FlhBc analyte injection are noted by each association. The 4 μM injection was repeated after all other injections to assure that regeneration of the surface did not affect reproducibility of binding.
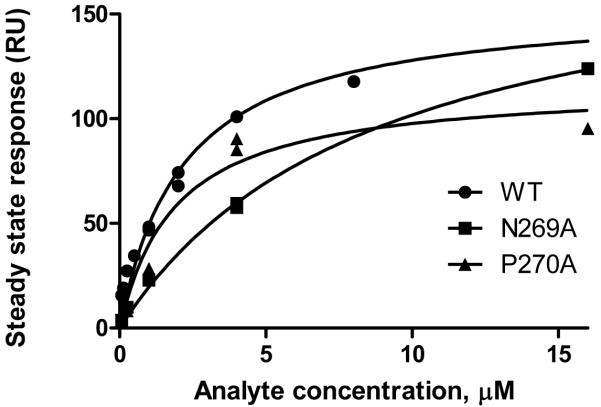
KDapp was thus measured by steady state fit to a saturation binding isotherm of response vs. analyte concentration (Fig. 3). Overall estimated affinities were similar: 3.2 μM for wild-type, 10.5 and 1.6 μM for FlhBc (N269A) and FlhBc (P270A), respectively. Confidence in the KDapp estimated for N269A is lower than the other two since the five concentrations examined did not allow for highly confident determination of Rmax.
Figure 3.
Steady state analysis to determine KDapp for FlhBc binding to FliK. Response at steady state is plotted versus analyte concentration for wild-type and both variants. A 28 μM point for wild-type FlhBc is not shown but was included in fitting and KDapp determination.
Other experiments examined time-of-association as a variable (Fig. 4). For all three variants, longer association times resulted in shorter amplitudes for the fast-off dissociation. Association times of 1, 4 and 9 min had increasingly prevalent slow off states for all FlhBcs. One interpretation of the phenomenon is a conformational change after binding (see discussion).
Figure 4.
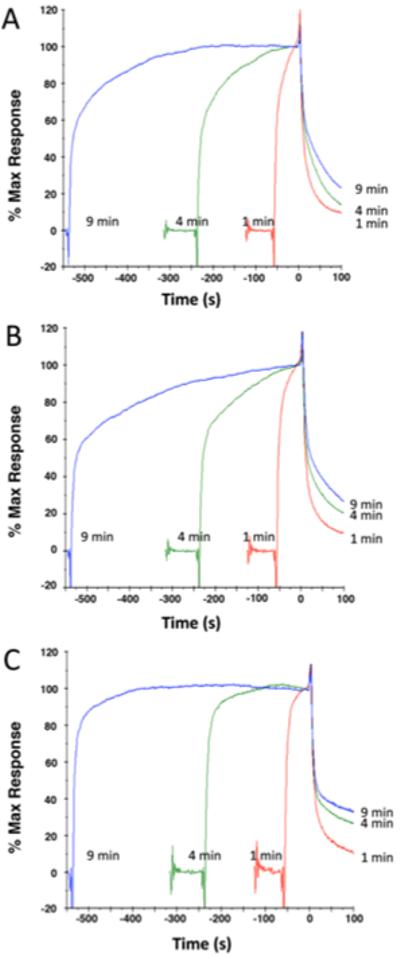
Time of association test for (A) FlhBc; (B) FlhBc(N269A); and (C) FlhBc(270A). Association times of 1, 4 and 9 min were followed by dissociation in buffer only. Data are normalized with respect to start of dissociation phase, which was defined as t=0 and Response=100.
BLI confirms complexity of the FliK-FlhBc interaction
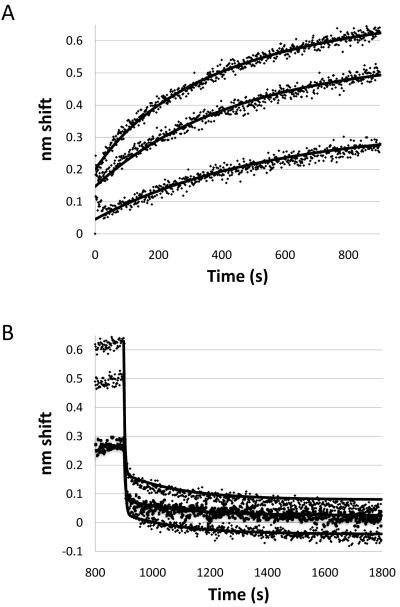
A second optical biosensing method, BLI, was used to model the interaction with wild-type FlhBc. In this case, biotinylated FliK was tethered to streptavidin sensors and analyte concentrations were higher due to the lower sensitivity of the interferometer. Complex interactions similar to those from the SPR experiments were observed and did not fit a simple 1:1 model. Association and dissociation experiments are shown in Fig. 5 along with simulation fits, for which the parameters are included in Table 1. At least two association components were observed; a third component too fast for the instrument to measure may account for the offsets. Two dissociation states were observed, for which simulation determined koffS of 0.263 s−1 and 5.0×10−3 s−1. Association rate constants could not be calculated because of the complexity of the interaction. Indeed, the differences between association and dissociation states are additional evidence for a conformational change after binding and differences among the concentrations could reflect weak dimerization of FlhBc (see supplemental materials). The instantaneous offset may reflect the very fast association observed in SPR, but could also result from discontinuity in measurement between baseline and association phases, a function of the instrument, or a combination of the two. Likewise, discontinuities between association and dissociation phases could have contributed error in determining the Yo used in simulation, but would produce no more than 10% error in the off rates.
Figure 5.
Biolayer interferometry analysis of FliK binding to FlhBc. Raw data with simulation fits are shown for 33, 17 and 9 μM FlhBc. (A) Association phase; (B) Dissociation phase, which began at 900 s; the last 100 s of the association phase are shown in both panels.
Table 1.
| A. BLI association phase simulation parameters | |||
|---|---|---|---|
| [FlhBc], μM | 33 | 17 | 9 |
| Yo (nm shift) | 0.200 | 0.147 | 0.045 |
| Amplitude, fast | 0.11 | 0.11 | 0.25 |
| Amplitude, slow | 0.39 | 0.30 | 0.07 |
| kobs, fast (s−1) | 6.7×10−3 | 3.3×10−3 | 2.0×10−3 |
| kobs, slow (s−1) | 1.8×10−3 | 9.1×10−4 | 4.5×10−4 |
| B. BLI dissociation phase simulation parameters | |||
|---|---|---|---|
| [FlhBc], μM | 33 | 17 | 9 |
| Yo (nm shift) | 0.630 | 0.518 | 0.295 |
| Amplitude, fast | 0.6 | 0.350 | 0.215 |
| Amplitude, slow | 0.070 | 0.090 | 0.055 |
| koff, fast (s−1) | 0.263 | 0.263 | 0.263 |
| koff, slow (s−1) | 5×10−3 | 5×10−3 | 5×10−3 |
Observation of succinimide intermediate demonstrates autocatalytic mechanism
The proposed mechanism of asparagine-mediated autocatalytic cleavage involves cyclization of the side chain through attack on the backbone carbonyl, forming a succinimide intermediate (20,21). This intermediate has a smaller mass (−18 Da) than the final product as the intermediate resolves to the final form containing asn or iso-asn (ref (20), Fig. 1) by addition of a water molecule through hydrolysis of the succinimide bond. Previous efforts to observe the succinimide were unsuccessful, perhaps due to its instability (19); however, an 18 Da difference can be resolved by mass spectrometry. To increase sensitivity in detecting this potentially rare intermediate, we performed LC-MS on FlhBc (P270A) and a shorter, N-terminally his-tagged version consisting of residues 257-383 (“Pep270A”) with or without base treatment to activate autocatalytic cleavage. Initial analysis of the uncleaved FlhBc (P270A) indicated a smaller than expected mass of 21,822.89 Da (Supplemental Figs. 1A and B), a discrepancy that was explained when resequencing of the FlhBc expression plasmid revealed a valine to alanine substitution 20 residues beyond the cleavage site. In agreement with the observed mass, the deduced isotopically-averaged molecular weight from the corrected sequence including vector-derived initiating methionine and C-terminal His-tag was 21,823.9 Da (Table 2). Similarly, auto-catalytic base cleavage produced an amino-terminal fragment containing the substitution with an observed mass of 14,711.3 Da (Supplemental Figs. 1A and C) and a carboxy-terminal fragment of 7,127.6 Da (Supplemental Fig. 1D) both in agreement with the expected masses (Table 2). In addition to the final carboxy-terminal product, pretreatment of P270A with NaOH immediately prior to injection also produced a lighter species (~18 Da) consistent with the expected mass of the succinimide intermediate as shown in the spectrum deconvoluted using MaxEnt calculations (Fig. 6A). Perhaps not surprisingly, given background and low signal, the MaxEnt calculations necessary to estimate the mass of the minor intermediate could not establish whether the mass difference was 17 or 18 Da. The second major species at 7143.50 Da is most likely FlhBcn with an oxidized methionine.
Table 2.
Anticipated and observed MW for FlhBc(P270A) species
| Species | P270A Anticipated MW (Da) |
P270A Observed MW (Da) |
|---|---|---|
| FlhBc,full length | 21.823.92 | 21,822.2 |
|
FlhBcc (residues 270-383,
plus vector-encoded His- tag) |
14,712.67 | 14,711.3 |
|
FlhBcn (residues 211-269,
plus vector-encoded Met) |
7,129.27 | 7,127.5 |
|
FlhBcn, intermediate
product |
7,111.25 | 7,109.5a |
From deconvoluted spectrum of base-treated sample.
Figure 6.
Immediate HPLC/MS analysis of the amino-terminal products produced by base treatment of FlhBc(P270A) and PEP270A. (A) Low resolution mass entropy deconvolution of ion chromatograms containing amino terminal products from base treated P270A eluted from a C-18 column (7,109.5 Da, succinimide species; 7,127.5 Da, expected product: 7,143.5 Da, oxidized product species). (B) High resolution mass entropy deconvolution of ion chromatograms containing amino-terminal products following base treatment of PEP270A. The mass difference between founding species of each product profile is indicated. For each fusion protein, treatment with NaOH produced approximately 50% cleavage (see supplemental Fig. 1 and 2).
Additional LC-MS data were collected on Pep270A subjected to base treatment in order to increase the abundance of the cyclic intermediate and provide additional evidence for its existence. To improve mass accuracy of the product and intermediate species, Pep270A was treated with NaOH (Supplemental Fig. 2A), injected onto the LC column and eluted rapidly for analysis by mass spectrometry. While the uncleaved protein was not observed (possibly due to irreversible binding to the C18 phase), both a carboxy-terminal peptide with a mass of 12,603.2 Da consistent with the expected mass including the alanine substitution (Supplemental Fig. 2B) and a carboxy-terminal product with a mass of 4355.8 Da (Supplemental Fig. 2C) were observed. As shown in a higher-resolution MaxEnt deconvolution (0.10 Da), the final product eluted near both a species ~18 Da lower in mass (Fig. 6B), consistent with the succinimide intermediate and the putative species with an oxidized methionine (+16 Da). Additionally, comparison of the elution profiles of intermediate and product show the suspected succinimide intermediate elutes slightly later than the product species (Supplemental Fig. 2D), which is consistent with increased hydrophobicity from the succinimide group. In contrast, untreated samples, which contain some cleaved peptide, show little signal trailing the product (Supplemental Fig. 2E). Unexpectedly, the mass observed for the carboxy-terminal product was 45.2 Da above the expected mass of 4310.6 deduced from the DNA sequence of fusion protein. Although we failed to identify the reason for this discrepancy, LC-MS analysis of trypsinized fusion protein identified the non-tag FlhB sequence with 75% sequence coverage, including the region around the cleavage site. These data and the ability of the peptide to autocatalyze suggest the mass discrepancy originates from the affinity tag region and is not important to our hypothesis.
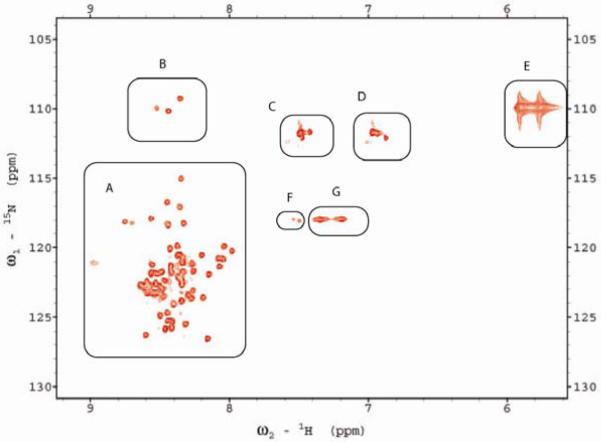
Although cleavage of the wild type protein during expression prevented MS analysis, evidence for the presence of an iso-asn product would indicate the same mechanism occurs in the wild-type. An 1H-15N-HSQC spectrum for wild-type FlhBc prepared under denaturing conditions to remove the carboxyl-terminal FlhBcc fragment so that only FlhBcn was present contains peaks that are consistent with the presence of iso-asn, one of the two potential products of succinimide hydrolysis (Fig. 7). Peaks due to urea, backbone amides and side chain nitrogens are all present as expected from a urea-denatured polypeptide with normal termini. Additional peaks, indicated in area F, may represent iso-asn. Efforts to express and purify an analogous FlhBcn-only construct were unsuccessful, perhaps due to instability of the N-terminal domain when expressed by itself, resulting in an inability to examine if the anomalous peaks were present in a parallel polypeptide that had not undergone cyclization. Future NMR endeavors will address these observations more fully.
Figure 7.
2D 1H-15N-HSQC spectrum of denatured wild-type FlhBcn. Peaks in regions of interest are indicated as A, backbone amides; B, glycine amides, C and D, Gln and asn side chains, E, urea, F, additional peaks that may represent iso-asn nitrogen-proton coupling systems, G, arg sidechains.
Discussion
The interaction between FliK and FlhBc is critical to specificity switching. Since two single residue substitutions in FlhBc result in switching defects with severe phenotypes, it was expected that the interaction between variant FlhBcs and FliK would be dramatically altered. Instead, highly similar binding was observed for all three FlhBcs. Binding is complex and does not fit a one-state model. Fast association and dissociation were beyond the ability of the SPR sensor to measure, meaning that ka(fast) was at least 107 M−1s−1 and kd(fast) at least 0.1 s−1. Given potential differences in active populations and experimental error in quantitation, it is likely that overall affinity does not vary significantly among the variants. The higher KDapp measured for N269A may be relevant as its polyhook phenotype is the more severe defect. This possibility is underscored by the dissociation phase of N269A (Fig. 2B), for which the fast component appears to be of greater amplitude than wild-type or P270A. However, for a relatively small difference to result in the phenotype, the underlying mechanism would have to be very sensitive to the individual molecular parameters. The greater kinetic similarity to wild-type of P270A, which also has a severe defect, suggests that any differences in binding between the variants and FliK are insufficient to explain the phenotypes.
Association studies showing a time-dependent stabilization of FliK-FlhBc binding (Fig. 4) suggest that rapid interaction is followed by slow conformational change leading to a more stable complex. This change may play a role in substrate specificity switching for the bound interaction as has been postulated (ref). However, the fact that FliK is exported after switching (28) argues that the FliK-FlhB interaction would be disrupted by morphogenic events, not stabilized.
SPR results are supported by BLI simulations for wild-type FlhBc binding to biotinylated FliK. The fast koff, 0.263 s−1, is indeed faster than the SPR sensor can measure. Other aspects of the simulation provide additional evidence for conformational change after initial binding. Assuming two parallel simple binding events, one of higher and one of lower affinity, KD would be calculated using kon(fast) and koff(slow) for the high affinity event. We would expect kobs to be the sum of kon and koff from the solution to the first order kinetics differential equation. However, there is no assignment of observed rates to binding components that consistently produces kobs>koff (Table 1). Thus, some other events, presumably conformational, occur.
The kinetic similarities among FlhBc variants observed in this study are informed by crystal structures of homologs in which the NPTH motif forms a β-turn (21, 29). Comparison of cleaved wild-type and uncleaved variants indicates little tertiary structure change. The overall structural similarities may explain the kinetic similiarities we observed, strengthening the hypothesis that other proteins must be directly involved in the specificity switching mechanism.
Observation of the succinimide intermediate in the cleavage of FlhBc(P270A) is direct evidence that the autocatalytic mechanism indeed occurs. Zarivach, et al. showed that crystals of EscU P263A, analogous to FlhBc(P270A), contain electron density commensurate with a stable intermediate in one of two molecules of the asymmetric unit (21). No intermediate was observed in LC-MS spectra of our untreated P270A. The succinimide observed on base treatment is likely more short-lived than it would be under neutral conditions, but sequence-specific effects may alter the stability of the succinimide, explaining why it was not previously observed (19).
FlhB and FliK have been qualitatively reported to bind each other as well a number of flagellar proteins by affinity blotting, but the biological relevance and ramifications of those interactions are largely unknown (30, 31). Herein we reported the first quantitative analysis of FliK-FlhBc binding. The interaction is complex and of micromolar affinity. How the complexity affects specificity switching remains to be determined but the similarities among variants suggest that other proteins are intimately involved. The recently postulated role of Flk and FlhA in perturbing the FliK-FlhB interaction is one strong possibility (32). It is also possible that interactions yet unidentified because of low affinity, high off-rates or other characteristics may be instrumental in the mechanism of substrate specificity switching and the function of the T3S apparatus itself.
Supplementary Material
Acknowledgement
We thank John Salerno for helpful discussions and assistance with the BLI simulations and Carol Chrestensen for critical reading of the manuscript.
Abbreviations used
- BLI
biolayer interferometry
- EDC
1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IPTG
isopropyl β-D-thiogalactopyranoside
- LC-MS
liquid chromatography-mass spectrometry
- NHS
N-hydroxysuccinimide
- NHS-LC-LC-biotin
succinimidyl-6-[biotinamido]-6-hexanamidohexanoate
- NMR
nuclear magnetic resonance
- SPR
surface plasmon resonance
- T3S
type III secretion
Footnotes
This work was supported by PHS Grant GM080701 from the National Institutes of Health and a Cottrell College Science Award (CC6900) from the Research Corporation (both to J.L.M.).
Supporting Information Available
Figures containing raw and deconvoluted spectra of full-length and fragmentary FlhBc(P270A) and Pep270A, SDS-PAGE analysis of base induced cleavage of PEP270A, ion chromatograms demonstrating that the succinimide species elutes later, consistent with slightly higher hydrophobicity and brief discussions of minor points. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kojima S, Blair DF. The bacterial flagellar motor: Structure and function of a complex molecular machine. Int. Rev. Cytology. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 2.Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol. Biosys. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- 3.Macnab RM. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature (London) 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 5.Minamino T, Namba K. Distinct roles of the flagellar ATPase and proton motive force in bacterial flagellar protein export. Nature (London) 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 6.Chevance FFV, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nature Rev. Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 8.Minamino T, Pugsley AP. Measure for measure in the control of type III secretion hook and needle length. Mol. Microbiol. 2005;56:303–308. doi: 10.1111/j.1365-2958.2005.04611.x. [DOI] [PubMed] [Google Scholar]
- 9.Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J. Mol. Biol. 2006;359:466–477. doi: 10.1016/j.jmb.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Makishima S, Komoriya K, Yamaguchi S, Aizawa S-I. Length of the flagellar hook and the capacity of the type III export apparatus. Science. 2001;291:2411–2413. doi: 10.1126/science.1058366. [DOI] [PubMed] [Google Scholar]
- 11.Hirano T, Shibata S, Ohnishi K, Tani T, Aizawa S-I. N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol. Microbiol. 2005;56:346–360. doi: 10.1111/j.1365-2958.2005.04615.x. [DOI] [PubMed] [Google Scholar]
- 12.Konishi M, Kanbe M, Nosaka Y, McMurry JL, Aizawa S-I. Flagellar formation in C-ring defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J. Bacteriol. 2009 doi: 10.1128/JB.00601-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erhardt M, Hirano T, Su Y, Paul K, Wee DH, Mizuno S, Aizawa S-I, Hughes KT. The role of the FliK molecular rules in hook-length control in Salmonella enterica. Mol. Microbiol. 2010;75:1272–1284. doi: 10.1111/j.1365-2958.2010.07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 15.Ferris HU, Minamino T. Flipping the switch: bringing order to flagellar assembly. Trends in Microbiol. 2006;14:519–525. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR. Bacterial injectisomes: Needle length does matter. Science. 2005;307:1278. doi: 10.1126/science.1107679. only. [DOI] [PubMed] [Google Scholar]
- 17.Williams AW, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, Macnab RM. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minamino T, Ferris HU, Moriya N, Kihara M, Namba K. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J. Mol. Biol. 2006;362:1148–1158. doi: 10.1016/j.jmb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J. Biol. Chem. 2005;280 doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 20.Voorter CEM, de-Haard-Hoekman WA, van den Oetelaar PJM, Bloemendal H, de Jong WW. Spontaneous bond cleavage in aging α–crystallin through a succinimide intermediate. J. Biol. Chem. 1988;263:19020–19023. [PubMed] [Google Scholar]
- 21.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NCJ. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature (London) 2008;453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- 22.Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 23.Concepcion J, Witte K, Wartchow C, Choo S, Yao D, Persson H, Wei J, Li P, Heidecker B, Ma W, Varma R, Zhao LS, Perillat D, Carricato G, Recknor M, Du K, Ho H, Ellis T, Gamez J, Howes M, Phi-Wilson J, Lockard S, Zuk R, Tan H. Label-free detection of biomolecular interactions using biolayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screen. 2009;12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 24.Robinson AB, McKerrow JH, Cary P. Controlled deamidation of peptides and proteins: an experimental hazard and a possible biological timer. Proceedings of the National Academy of Sciences of the United States of America. 1970;66:753–757. doi: 10.1073/pnas.66.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 26.Stephenson RC, Clarke S. Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 1989;264:6164–6170. [PubMed] [Google Scholar]
- 27.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biolmol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Minamino T, González-Pedrajo B, Yamaguchi K, Aizawa S-I, Macnab RM. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 29.Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, Lea SM. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol. Microbiol. 2008;69:267–276. doi: 10.1111/j.1365-2958.2008.06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate-specificity switching. J. Bacteriol. 2000;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minamino T, Saijo-Hamano Y, Furukawa Y, González-Pedrajo B, Macnab RM, Namba K. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J. Mol. Biol. 2004 doi: 10.1016/j.jmb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Hirano T, Mizuno S, Aizawa S-I, Hughes KT. Mutations in flk, flgG, flhA and flhE that affect the flagellar type III secretion specificity switch in Salmonella enterica. J. Bacteriol. 2009;191:3938–3949. doi: 10.1128/JB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.