Abstract
Multidrug membrane transporters (efflux pumps) in both prokaryotes and eukaryotes are responsible for impossible treatments of a wide variety of diseases, including infections and cancer, underscoring the importance of better understanding their structures and functions for design of effective therapies. In this study, we designed and synthesized two silver nanoparticle (Ag NP) solution with average diameters of 13.1 ± 2.5 nm (8.1–38.6 nm) and 91.0 ± 9.3 nm (56–120 nm), and used the size-dependent plasmonic spectra of single NPs to probe the size-dependent transport kinetics of MexAB-OprM (multidrug transporter) in Pseudomonas (P.) aeruginosa in real-time at nanometer resolution. We found that the accumulation of intracellular NPs in wild-type (WT) cells was higher than in nalB1 (over-expression of MexAB-OprM), but less than ΔABM (deletion of MexAB-OprM). In the presence of proton ionophores (CCCP, inhibitor of proton-motive-force), we found that intracellular NPs in nalB1 were nearly doubled. These results suggest that the MexAB-OprM is responsible for the extrusion of NPs out of cells and NPs (orders of magnitude larger than conventional antibiotics) are the substrates of the transporter, which indicates that the substrates may trigger the assembly of the efflux pump optimized for the extrusion of the encountered substrates. We found that the smaller NPs stayed longer inside the cells than larger NPs, suggesting the size-dependent efflux kinetics of the cells. This study shows that multi-sized NPs can be used to mimic various sizes of antibiotics for probing the size-dependent efflux kinetics of multidrug membrane transporters in single living cells.
Keywords: Bacteria, efflux pump, single living cell imaging, P. aeruginosa, multidrug resistance (MDR), multi-antibiotic resistance, membrane transporter, single nanoparticle plasmonic optics
Multi-substrate extrusion systems (membrane transporters or efflux pumps) have been found in both prokaryotes and eukaryotes (1, 2). Notably, some of these extrusion transport apparatus allow cells to achieve cellular self-defense mechanisms and resistance to noxious compounds, leading to multi-drug resistance (MDR) (2–6). For instance, the efflux pumps of Pseudomonas (P.) aeruginosa can selectively extrude a variety of structurally and functionally diverse substrates (e.g., chemotoxics, dyes, antibiotics), causing MDR (4, 7–10). P. aeruginosa is a ubiquitous gram-negative bacterium, and has emerged as a major opportunistic human pathogen and the leading cause of nosocomial infections in cancer, transplant, burn, and cystic fibrosis patients (8, 11–14). These infections are difficult to treat, due in part to its intrinsic resistance to a wide spectrum of structurally and functionally unrelated antibiotics (7, 15). MDR is one of leading causes of ineffective therapies and the primary reason for using high doses of therapeutic agents to treat a variety of diseases (e.g., infections, cancer), leading to severe side effects.
P. aeruginosa possesses several multidrug membrane transporters (efflux pumps) (6, 10, 15–18). The MexAB-OprM is the primary membrane transporter in wild-type (WT) of P. aeruginosa and consists of two inner membrane proteins (MexA and MexB) and one outer membrane protein (OprM) (19–21). This efflux pump can extrude a wide spectrum of structurally and functionally unrelated antibiotics and substances using the drive-force generated by proton gradients across the cellular membrane (22–25). For instance, the MexAB-OprM of P. aeruginosa can extrude dye molecules (e.g., EtBr) and antibiotics such as azthreonam, rifampicin, chloramphenicol, and gentamicin (26). The sizes and structures of these pump substrates vary tremendously. The interplay between the MexAB-OprM efflux system and the outer membrane barrier plays an important role in MDR (16, 27, 28). Despite extensive studies, molecular mechanisms of multi-substrate or multi-drug efflux pump remain not yet fully understood (4, 8, 9). It is very likely that membrane proteins are triggered by pump substrates to assemble membrane transporters optimized for the extrusion of encountered substrates. Therefore, real-time measurements of the size change of efflux pumps at the molecular level are crucial to better understand such universal cellular extrusion defense mechanisms.
Currently, the primary methods for the study of transport kinetics of efflux pumps in bacteria include using radioactively labeled (14C and 3H) (29), and fluorescent quinolones (e.g., EtBr) as probes to study the accumulation rates of substrates in cells (30–33). Even though fluorescence microscopy and spectroscopy has been used to probe the efflux kinetics of single membrane transporters of single living cells in real-time (30, 32, 33), most reported studies probe the ensemble accumulation kinetics of bulk cells (9, 31, 34). The ensemble measurement does not represent the accumulation kinetics of single membrane transporters of single cells, because individual transporters and cells have unsynchronized membrane transport kinetics (30, 32). Notably, these current methods, with the use of either radioactive or fluorescence probes, cannot provide insights into the change of membrane permeability and pore sizes of membrane transporters of single living cells in real time.
The primary method to measure the sizes of membrane transporters at atomic resolution is X-ray crystallography and cryoTEM (19–21, 35, 36). Crystallization of membrane proteins is difficult, limiting the application of crystallography. Furthermore, x-ray crystallography and cryoTEM cannot provide real-time transport dynamics of substrates and self-assembly of pump proteins in living cells. Therefore, even with the recent success of solving the structures of membrane transporters at atomic resolution (19–21), the molecular mechanisms and functions of multidrug efflux pumps remain elusive (8, 9).
Noble metal (e.g., Ag) nanoparticles (NPs) possess exceptionally high quantum yield of Rayleigh scattering that are orders of magnitude higher than fluorophors (e.g., R6G) (37, 38),allowing them to be directly imaged and characterized using dark-field optical microscopy and spectroscopy (DFOMS) (39–43). Unlike fluorescent probes and semiconductor quantum dots (QDs), these noble metal NPs show superior photostability (non-photobleaching and non-blinking), enabling them to serve as photostable nanophotonic optical probes for sensing and imaging molecules and dynamics events of interest in single living cells and in embryos for desired period of time (40–42, 44–48). More importantly, the size-dependent plasmonic spectra (color), localized surface plasmon resonance (LSPR) spectra, of single NPs allow us to image the sizes of NPs in solution, single living cells and embryos in real time (40–42, 44–49). This approach enables us to use the color (LSPR spectra) index of the multicolor NPs as a nanometer-size index to directly measure sizes of single NPs as they are transported in and out of single living cells, and to determine the sizes of membrane pores at the nanometer scale in real-time (45–47).
In our previous studies, we have demonstrated the feasibility of using intrinsic optical properties of the noble metal NPs (e.g., Au, Ag) for probing sizes and transport dynamics of single membrane transporters, and the transformation of pore sizes of cellular membranes of single living cells induced by antibiotics (e.g., aztreonam, chloramphenicol) at sub-100 nm spatial resolution and millisecond temporal resolution (45–47). In this study, we synthesized two groups of Ag NPs, ranging from 8.1 to 38.6 nm, and 56 to 120 nm, characterized size-dependent LSPR spectra of individual NPs, and imaged the sizes of single NPs in and out of single living cells at nm resolution in real-time. We studied the dependence of accumulation and efflux kinetics of single NPs in single living cells (P. aeruginosa) on the expression level of MexAB-OprM, the presence of proton ionophores (inhibitor of proton-motive-force), and sizes and concentrations of NPs (substrates).
Materials and Methods
Reagents
Sodium citrate (99%), AgClO4 (99%), NaBH4 (98%), NaCl, NaH2PO4, Na2HPO4, ethidium bromide (EtBr) (≥ 99%), bacto-tryptone, bacto-yeast extract, and CCCP (carbonyl cyanide-m-chlorophenylhydrazone, ≥ 97%) were purchased from Sigma-aldrich, and live/dead bacLight viability and counting assay was purchased from invitrogen. All reagents were used as received. The nanopure deionized (DI) water (18 MΩ water, Barnstead) was used to prepare all solutions and rinse glassware.
Synthesis and Characterization of Ag NPs
We synthesized Ag NPs (13.1 ± 2.5 nm), as we described previously (39, 41, 42). Briefly, AgClO4 (2.5 mL, 10 mM, ice-cold) was rapidly added into the stirring mixture (247.5 mL) of ice-cold sodium citrate (3 mM) and NaBH4 (10 mM), and left stirring at room temperature for 4 h. The solution was then filtered using 0.22 µm filters (Whatman) and washed twice with nanopure DI water using ultra-centrifugation at 15,000 rcf (relative centrifugal force), to prepare stable and purified Ag NPs. We synthesized 91.0 ± 9.3 nm Ag NPs by adding sodium citrate (10 mL, 34 mM) into a refluxing (100 °C) stirring aqueous solution of AgNO3 (500 mL, 3.98 mM), and stirring for additional 35 min. We then stopped heating and allowed it to cool to room temperature without stirring. Additional sodium citrate (2.5 mM) was added to further stabilize the NPs in solution. The solution was filtered using 0.22 µm filters, and washed three times using nanopure DI water via centrifugation.
Both washed Ag NPs were resuspended in nanopure DI water and stored at 4°C in the dark until use. The molar concentration of Ag NPs was determined and calculated as described in our previous studies (41, 44, 50). We characterized the concentrations of NPs using UV-Vis spectroscopy (Hitachi U-2010), the LSPR images and spectra of single NPs using dark-field optical microscopy and spectroscopy (DFOMS), and their sizes using high resolution transmission electron microscopy (HRTEM, FEI Tecnai G2 F30 S-Twin) and dynamic light scattering (DLS, Nicomp 380ZLS).
We have fully described the design and construction of our DFOMS (also named as SNOMS by us) for imaging of single Ag and Au NPs in solution, in single living cells, and in single embryos in real-time (30, 32, 39–42, 44–47). In this study, we imaged single Ag NPs in solution and single live cells, and measure the LSPR spectra of single Ag NPs using dark-field optical microscope (DFOM) equipped with CCD camera (5 MHz Micromax, Roper Scientific) and hidefinition color camera, and Nuance multi-spectral imaging system (CRI), respectively. The DFOM is equipped with a dark-field condenser (Oil 1.43-1.20, Nikon) and a 100x objective (Nikon Plan fluor 100x oil, iris, SL. N.A. 0.5–1.3, W.D. 0.20 mm) with the depth of field (focus) of 190 nm.
Cell Culture and Preparation
Three strains of gram-negative bacterial cells (Pseudomonas aeruginosa): WT (PA04290, normal expression level of MexAB-OprM), nalB1 (over expression mutant of MexAB-OprM), and ΔABM (deletion of MexAB-OprM), were generously provided by Dr. Hiroshi Yoneyama, and used for this study (31, 51). The cells were cultured using L-broth medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl, pH 7.2) in a rotary shaker (Lab-line Orbit Envivon-Shaker, 120 rpm and 37 °C). Cells were first pre-cultured for 12 h and then cultured using the fresh medium for another 8 h. The cells were harvested by centrifugation (Beckman JA-14, 7500 rpm), and rinsed with the PBS buffer (0.5 mM phosphate buffer, 1.5 mM NaCl at pH 7.0) three times via centrifugation. The cell concentration (OD600 nm = 0.1) was adjusted and used for all experiments (30, 32, 33, 45–47)
Imaging of Single Ag NPs in and out of Single Living Cells and Probing Cellular Viability
The cells (OD600 nm = 0.1) were incubated with Ag NPs in the PBS buffer (0.5 mM phosphate buffer, 1.5 mM NaCl, pH 7.0) and the timer was started to record the incubation time as Ag NPs were added into the cell suspension. The mixture was transferred to a freshly prepared microchamber and imaged using DFOMS, as we described previously (30, 32, 33, 45–47). We continuously imaged the transport of single Ag NPs in and out of single living cells in real-time using DFOMS equipped with color digital camera and CCD camera with temporal resolution of one second. We could achieve sufficient signal to noise ratios with a 5 ms temporal resolution. To track the transport of single NPs in and out of single living cells for hours continuously with 5 ms temporal resolution, one experiment would create each single data file with enormous sizes (> 100 GB) that were hard to handle and analyze. Fortunately, we found that the transport of single NPs in and out of single living cells was not a rapid process and a temporal resolution of 1 s was sufficient to measure their transport kinetics in real time.
We also performed the control experiments in the presence of 100 µM CCCP, which is an inhibitor of proton-motive-force, allowing us to determine whether the amounts of intracellular NPs were directly related to the proton-motive efflux pumps (MexAB-OprM) of live cells.
We also studied the intracellular NPs in the large amount of cells, aiming to determine the intracellular NPs in bulk cells at single cell resolution. Instead of imaging single NPs in and out of the same set of single living cells in the microchamber in real-time for 2 h, we imaged 5 different sets of single living cells each 5 min, and sampled the mixture of the cells and NPs to the freshly prepared micro-chamber every 20 min over 2 h. Using the same approaches, we imaged intracellular and extracellular NPs of single living cells using DFOMS over time (45–47). We determined intracellular NPs and plotted them versus time to measure the accumulation rates of single NPs over time (accumulation rate = slope of the plot). This approach allowed us to image massive amount of cells (3000 cells) for each sample to gain sufficient statistics for probing the accumulation rates of bulk cells at single cell resolution.
By the end of each experiment, we studied the viability of the cells using live/dead BacLight bacterial viability and counting assay, as described in the assay manual. The green fluorescence (peak wavelength of fluorescence spectra, λmax = 520 nm) cells and red fluorescence (λmax = 610 nm) cells were counted as live and dead cells, respectively.
Fluorescence Spectroscopic Measurements
Fluorescence intensity of EtBr from the cell suspension (OD600 nm = 0.1) containing 4 µM EtBr was continuously measured in real-time (3 s time interval) for 2 h using a fluorescence spectrometer (Perkin-Elmer LS50B) with the excitation and emission wavelengths at 488 and 590 nm, respectively (30, 32, 33).
Data Analysis and Statistics
For each measurement of the sizes of single Ag NPs using HRTEM and LSPR spectra (colors) of single Ag NPs, a minimal of 100 Ag NPs were imaged and characterized. The measurement was repeated three times and a minimal of 300 Ag NPs were studied for each sample. For realtime imaging of single NPs in and out of single living cells for 2 h, ~ 25 bacterial cells were acquired in each image and studied for each measurement. Each experiment was repeated at least three times. Thus, the minimal of 75 cells were studied for each sample.
For study of accumulation rates of intracellular NPs in single living cells, a minimal of ~3000 cells were imaged at each 20 min, and 18,000 cells were studied over 2 h for each measurement. Each experiment was repeated three times. Thus, 54,000 cells were studied for each sample to gain sufficient statistics to achieve probing of accumulation rates of bulk cells at single cell resolution. We analyzed the number of intracellular NPs in 800 cells at each 20 min, plotted them over time, and used the slope of the plots to determine the accumulation rates of intracellular NPs in each type of the cells incubated with given sizes and concentrations of NPs. The equilibrium times of accumulation of NPs in the cells were determined at the times when the accumulation rates of intracellular NPs remained unchanged over time. For the study of viability of single cells, a minimal of 300 cells was assayed for each measurement. Each measurement was repeated three times. Thus, 900 cells were assayed for each sample.
Results and Discussion
Synthesis and Characterization of Plasmonic NP Probes
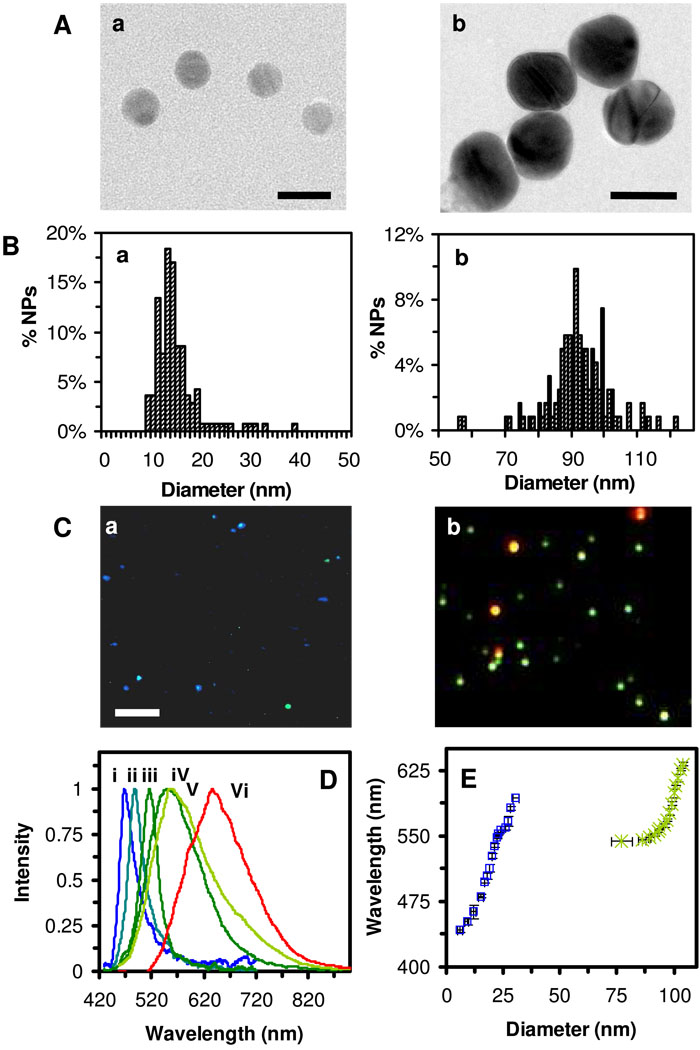
We synthesized two Ag NP solutions, as described in Methods. Representative high-resolution TEM (HRTEM) images of these Ag NPs and their size distributions (histograms) in Figures 1A and B show the spherical NPs with average diameters of 13.1 ± 2.5 nm, ranging from 8.1 to 38.6 nm, and spheroidal NPs (an aspect ratio of 1.4 ± 0.4) with average diameters of 91.0 ± 9.3 nm, ranging from 56 to 120 nm, respectively. The optical imagesshow predominate plasmonic blue NPs, with some being cyan and light green NPs for 13.1 ± 2.5 nm Ag NPs in Figure 1C-a, and primary green NPs with some being yellow and red NPs for 91.0 ± 9.3 nm in Figure 1C-b. Histograms of the color distribution of individual NPs in nanopure DI water (Figure 1S in supporting information) show 73% of blue, 21% of green and 6% of red NPs with the peak wavelengths (λmax) of representative LSPR spectra at 468, 488, and 577 nm (Figure 1D: i-iii) for 13.1 ± 2.5 nm Ag NPs. In contrast, it shows 68% of green, 16% of yellow and 16% of red NPs with λmax of representative LSPR spectra at 552, 560, and 637 nm (Figure 1D: iv-vi) for 91.0 ± 9.3 nm Ag NPs. The results show the red shifts of LSPR spectra as sizes of NPs increase, which agrees with those described in the literature (38, 52–54).
Figure 1. Characterization of the size and plasmonic optical properties of single Ag NPs.
(A) HRTEM images and (B) Histograms of size distributions of Ag NPs show nearly spherical NPs with narrow size distribution at the diameters of (a) 13.1 ± 2.5 nm and (b) 91.0 ± 9.3 nm.
(C) Their dark-field optical images show plasmonic colors with (a) 73% of blue, 21% of green and 6% of red NPs; and (b) 68% of green, 16% of yellow and 16% of red NPs, respectively. See Figure 1S in supporting information.
(D) Representative LSPR spectra of single Ag NPs in (a) and (b) show the peak wavelength (λmax) of (a): (i) 468, (ii) 488 and (iii) 577 nm; and (b): (iv) 552, (v) 560 and (vi) 637 nm, respectively.
(E) Correlation of the λmax of LSPR spectra of single Ag NPs with their sizes shows linear calibration curves for Ag NPs in (a) and (b), allowing us to determine the sizes of NPs in solution and in living cells in real-time using DFOMS. Scale bars in (A-a), (A-b), and (C) = 25 and 90 nm, and 4 µm, respectively. The scale bar in (C) shows the distances among NPs, but not the sizes of NPs, because they are imaged under optical diffraction limit (~200 nm).
The plots of λmax of LSPR spectra of individual NPs measured using DFOMS versus their sizes determined using HRTEM in Figure 1E show nearly linear calibration curves for each Ag NP solution, which allows us to get around the optical diffraction limit of optical microcopy and determine the sizes of single NPs at the nanometer (nm) scale using DFOMS. Nonetheless, we observed a small overlap of wavelength ranges of λmax of LSPR spectra of single NPs in 13.1 ± 2.5 nm and 91.0 ± 9.3 nm Ag NP solutions. Such an overlap may be attributed to the different shapes of Ag NPs and various surface dielectric constants of NPs in two different NP solutions due to the different amounts of synthetic reagents. Fortunately, the slight overlap occurs between the two NP solutions, and the λmax of LSPR spectra of single NPs are nearly proportional to its sizes for each NP solution. Each NP solution was incubated with cells for probing the transport kinetics of the efflux pump of single living cells, separately. Therefore, the small overlap between two NP solutions does not create any problem for us to determine the sizes of single NPs using the λmax of LSPR spectra of single NPs. Notably, these interesting observations emphasize the importance of calibration of λmax of LSPR spectra of single Ag NPs with their sizes for each NP solution, in order to use them as size-dependent plasmonic NP probes and determine their sizes at nanometer scale using their LSPR spectra via DFOMS.
Single living cells (P. aeruginosa) need to be suspended in the PBS buffer (10 mM phosphate, 1.5 mM NaCl, pH =7.0) to sustain cellular viability over time. To study the function of membrane transporters of single living cells using size-dependent plasmonic NPs, it is essential to maintain and characterize the stability (non-aggregation) of individual Ag NPs in the buffer during the duration of experiments. The results (Figure 2S-A in supporting information) illustrate that UV-vis spectra of both Ag NP solutions (suspended in the buffer) remain unchanged over 12 h of incubation. Their average diameters measured using dynamic light scattering (DLS) in Figure 2S-B of supporting information and the histograms of color distribution of single NPs imaged using DFOMS in Figure 2S-C of supporting information remain essentially constant, suggesting that the Ag NPs are stable in the buffer for 12 h. Notably, the sizes of NPs were determined under the vacuum using HRTEM and in solution using DLS. Therefore, the diameters of NPs measured using DLS in Figure 2S-B differ slightly from those measured using HRTEM, which may be attributed to the solvation of NPs and exchange of surface adsorbates (e.g., citrate ions) of NPs with the ions of the buffer solution.
Real-time Probing of Single Membrane Transporter of Single Living Cells
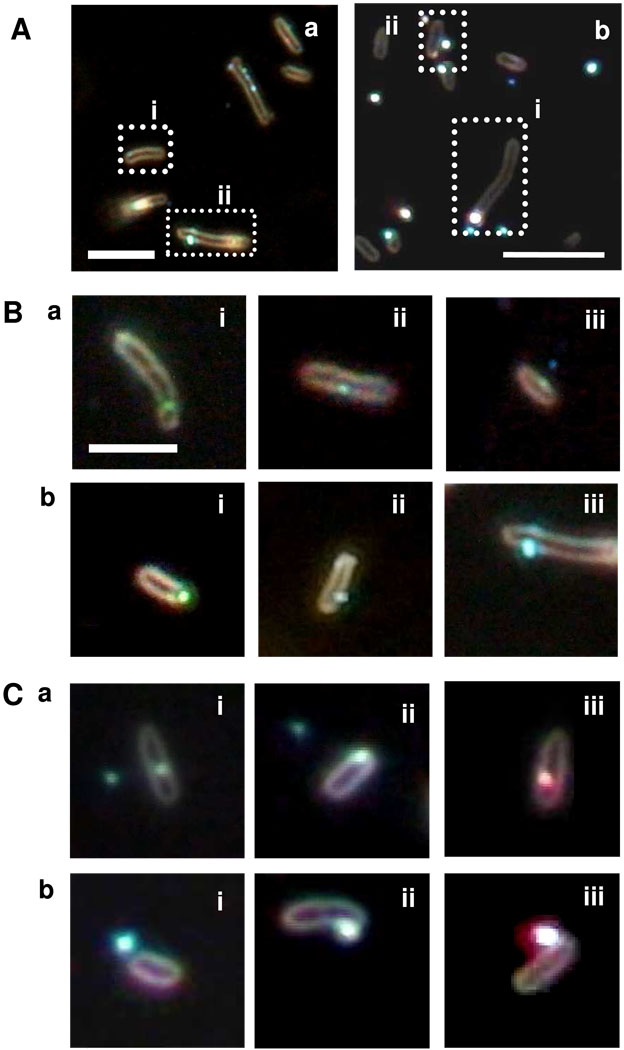
Optical images of single living cells incubated with 770 pM of 13.1 ± 2.5 nm Ag NPs and 3.7 pM of 91.0 ± 9.3 nm Ag NPs in Figure 2A (a) and (b) illustrate intracellular and extracellular Ag NPs, as squared in (i) and (ii), respectively. The depth of field (focus) of our DFOMS imaging system is 0.19 µm, as described in Methods. The cross-section of rod-shape bacterial cells with a length of 2 µm and diameter of 0.5 µm in Figure 2 demonstrates that the focal plane (depth of field) of dark-field microscopy indeed allows us to image thin-layer sections of single bacterial cells. The top and bottom membranes of the cells are invisible under dark-field illumination. Therefore, NPs on the surface of top or bottom membranes of the cells are invisible under dark-field illumination, because they are out of the focal plane of the dark-field microscopy.
Figure 2. Imaging of single intracellular and extracellular Ag NPs in single living cells.
(A) Color images of individual cells (WT) incubated with (a) 770 pM of 13.1 nm Ag NPs and (b) 3.7 pM of 91.0 nm Ag NPs show: (i) intracellular and (ii) extracellular NPs, as squared by dash-lines.
(B-C) (a) intracellular and (b) extracellular NPs with single cells, selected from the solution and images as those in (A-a) and (B-a), respectively. The scale bars in (A-C) are 4 µm.
The illumination of dark-field microscopy needs to penetrate into the cellular membrane in order to irradiate intracellular NPs, and light scattering of intracellular NPs must transmit through the membrane in order to reach the detector. The cellular membrane absorbs photons, leading to lower intensity of intracellular NPs (as they are dimmer). The focal plane of dark-field microscopy is centered on the cross-section of the membrane. Intracellular NPs are situated inside the cellular membrane, leading to out of the focus plane and blurry images. In contrast, the scattering intensity of the extracellular NPs on the membrane is the sum of the scattering intensity of NPs and cellular membrane, leading to radiating imaging and higher intensity.
Therefore, the intracellular NPs are dimmer and more blurry than NPs in solution, as those shown in the zoom-in images of intracellular Ag NPs in (a) of Figure 2B and C. In contrast, the extracellular NPs are radiating and much brighter than NPs in solution, as shown in (b) of Figure 2B and C. These optical imaging approaches for determining intracellular NPs have been verified by imaging intracellular NPs using TEM, as we reported previously (45–47).
We distinguished intracellular and extracellular NPs using the scattering intensity of single NPs. We measured scattering intensity of single NPs with single living cells over time and subtracted the scattering intensity of the cells in the absence of NPs, once the NPs was extruded out of the cells. Notably, the same NPs and cells were tracked over the entire extrusion process. This approach allowed us to measure the scattering intensity of intracellular and extracellular single NPs over time. We found that the intensity of extracelluar NPs is about 1.2–1.9 times higher than intracellular NPs, depending upon the sizes and locations of NPs. Generally, the scattering intensity of a single intracellular NP increased about 8.7-62%, as it was extruded out of the cells.
Notably, the sizes of NPs and the thickness of bacterial membrane (36 nm) are under the optical diffraction limit (~200 nm), and they cannot be resolved spatially using optical microscopy. However, the sizes of single NPs were determined at nm resolution using their LSPR spectra, as illustrated in the calibration curves (Figure 1E). The higher reflectivity of Ag NPs than the cellular membrane makes intracellular NPs appear to stick out of the membrane (Figure 2).
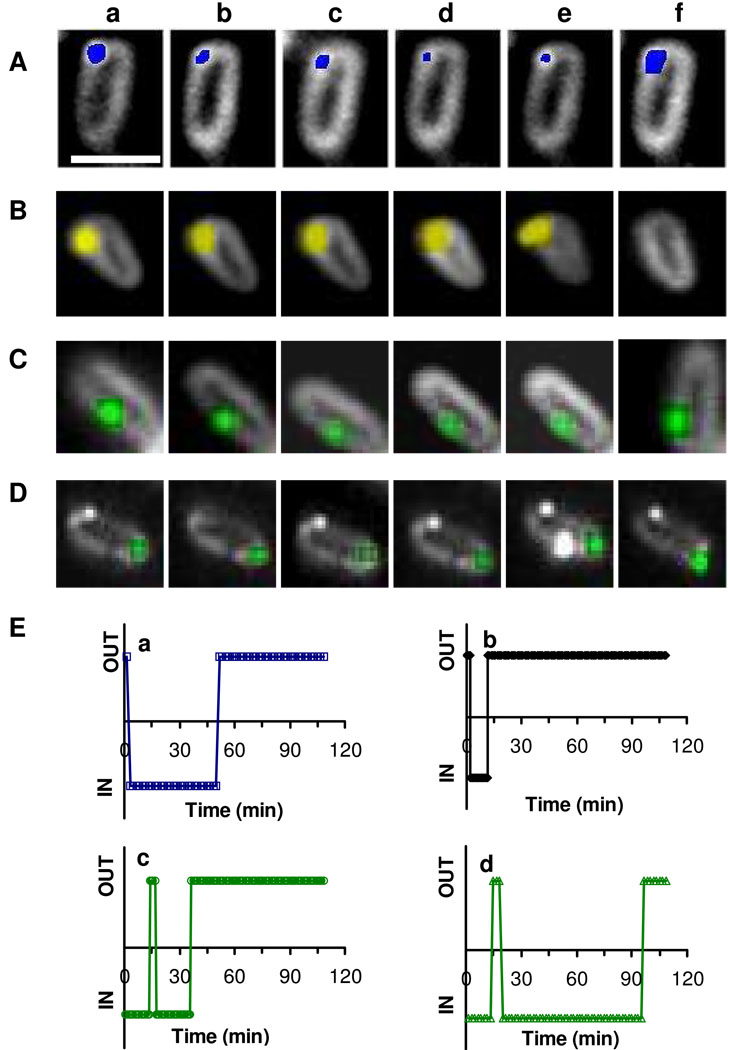
These approaches allow us to track the transport of single NPs in and out of single living cells in real-time and to determine the sizes of individual NPs and the durations of time for them to stay inside and outside of the cells. Real-time videos (Movies 1–4 with 1 s frame interval in supporting information) and their snap shots in Figure 3 show that single blue, yellow and green NPs are in and out of single living cells (nalB1, overexpression of MexAB-OprM) in the absence and presence of a proton ionophore (CCCP).
Figure 3.
Real time Probing of efflux pump (MexAB-OprM membrane transporter) of single living cells (nalB1) using single NP probes. Snap shots of sequential images of (A) a single blue NP (9.7 nm in diameter) in and out of single live cells at (a) 2.55, (b) 5.1, (c) 17.85, (d) 33.15, (e) 52.22, and (f) 54.77 min.
(B) a single yellow NP (88 nm) in and out of single live cells at (a) 2.63, (b) 5.25, (c) 6.3, (d) 11.03, (e) 11.55, and (f) 12.07 min.
(C) a single green NP (17 nm) in and out of single live cells at (a)13.3, (b) 16.3, (c) 20, (d) 26.7, (e) 32.6, and (f) 36.7 min.
(D) a single green NP (17 nm) in the presence of CCCP (100 µM) in and out of single live cells at (a) 17.5, (b) 19.5, (c) 24, (d) 48, (e) 94.5, and (f) 145 min.
(E) Plots of imaging of in and out of single NPs in (A-D) versus time shows that the duration of single NPs stays inside the cells: (a) 47.1, (b) 5.7, (c) 18.3, and (d) 75 min, respectively. The scale bars in (A-D) are 2 µm for all images. Real-time videos of (A-D) with a frame interval of 1 s are shown in movie 1–4 in supporting information, with the observed cells that are highlighted by squares in the videos.
Using the size-dependent plasmonic NPs and the size-dependent scattering intensity, we determined that the sizes of blue, yellow and green NPs in Figure 3 were 9.7, 88, and 17 nm in diameters, respectively. The blue NP (9.7 nm) stayed inside the cells for 47.1 min, while the green NP (17 nm) and yellow NP (88 nm) stayed inside the cells for 18.3 and 5.7 min, respectively. In the presence of CCCP (pump inhibitor), a single green NP (17 nm) stayed inside the cells for 75.0 min, much longer than 18.3 min observed in the absence of CCCP. The CCCP decreases or eliminates proton concentration gradients across the cellular membranes of living cells, disabling the proton-motive transporters (e.g., MexAB-OprM) and leading to the slow efflux rate (31, 55). Therefore, the results suggest that the NPs are indeed extruded by the proton-motive pumps.
Similar sizes of single NPs were studied for at least three times. In the absence of CCCP, we found that single NPs with diameters of 9.5 ± 0.7, 16.1 ± 0.5, and 84.1 ± 3.9 nm stayed inside the cells for 55.7 ± 12.7, 27.0 ± 6.5, and 8.9 ± 3.1 min, respectively. In the presence of CCCP, the NPs with diameters of 16.8 ± 0.7 nm stayed inside the cells for 82.0 ± 6.6 min. Notably, colors (LSPR spectra) of single intracellular and extracellular NPs remain essential the same, indicating that the NPs are stable (not aggregated) inside the cells.
It is worth noting endocytosis, pinocytosis and exocytosis that are widely reported in eukaryotes, do not exist in prokaryotes (bacterial cells). Thus, the transport of NPs in and out of the bacterial cells (P. aeruginosa) that we observed in this study is not attributable to these processes.
Taken together, the results show the size-dependent efflux kinetics of single living cells and the inhibitory effect of CCCP on the efflux kinetics of single NPs, suggesting that the NPs are indeed extruded by the proton-motive pumps (MexAB-OprM), and the multi-sized NPs are suitable substrates for probing the efflux pump (MexAB-OprM). Notably, the pump responds to the substrates up to 88 nm, which are nearly two orders of magnitude larger than conventional antibiotics. It suggests the possibility that the substrates trigger three membrane proteins (MexA, MexB, OprM) to assemble an efflux pump for extrusion of the substrates. The smaller NPs stay inside the cells longer than larger NPs, suggesting that they are more biocompatible with the cells, and they can serve as effective carriers for drug delivery.
Study of Dependence of Membrane Transport of NPs on Express Level of MexAB-OprM
To further investigate whether the membrane transporter (MexAB-OprM) is indeed responsible for the efflux of NPs out of living cells, we studied the dependence of membrane transport kinetics of NPs on the expression level of MexAB-OprM. We selected the WT of P. aeruginosa with normal expression level of MexAB-OprM and their two mutants, nalB-1 (over expression of MexAB-OprM) and ΔABM (deletion of MexAB-OprM). We incubated these three strains of cells (OD 600 nm = 0.1) with two different sizes of Ag NPs, 13.1 ± 2.5 nm (770 pM) and 91.0 ± 9.3 nm (3.7 pM), over time, and studied the accumulation rates of intracellular NPs in the cells.
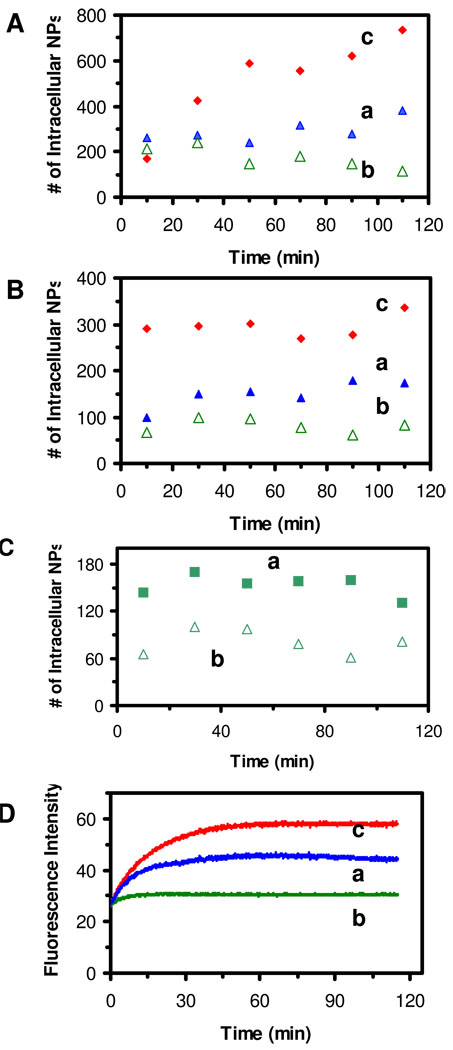
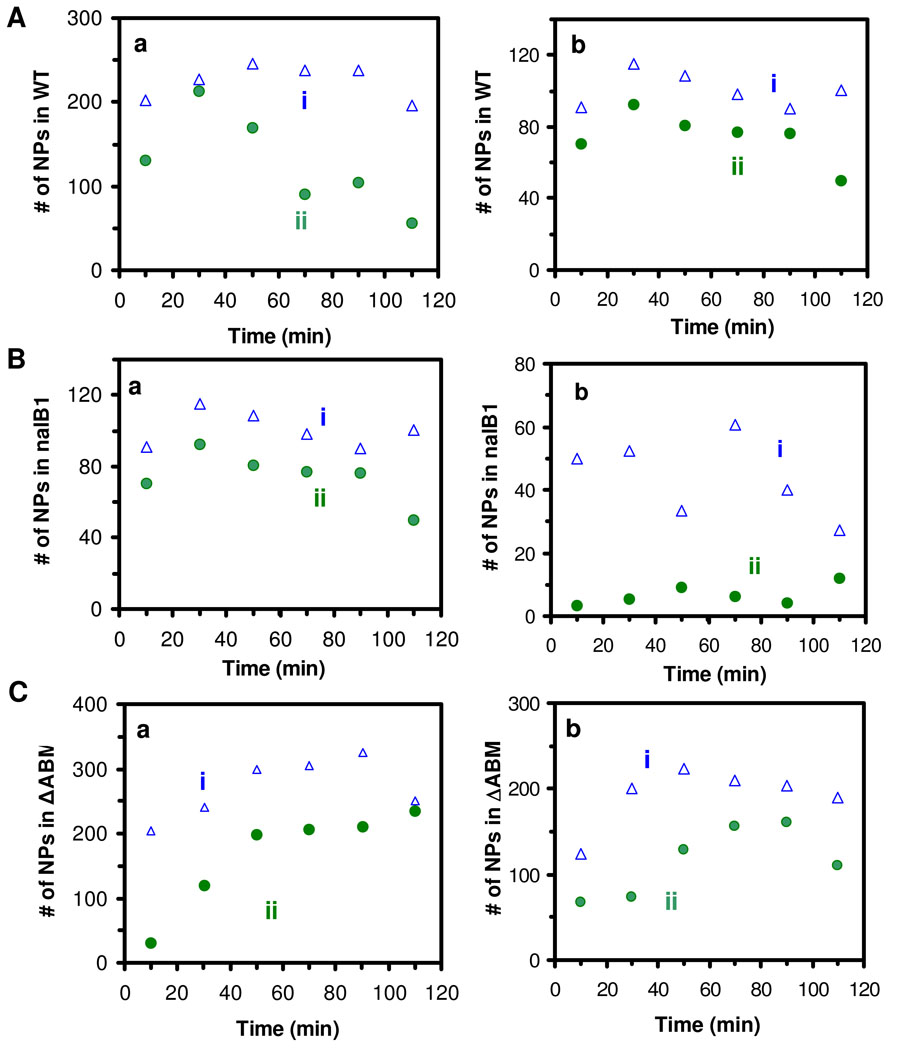
We imaged intracellular NPs over time and plotted the number of intracellular NPs versus time. The results in Figure 4A and B show that WT cells accumulate more intracellular NPs in (a) than nalB1 cells in (b), but less intracellular NPs than ΔABM cells in (c). In other words, ΔABM cells accumulate the highest number of intracellular NPs with the highest accumulation rate, while the nalB1 cells accumulate the lowest number of NPs with the lowest accumulation rate. These results show that the accumulation rates of intracellular NPs highly depend upon the expression level of a membrane transporter (MexAB-OprM), and suggest that the lowest accumulation of intracellular NPs in the cells with the highest expression of MexAB-OprM (nalB1) is attributed to the efflux of NPs by the pump. The absence of MexAB-OprM in ΔABM cells disables the extrusion of NPs, leading to the highest accumulation of intracellular NPs. At the same incubation time, the cells accumulate more small NPs (Figure 4A) than large NPs (Figure 4B), which agrees with what we observed in Figure 3 that small NPs stay inside the cells longer. These observations may be attributed to the higher membrane permeability and higher concentration of small NPs.
Figure 4. Study of expression-level dependent transport kinetics of efflux pump of single living cells.
(A-B) Plots of intracellular NPs in the cells: (a) WT, (b) nalB1 and (c) ΔABM, versus time, for the cells incubated with (A) 770 pM of 13.1 ± 2.5 nm NPs and (B) 3.7 pM of 91.0 ± 9.3 nm NPs for 2 h, show that accumulation of intracellular NPs depends upon the expression levels of MexAB-OprM and sizes of NPs.
(C) Plots of intracellular NPs versus time, for the cells (nalB1) that are incubated with 3.7 pM of 91.0 nm Ag NPs (a) in the presence and (b) absence of CCCP, show that the number of intracellular NPs is nearly doubled in the presence of CCCP.
(D) Time-courses of fluorescence intensity of intracellular EtBr for the cells: (a) WT, (b) nalB1 and (c) ΔABM cells, incubated with 4 µM EtBr for 2 h, show that accumulation of intracellular EtBr depends upon the expression levels of MexAB-OprM.
In the presence of CCCP (pump inhibitor), we found that the amount of intracellular NPs in nalB1 is nearly doubled (Figure 4C), suggesting that the CCCP (proton ionophore) decreased or eliminated the proton gradients across the membrane of living cells, which removed the drive-force of the membrane efflux pump (31, 55) and led to the accumulation of the higher amount of intracellular NPs. It indicates that, in the absence of CCCP (Figure 4A and B), the efflux pump (MexAB-OprM) extrudes the NPs out of the cells, leading to the lower accumulation of intracellular NPs.
The sizes of NPs used here are orders of magnitude larger than the conventional substrates of the pump. It seems almost inconceivable that these NPs can permeate into the cells and be extruded out by the efflux pump. To determine any possible steric effects of NPs and the proper functioning of efflux pump in living cells, we used a well-known fluorescence probe (EtBr) to characterize the function of MexAB-OprM pump using fluorescence spectroscopy (30–33), while we carried out the study of the membrane transporter using single NPs. The fluorescence intensity of EtBr molecules increases up to 25-fold, as they enter the cells and intercalate with DNA, leading to higher quantum efficiency of EtBr due to hydrophobic intracellular environments (56). Thus, one can use the fluorescence intensity of EtBr to monitor its accumulation in cells (30–33). The results in Figure 4D show that the accumulation rate of intracellular EtBr in WT cells is lower than ΔABM cells, but higher than nalB1. In other words, the highest accumulation rate is observed in ΔABM, while the lowest accumulation rate is found in nalB1. These results (Figure 4D) are very similar to those observed using single NP probes (Figure 4A and B), suggesting that large sizes of NPs did not hinder their transport into the cells and their extrusion by the pump. Therefore, multiple sized NPs can be used to mimic various sized antibiotics (drugs) for probing the size-dependent transport kinetics of multidrug efflux pumps of single living cells in real-time at the nanometer resolution.
Probing of Dependence of Membrane Transport of NPs on their Size and Concentration
We further studied the dependence of accumulation and efflux rates of NPs on their sizes and concentrations (doses), aiming to determine their molecular mechanisms and compare them with those observed for antibiotics. We used two concentrations for each size of NPs and studied their accumulation rates in WT, nalB-1 and ΔABM. The results in Figure 5 show the dependence of the number of intracellular NPs on NP concentrations for all three strains of cells and the number of intracellular NPs increases as the concentration of NPs increases, suggesting that passive diffusion may play the role in transport of extracellular NPs into the cells, similar to those transport mechanisms of antibiotics into the cells.
Figure 5.
Study of concentration-dependent transport rates of Ag NPs in single living cells. Plots of intracellular NPs in (A) WT, (B) nalB1 and (C) ΔABM versus time, for the cells that are incubated with (a) 13.1 ± 2.5 nm NPs at concentrations of (i) 190 pM and (ii) 96.3 pM; and (b) with 91.0 ± 9.3 nm NPs at concentrations of (i) 1.85 pM and (ii) 0.93 pM, respectively. We analyzed 800 cells at each 20 min over 2 h for each data point.
Notably, the accumulation rates in Figure 5 depend upon the expression level of MexAB-OprM and sizes of NPs, similar to those in Figure 4. The accumulation of intracellular NPs in WT (Figure 5A) is higher than nalB-1 (Figure 5B), but lower than ΔABM (Figure 5C). The highest number of intracellular NPs and accumulation rates are found in ΔABM, while the lowest number of intracellular NPs and lowest accumulation rates are observed in nalB1. The results suggest that the absence of MexAB-OprM in ΔABM leads to the highest accumulation of intracellular NPs, while the over-expression of MexAB-OprM in nalB1 enables the effective extrusion of intracellular NPs, which leads to the lowest accumulation rates and lowest number of intracellular NPs.
We summarize quantitative accumulation rates and equilibrium times of accumulation of intracellular NPs in living cells (Figure 4 and Figure 5) in Table 1, which shows their dependence on the expression level of MexAB-OprM, and sizes and concentrations of NPs. For 13.1 ± 2.5 nm NPs, the accumulation rates of ΔABM is about 2.3 times higher than nalB-1 and 1.9 times higher than WT for 770 pM NPs, and 1.6 and 1.5 times for 190 pM NPs, and 1.4 and 1.1 times for 96.3 pM NPs, respectively. For 91.0 ± 9.7 nm NPs, the accumulation rates of ΔABM is about 5.6 times higher than nalB-1, and 1.7 times higher than WT for 3.7 pM NPs, and 2.1 and 1.6 times for 1.85 pM NPs, and 2.9 and 1.6 times for 0.93 pM NPs, respectively.
Table 1.
Summary of Accumulation Rates and Equilibrium Times of Single NPs in Single Living Cells
| Diameter (nm) | C Ag NPs (pM) |
Accumulation Rate (NPs/min) |
Equilibrium Time (min) |
||||
|---|---|---|---|---|---|---|---|
| WT | nalB1 | ΔABM | WT | nalB1 | ΔABM | ||
| 13.1 ± 2.5 | 770 | 1.721 | 0.921 | 2.137 | 27.5 | 17.5 | 47.5 |
| 190 | 1.218 | 0.802 | 1.295 | 22.5 | 12.5 | 42.5 | |
| 96.3 | 0.844 | 0.751 | 1.044 | 17.5 | 12.5 | 37.5 | |
| 91.0 ± 9.7 | 3.7 | 1.652 | 0.975 | 5.43 | 32.5 | 22.5 | 37.5 |
| 1.85 | 1.156 | 0.735 | 1.535 | 22.5 | 17.5 | 27.5 | |
| 0.93 | 0.365 | 0.233 | 0.669 | 17.5 | 12.5 | 27.5 | |
Characterization of Viability of Single Cells
Studies show that Ag NPs inhibit bacterial growth in a dose-dependent manner (57, 58). Therefore, we characterized the viability of cells to ensure that the doses of NPs that we used to probe the efflux pump of P. aeruginosa did not affect the function and viability of the cells.
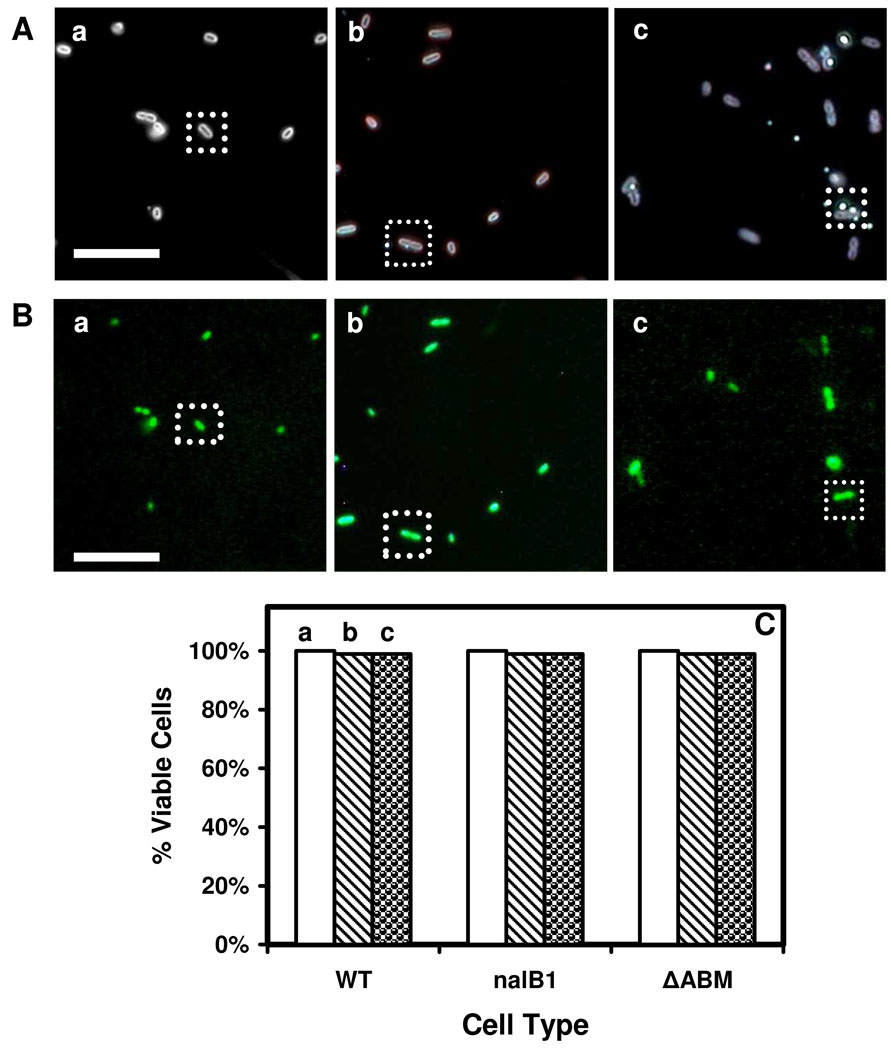
The viability of cells (WT, nalB1, ΔABM) incubated with NPs for 12 h were characterized using live/dead bacLight viability and counting assay, which determines the viability of cells by detecting both live and dead cells using SYTO9 nucleic acid stain and propidium iodide, respectively (59). Observation of the green fluorescence (λmax = 520 nm) of SYTO9 in bacterial cells indicates the viable cells, while the display of red fluorescence (λmax = 610 nm) of propidium iodide in bacterial cells show the dead cells, allowing us to count the number of live and dead cells and determine the percentage of the viable cells. Optical images of the cells (WT) incubated with the PBS buffer (blank control, in the absence of NPs), 770 pM 13.1 ± 2.5 nm Ag NPs, and 3.7 pM of 91.0 ± 9.3 nm Ag NPs, for 12 h (Figure 6A: a-c), show that the cells without NPs in (a), and cells with intracellular and extracellular NPs in (b-c). Their fluorescence images in Figure 6B illustrate that the cells with and without NPs, as those squared, are alive.
Figure 6.
Characterization of cellular viability: (A) Optical images of cells (WT, nalB1, ΔABM) that are incubated with (a) the buffer (blank control), (b) 3.7 pM of 91.0 nm NPs and (c) 770 pM of 13.1 nm NPs for 12 h; (B) Fluorescence images of the cells in (A), that are assayed using live/dead bacLight assays, show that the cells are alive. The scale bars in (A-B) are 8 µm. (C) More than 99% of the cells (WT, nalB1, ΔABM) in (A) are viable.
We determined the number of live and dead cells and plotted the percentage of viable cells. The results in Figure 6C show that more than 99% of the cells (WT, nalB1, ΔABM) incubated with the buffer, 770 pM 13.1 ± 2.5 nm Ag NPs, and 3.7 pM of 91.0 ± 9.3 nm Ag NPs, for 12 h, are alive, demonstrating that we indeed studied the efflux pump of single living cells. The results demonstrate that the NPs at these selected concentrations are biocompatible to the cells, showing that the Ag NPs are well suited for probing of membrane transporters of single living cells.
Summary
In summary, we have designed and synthesized Ag NPs with the average diameters of 13.1 ± 2.5 nm and 91.0 ± 9.3 nm, and used their size-dependent plasmonic spectra to image and determine the sizes of single NPs in and out of single living bacterial cells. The NPs (770 pM of 13.1 nm NPs and 3.7 PM of 91 nm NPs) are stable (non-aggregated) in the PBS buffer and in cells, and the cells incubated with these concentrations of NPs for 12 h are viable. We found that NPs transported into the cells in a concentration-dependent manner, suggesting that the passive diffusion driven by concentration gradients is the primary mechanism for NPs to enter the cells, similar to those observed for antibiotics. The accumulation of intracellular NPs depends upon the expression level of MexAB-OprM, sizes and concentrations of NPs, and the presence of proton ionophores (CCCP) (pump inhibitor). The highest number of intracellular NPs was found in ΔABM (deletion of MexAB-OprM), while the lowest number was observed in nalB1 (over-expression of MexAB-OprM). In the presence of CCCP, we observed the higher number of intracellular NPs in nalB1. These results show that the accumulation and efflux of intracellular NPs are directly associated with the expression level and function of MexAB-OprM, suggesting that the efflux pump (MexAB-OprM) is responsible for the extrusion of NPs out of cells. The smaller NPs stay longer inside the cells than larger NPs, suggesting size-dependent efflux kinetics of the membrane transporter. Taken together, the results show that size-dependent plasmonic NP probes can be used to study the multidrug membrane transporters in single living cells, offering the possibility of better understanding of MDR. Work is in progress to probe molecular mechanisms of the membrane transporter.
Supplementary Material
Acknowledgements
We thank Hiroshi Yoneyama at Tohoku University, Japan, for providing us three strains of Pseudomonas aeruginosa, WT, nalB-1 and ΔABM, and CharFac of U. of Minnesota (a NNIN site funded by NSF) for their assistance to characterize Ag nanoparticles using HRTEM.
Abbreviations
- Ag
silver
- CCCP
carbonyl cyanide-m-chlorophenylhydrazone
- EtBr
ethidium bromide
- MDR
multi-drug resistance
- NP
nanoparticles
- nalB
strain of Pseudomonas aeruginosa with over-expression of MexAB and OprM
- nm
nanometer
- OD
optical density
- P
Pseudomonas
- LSPR
localized surface plasmon resonance
- TEM
transmission electron microscope
- WT
wide-type
- ΔABM
strain of Pseudomonas aeruginosa with deletion of MexAB and OprM
Footnotes
This work is supported in part by NSF (NIRT: BES 0507036) and NIH (R01 GM076440). Nallathamby is grateful for the support of Dominion Scholar Fellowship.
This material is available free of charge via the internet at http://pubs.asc.org
REFERENCES
- 1.Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol. 2009;12:512–519. doi: 10.1016/j.mib.2009.07.003. and references therein. [DOI] [PubMed] [Google Scholar]
- 2.Pfister O, Liao R. Pump to survive: novel cytoprotective strategies for cardiac progenitor cells. Circ. Res. 2008;102:998–1001. doi: 10.1161/CIRCRESAHA.108.176453. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. and references therein. [DOI] [PubMed] [Google Scholar]
- 4.Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 2007;13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. and references therein. [DOI] [PubMed] [Google Scholar]
- 5.Seeger MA, Diederichs K, Eicher T, Brandstätter L, Schiefner A, Verrey F, Pos KM. The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance. Curr. Drug Targets. 2008;9:729–749. doi: 10.2174/138945008785747789. [DOI] [PubMed] [Google Scholar]
- 6.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39:162–176. doi: 10.1080/07853890701195262. and references therein. [DOI] [PubMed] [Google Scholar]
- 7.Alibert-Franco S, Pradines B, Mahamoud A, Davin-Regli A, Pagès JM. Efflux mechanism, an attractive target to combat multidrug resistant Plasmodium falciparum and Pseudomonas aeruginosa. Curr. Med. Chem. 2009;16:301–317. doi: 10.2174/092986709787002619. and references therein. [DOI] [PubMed] [Google Scholar]
- 8.Li X-Z, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–1623. doi: 10.2165/11317030-000000000-00000. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagès J-M, Amaral L. Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of Gram-negative bacteria. Biochimica et Biophysica Acta. 2009;1794:826–833. doi: 10.1016/j.bbapap.2008.12.011. and references therein. [DOI] [PubMed] [Google Scholar]
- 10.Pietras Z, Bavro VN, Furnham N, Pellegrini-Calace M, Milner-White EJ, Luisi BF. Structure and mechanism of drug efflux machinery in Gram negative bacteria. Curr. Drug Targets. 2008;9:719–728. doi: 10.2174/138945008785747743. and references therein. [DOI] [PubMed] [Google Scholar]
- 11.Aeschlimann JR. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy. 2003;23:916–924. doi: 10.1592/phco.23.7.916.32722. and references therein. [DOI] [PubMed] [Google Scholar]
- 12.Giamarellou H, Poulakou G. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs. 2009;69:1879–1901. doi: 10.2165/11315690-000000000-00000. and references therein. [DOI] [PubMed] [Google Scholar]
- 13.Rice LB. The clinical consequences of antimicrobial resistance. Curr. Opin. Microbiol. 2009;12:476–481. doi: 10.1016/j.mib.2009.08.001. and references therein. [DOI] [PubMed] [Google Scholar]
- 14.Vergidis PI, Falagas ME. Multidrug-resistant Gram-negative bacterial infections: the emerging threat and potential novel treatment options. Curr. Opin. Investig. Drugs. 2008;9:176–183. [PubMed] [Google Scholar]
- 15.Page MG, Heim J. Prospects for the next anti-Pseudomonas drug. Curr. Opin. Pharmacol. 2009;9:558–565. doi: 10.1016/j.coph.2009.08.006. and references therein. [DOI] [PubMed] [Google Scholar]
- 16.Germ M, Yoshihara E, Yoneyama H, Nakae T. Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 1999;261:452–455. doi: 10.1006/bbrc.1999.1045. [DOI] [PubMed] [Google Scholar]
- 17.Maseda H, Yoneyama H, Nakae T. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000;44:658–664. doi: 10.1128/aac.44.3.658-664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000;44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Yoneyama H, Narita S, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 20.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 21.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NC. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakae T. Multiantibiotic resistance caused by active drug extrusion in Pseudomonas aeruginosa and other gram-negative bacteria. Microbiologia. 1997;13:273–284. [PubMed] [Google Scholar]
- 23.Ryan BM, Dougherty TJ, Beaulieu D, Chuang J, Dougherty BA, Barrett JF. Efflux in bacteria: what do we really know about it? Expert Opin. Investig. Drugs. 2001;10:1409–1422. doi: 10.1517/13543784.10.8.1409. and references therein. [DOI] [PubMed] [Google Scholar]
- 24.Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev. Anti. Infect. Ther. 2010;8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen IT, Brown MH, Skurray RA. Proton-Dependent Multidrug Efflux Systems. Microbiol. Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. and references therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenwood D. Modes of action in antibiotic and chemotherapy. In: O’Grady F, Lambert HP, Finch RG, Greenwood D, editors. Antibiotic and Chemotherapy: Anti-infectiVe Agents and Their Use in Therapy. New York: Churchill, Livingstone; 1997. pp. 1–22. and references therein. [Google Scholar]
- 27.Nakajima A, Sugimoto Y, Yoneyama H, Nakae T. High-level fluoroquinolone resistance in Pseudomonas aeruginosa due to interplay of the MexAB-OprM efflux pump and the DNA gyrase mutation. Microbiol. Immunol. 2002;46:391–395. doi: 10.1111/j.1348-0421.2002.tb02711.x. [DOI] [PubMed] [Google Scholar]
- 28.Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 29.Mortimer PG, Piddock LJ. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 1991;28:639–653. doi: 10.1093/jac/28.5.639. [DOI] [PubMed] [Google Scholar]
- 30.Kyriacou SV, Nowak ME, Brownlow WJ, Xu X-HN. Single live cell imaging for real-time monitoring of resistance mechanism in Pseudomonas aeruginosa. J. Biomed. Opt. 2002;7:576–586. doi: 10.1117/1.1506707. [DOI] [PubMed] [Google Scholar]
- 31.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J. Biol. Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 32.Xu X-HN, Brownlow WJ, Huang S, Chen J. Real-time measurements of single membrane pump efficiency of single living Pseudomonas aeruginosa cells using fluorescence microscopy and spectroscopy. Biochem. Biophys. Res. Commun. 2003;305:79–86. [Google Scholar]
- 33.Xu X-HN, Wan Q, Kyriacou SV, Brownlow WJ, Nowak ME. Direct observation of substrate induction of resistance mechanism in Pseudomonas aeruginosa using single live cell imaging. Biochem. Biophys. Res. Commun. 2003;305:941–949. doi: 10.1016/s0006-291x(03)00874-x. [DOI] [PubMed] [Google Scholar]
- 34.Viveiros M, Martins M, Couto I, Rodrigues L, Spengler G, Martins A, Kristiansen JE, Molnar J, Amaral L. New methods for the identification of efflux mediated MDR bacteria, genetic assessment of regulators and efflux pump constituents, characterization of efflux systems and screening for inhibitors of efflux pumps. Curr. Drug Targets. 2008 Sep;9(9):760–778. doi: 10.2174/138945008785747734. 9, 760-78. [DOI] [PubMed] [Google Scholar]
- 35.Chiu W, Moffat K. Biophysical methods: structure, dynamics and gorgeous images. Curr. Opin. Struct. Biol. 2007;17:546–548. doi: 10.1016/j.sbi.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl. Acad. Sci. USA. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain PK, Huang X, El-Sayed IH, El-Sayed MA. Noble metals on the nanoscale: optical and photothermal properties and some applications in imaging, sensing, biology, and medicine. Acc. Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 38.Kreibig U, Vollme M. Optical Properties of Metal Clusters. Berlin: Springer; 1995. [Google Scholar]
- 39.Huang T, Nallathamby PD, Gillet D, Xu X-HN. Design and synthesis of single nanoparticle optical biosensors for imaging and characterization of single receptor molecules on single living cells. Anal. Chem. 2007;79:7708–7718. doi: 10.1021/ac0709706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang T, Nallathamby PD, Xu X-HN. Photostable single-molecule nanoparticle optical biosensors for real-time sensing of single cytokine molecules and their binding reactions. J. Am. Chem. Soc. 2008;130:17095–17105. doi: 10.1021/ja8068853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee KJ, Nallathamby PD, Browning LM, Osgood CJ, Xu X-HN. In vivo imaging of transport and biocompatibility of single silver nanoparticles in early development of zebrafish embryos. ACS Nano. 2007;1:133–143. doi: 10.1021/nn700048y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nallathamby PD, Lee KJ, Xu X-HN. Design of stable and uniform single nanoparticle photonics for in vivo dynamics imaging of nanoenvironments of zebrafish embryonic fluids. ACS Nano. 2008;2:1371–1380. doi: 10.1021/nn800048x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz S, Smith DR, Mock JJ, Schultz DA. Single-target molecule detection with nonbleaching multicolor optical immunolabels. Proc. Natl. Acad. Sci. U S A. 2000;97:996–1001. doi: 10.1073/pnas.97.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning LM, Lee KJ, Huang T, Nallathamby PD, Lowman J, Xu X-HN. Random walk of single gold nanoparticles in zebrafish embryos leading to stochastic toxic effects on embryonic developments. Nanoscale. 2009;1:138–152. doi: 10.1039/b9nr00053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyriacou SV, Brownlow WJ, Xu XHN. Using nanoparticle optics assay for direct observation of the function of antimicrobial agents in single live bacterial cells. Biochemistry. 2004;43:140–147. doi: 10.1021/bi0351110. [DOI] [PubMed] [Google Scholar]
- 46.Xu X-HN, Brownlow WJ, Kyriacou SV, Wan Q, Viola JJ. Real-time probing of membrane transport in living microbial cells using single nanoparticle optics and living cell imaging. Biochemistry. 2004;43:10400–10413. doi: 10.1021/bi036231a. [DOI] [PubMed] [Google Scholar]
- 47.Xu X-HN, Chen J, Jeffers RB, Kyriacou SV. Direct Measurement of Sizes and Dynamics of Single Living Membrane Transporters Using Nano-Optics. Nano Letters. 2002;2:175–182. [Google Scholar]
- 48.Xu X-HN, Song Y, Nallathamby PD. Probing Membrane Transport of Single Live Cells Using Single Molecule Detection and Single Nanoparticle Assay. In: Xu X-HN, editor. New Frontiers in Ultrasensitive Bioanalysis: Advanced Analytical Chemistry Applications in Nanobiotechnology, Single Molecule Detection, and Single Cell Analysis. New Jersey: Wiley; 2007. pp. 41–65. [Google Scholar]
- 49.Nallathamby PD, Xu X-HN. Study of cytotoxic and therapeutic effects of stable and purified silver nanoparticles on tumor cells. Nanoscale. 2010 doi: 10.1039/c0nr00080a. DOI: 10.1039/c0nr00080a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X-HN, Huang S, Brownlow W, Salatia K, Jeffers R. Size and temperature dependence of surface plasmon absorption of gold nanoparticles induced by Tris(2,2'-Bipyridine)Ruthenium(II) J. Phys. Chem. B. 2004;108:15543–15551. [Google Scholar]
- 51.Yoneyama H, Ocaktan A, Tsuda M, Nakae T. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 1997;233:611–618. doi: 10.1006/bbrc.1997.6506. [DOI] [PubMed] [Google Scholar]
- 52.Mie G. Beiträg Zur Optik Trüber Medien, Speziell Kolloidaler Metrallősungen. Ann. Phys. 1908;25:377–445. [Google Scholar]
- 53.Kelly KL, Coronado E, Zhao LL, Schatz GC. The optical properties of metal nanoparticles: the influence of size, shape, and dielectric environment. J. Phys. Chem. B. 2003;107:668–677. [Google Scholar]
- 54.Pastoriza-Santos I, Liz-Marzan LM. Colloidal silver nanoplates. state of the art and future challenges. J. Mat. Chem. 2008;18:1724–1737. [Google Scholar]
- 55.Ikonomidis A, Tsakris A, Kanellopoulou M, Maniatis AN, Pournaras S. Effect of the proton motive force inhibitor Carbonyl Cyanide-m-Chlorophenylhydrazone (CCCP) on Pseudomonas aeruginosa biofilm development. Letters Appl. Microbiol. 2008;47:298–302. doi: 10.1111/j.1472-765x.2008.02430.x. [DOI] [PubMed] [Google Scholar]
- 56.Morgan AR, Lee JS, Pulleyblank DE, Murray NL, Evans DH. Review: ethidium fluorescence assays. part 1. physicochemical studies. Nucleic Acids Res. 1979;7:547–569. doi: 10.1093/nar/7.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kyriacou SV. Department of Chemistry and Biochemistry. Norfolk, VA: Old Dominion University; 2003. [Google Scholar]
- 58.Vaidyanathan R, Kalishwaralal K, Gopalram S, Gurunathan S. Nanosilver--the burgeoning therapeutic molecule and its green synthesis. Biotechnol. Adv. 2009;27:924–937. doi: 10.1016/j.biotechadv.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Berney M, Hammes F, Bosshard F, Weilenmann HU, Egli T. Assessment and interpretation of bacterial viability by using the live/dead baclight kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007;73:3283–3290. doi: 10.1128/AEM.02750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.