Abstract
The scaffold protein IQGAP1 shows elevated levels in several cancer types, but its expression in hepatocellular carcinoma is unknown. We found that 58% of human hepatocellular carcinoma tissue samples had increased IQGAP1 expression compared to adjacent normal tissue. Overexpressing IQGAP1 raised the in vivo tumorigenicity of hepatocellular carcinoma cells, and forced overexpression of IQGAP1 in vitro stimulated cell proliferation. Cell growth was reduced by knockdown or mutation of IQGAP1, or by treatment of cells with a phosphotidylinositol 3-kinase inhibitor. To determine the mechanism by which IQGAP1 overexpression affected hepatocellular carcinoma cells, we confirmed its interaction in these cells with mammalian target of rapamycin (mTOR), a serine/threonine kinase that integrates signals about nutrient and energy status with downstream effectors that influence cell division. In addition, we discovered a new interaction involving IQGAP1, mTOR and Akt, which is a downstream target of mTOR. Akt phosphorylation on Ser-473, which is catalyzed by mTOR and required for Akt activation, increased with increasing amounts of IQGAP1, and decreased with IQGAP1 mutation. We hypothesize that IQGAP1 is a scaffold that facilitates mTOR and Akt interaction.
Keywords: carcinoma, hepatocellular; IQ motif containing GTPase activating protein; mTOR protein; oncogenes; proto-oncogene proteins c-akt
Introduction
The IQGAP family of proteins is found in eukaryotes from yeast to mammals. Named for their isoleucine-glutamine (IQ) and GTP-activating protein (GAP) domains, IQGAP family members contain multiple protein-protein interaction motifs. Of the three human IQGAPs, IQGAP1 is the most well characterized. In addition to four IQ domains, IQGAP1 contains a calponin homology domain, a polyproline interaction domain, a RasGAP-related domain (GRD), and a RasGAP_C carboxy-terminal sequence common to IQGAP family members (Briggs and Sacks, 2003; White et al., 2009). IQGAP1 is ubiquitously expressed in human tissues, and is found in the cytoplasm, where it acts as a scaffolding protein, and interacts with the GTP-bound form of Rac1 and Cdc42, which are Rho/Rac family small GTPases. This interaction, and interaction with actin, is thought to regulate cell-cell adhesion and cell migration, processes that are particularly relevant to cancer progression, specifically metastasis (Noritake et al., 2005; Matazara et al., 2007; Owen et al., 2008). IQGAP1 may also be involved in crosstalk with the Erk pathway (Roy et al., 2005). IQGAP1 participates as a scaffolding protein in MEK-ERK signaling. IQGAP1 and MEK associate both in vitro and in intact MCF-7 human breast epithelial cells. IQGAP1 also binds directly to ERK2 and regulates its activity (Roy et al., 2004).
IQGAP1 interacts with the mammalian target of rapamycin (mTOR), a serine-threonine kinase that coordinates information about nutrient, redox and energy status, with cell growth processes like translation, growth, and cell motility (Wang et al., 2009). One of the substrates of mTOR is the Akt serine/threonine kinase. Phosphorylation of Ser-473 on Akt by mTOR contributes to Akt activation (Sarbassov et al., 2005). Activation of Akt also requires the activity of phosphotidylinositol 3-kinase (PI3K), which generates the phosphotidylinositol triphosphate that is bound by Akt, to direct it to the membrane for mTOR activation. Downstream effects of Akt activation include inhibition of apoptosis (Franke, 2008), which has implications for cancer progression (Zhou et al., 2007; Dubrovska et al., 2009).
Since IQGAP1 is overexpressed in several kinds of cancer (Nabeshima et al., 2002; Dong et al., 2006; Jadeski et al., 2008), we investigated IQGAP1 expression levels in vitro, in hepatocellular carcinoma cell lines, and in vivo, in hepatocarcinoma tissue samples. As a scaffold protein, IQGAP1 binds multiple proteins and facilitates their interaction. We present evidence that, in addition to the previously known functions of IQGAP1 as a platform for protein-protein interaction, it also appears to facilitate mTOR and Akt interaction. These results add to our understanding of how overexpression of IQGAP1 may contribute to the hyperproliferation of cancer cells.
Results
IQGAP1 is overexpressed in human hepatocellular carcinoma tissue
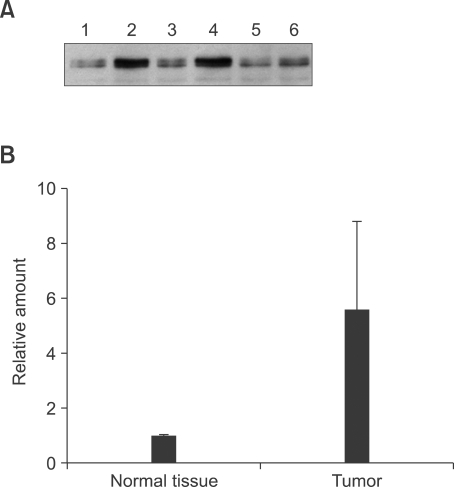
IQGAP1 levels were analyzed in tumor samples from 55 hepatocellular carcinoma patients, and compared to adjacent normal tissue from each patient. Detection of IQGAP1 protein showed that expression of IQGAP1 was significantly increased in 58% of analyzed carcinoma tissue samples, compared to adjacent normal controls (Supplemental Data Table S1). Akt and mTOR expressions were not detectably altered in tumor tissue compared to normal tissues (data not shown). Three representative examples are shown in Figure 1. These results indicated that IQGAP1 might play an important role in the tumorigenesis of hepatocellular carcinoma.
Figure 1.
Expression of IQGAP1 in human hepatocellular carcinoma tissue. (A) Equal amounts of protein lysate from hepatocellular carcinoma tissue (lanes 2, 4 and 6) and paired normal liver (lanes 1, 3 and 5) from three different patients, were analyzed by Western blot using monoclonal anti-IQGAP1. (B) Quantification by TotalLab software showing the average of the three samples in A. Error bars show standard deviation.
IQGAP1 is crucial for HepG2 proliferation
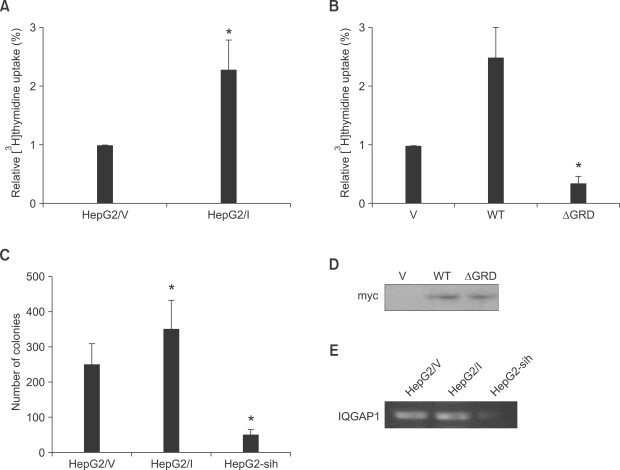
To determine the relationship between the observed in vivo increase in IQGAP1 levels, and cell proliferation in hepatocellular carcinoma, we investigated the role of IQGAP1 in the in vitro tumorigenic growth of the HepG2 human hepatocellular carcinoma line. Expression levels of IQGAP1 were altered using stable transfection with IQGAP1 overexpressing vectors, or vectors expressing shRNA that targeted IQGAP1. As measured by DNA replication, HepG2 cell proliferation increased when IQGAP1 was overexpressed, and decreased when IQGAP1was knocked down with shRNA. When HepG2 cells were transfected with IQGAP1ΔGRD, a dominant negative mutant of IQGAP1 (Jadeski et al., 2008), proliferation of the hepatocellular carcinoma cells decreased (Figures 2A and 2B). In a separate assay for cell growth, transfected cells formed more colonies by soft agar colony formation assay when overexpressing IQGAP1, and fewer when IQGAP1 was knocked down (Figure 2C).
Figure 2.
Effect of IQGAP1 on proliferation of HepG2 cells. (A) HepG2 cells (2 × 105) stably expressing control vector pcDNA3 (HepG2/V) or Myc-tagged IQGAP1 (HepG2/I), were seeded into 24-well culture dishes. After 24 h, [3H]thymidine was added for 18 h, and incorporation measured. All assays were performed in triplicate. The means ± S.D. are shown (n = 12 replicates/group). *P < 0.01. (B) Equal numbers of HepG2 cells were transiently transfected with 10 µg of vector (V), wild type (WT) IQGAP1, or IQGAP1ΔGRD (ΔGRD). Cell proliferation was quantified as described for A. Data are expressed as means ± S.D. [3H]thymidine uptake relative to V cells, (n = 3, in quadruplicate). *P < 0.01; **P < 0.05. (C) HepG2/V, HepG2/I, and HepG2-sih (1 × 104) cells were assessed by soft agar assay after 14 days at 37℃. Colonies > 0.2 mm were counted using a light microscope. The data are expressed as means ± S.D, n = 2, performed in triplicate. *P < 0.01 relative to HepG2/V-derived colonies. (D) Expression level of transfected IQGAP1 in HepG2 cells that transfected with 10 µg of vector (V), wild type (WT) IQGAP1, or IQGAP1ΔGRD (ΔGRD) was detected by Western blot using myc antibody. (E) The mRNA level of IQGAP1 in HepG2/V, HepG2/I, and HepG2-sih (1 × 104) cells was accessed by RT-PCR.
IQGAP1 is crucial for HepG2 proliferation in vivo
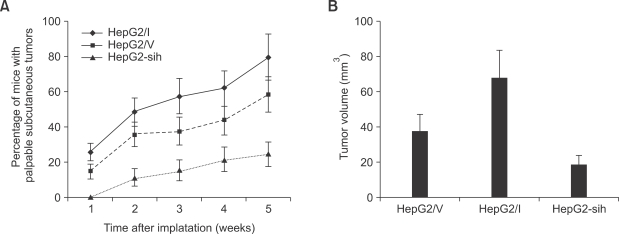
To test the effects of overexpressing IQGAP1 on in vivo tumorigenesis, cells overexpressing IQGAP1, or with the shRNA construct to knockdown IQGAP1 expression, were introduced into immunocompromised nude mice. Tumor formation was compared to mice receiving control cells transfected with vector only. More mice formed tumors when injected with cells overexpressing IQGAP1, than when injected with control or IQGAP1-knockdown cells. Tumors were also larger in mice receiving IQGAP1-overexpressing cells (Figure 3).
Figure 3.
IQGAP1 promotes in vivo tumorigenic growth of HepG2 cells. (A) Immunocompromised nude mice received subcutaneous injections of equal numbers (1 × 105) of HepG2/V, HepG2/I, and HepG2-sih cells and the rate of appearance of palpable primary tumors was examined. (B) Final volume of tumors derived from subcutaneous implants. Primary tumor volumes were measured 60 days after transplantation of HepG2/V, HepG2/I, and HepG2-sih cells. Data represent the mean ± S.D, n = 8 animals/group.
IQGAP1 affects HepG2 proliferation by regulation of PI3K/Akt
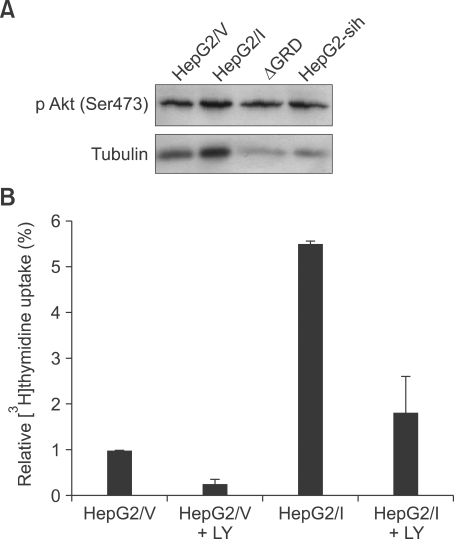
The mechanism by which IQGAP1 overexpression stimulated HepG2 cell growth was investigated by analyzing Akt phosphorylation in the cell lines in which IQGAP1 was knocked down or IQGAP1 was mutated. Steady-state levels of Akt phosphorylated on Ser-473 increased when IQGAP1 was overexpressed, and decreased when IQGAP1 was knocked down or mutated (Figure 4A). PI3K is required for Akt activation, so we tested if it was involved in IQGAP1 promotion of hepatocarcinoma. Regulation appeared to be through PI3K, because treatment with the PI3K inhibitor LY294002 reduced cell proliferation in both control cells, and cells overexpressing IQGAP1 (Figure 4B).
Figure 4.
IQGAP1 regulates HepG2 cells proliferation through the PI3K/Akt pathway. (A) equal numbers of HepG2 cells were stably transfected with pcDNA3 vector (HepG2/V), Myc-tagged IQGAP1 (HepG2/I), IQGAP1ΔGRD (ΔGRD) or IQGAP1-sih (HepG2-sih) and analyzed by Western blot. (B) proliferation of HepG2/V cells, HepG2/I cells, or cells incubated with 50 µM PI3K inhibitor LY294002 (LY).
IQGAP1 is a scaffold for mTOR and Akt interaction
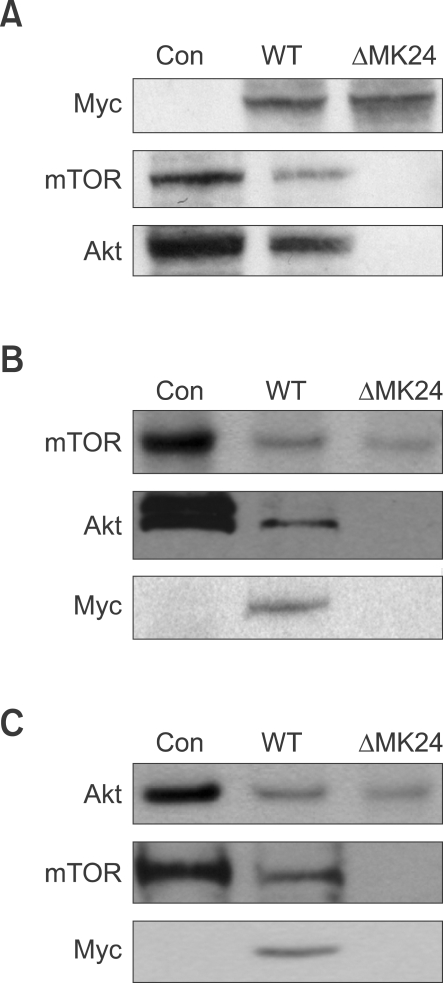
Since IQGAP1 is a scaffold for protein-protein interactions, and mTOR phosphorylates Akt, we tested for IQGAP1-mediated interaction between mTOR and Akt by co-immunoprecipitation. When HEK293 cells were transfected with Myc-IQGAP1, immunoprecipitation of IQGAP1 also precipitated mTOR and Akt. When cells were transfected with vector only, or with the Cdc42/Rac1 binding-site mutant Myc-IQGAP1ΔMK24, no interaction between IQGAP1 and mTOR and Akt was observed (Figure 5). These data suggested that IQGAP1 might be a scaffold for mTOR and Akt interaction, either directly, or through indirect interaction with Rac1 or Cdc42.
Figure 5.
IQGAP1 is a scaffold protein that facilitates mTOR interaction with Akt. HEK-293 cells were transiently transfected with equal amounts of Myc-IQGAP1(WT), Myc-IQGAP1ΔMK24. After 36 h, cells were lysed. (A) Immunoprecipitated with anti-Myc antibody for IQGAP1, and subjected to Western blot with anti-Myc, anti-mTOR, or anti-Akt. Con indicates total lysates from non-transfected cells. (B) Immunoprecipitated with anti-mTOR antibody, and subjected to Western blot with anti-Myc, anti-mTOR, or anti-Akt. (C) Immunoprecipitated with anti-Akt antibody, and subjected to Western blot with anti-Myc, anti-mTOR, or anti-Akt.
Discussion
IQGAP1 protein is overexpressed in vivo, in a number of different cancer types, including breast cancer (Jadeski et al., 2008), ovarian carcinoma (Dong et al., 2006), and colorectal carcinoma (Nabeshima et al., 2002). Here we show that, as expected from previous findings, it was also overexpressed in liver tumor tissue samples, relative to adjacent normal tissues. In vitro, forced overexpression of IQGAP1 increased cell proliferation, as measured by two different assays, and either mutation or knockdown decreased cell growth. The results of in vivo tumor formation using the overexpressing or knockdown cells were consistent with the in vitro growth assays, suggesting a crucial effect of IQGAP1 in human hepatocarcinoma. Because a loss-of-function IQGAP1 mutation, or downregulation of IQGAP1 reduced cell growth, we hypothesize that IQGAP1 overexpression is crucial for the proliferation of liver cancer cells. In support, Zhou et al. (2009) showed that treatment of HepG2 cells with the flavonoid quercetin inhibited both IQGAP1 expression at the RNA and protein levels, and also reduced proliferation and migration of the cells.
Roy reported (2005) that IQGAP1 mediates the coupling between the EGF receptor and its downstream signaling partners Mek and Erk. EGF increases Mek1 binding to IQGAP1, further augmenting Mek1/Erk1 signaling. For Mek2, EGF reduced the binding of Mek2 to IQGAP1, while both Erk1 and Erk2 increased the IQGAP1-Mek2 interaction. Results above indicate the scaffold function of IQGAP1 on MAPK pathways. How are functions of IQGAP1 on PI3K/Akt pathway, which is also important for cell proliferation and survival, particularly in hepatocellular carcinoma cells? Akt is a crucial molecule for cell proliferation, and its activation inhibits apoptosis. Phosphorylation on Ser-473 by mTOR is a critical step in its activation, and Akt phosphorylation levels corresponded to IQGAP1 levels. By co-immunoprecipitation method, we confirmed interaction between IQGAP1 and mTOR in hepatocellular carcinoma cell lines, and found that IQGAP1 binds Akt, either directly, or indirectly through mutual interaction with other proteins. This novel interaction has not been reported before. Overexpression of IQGAP increased this interaction, and was abolished by mutation of the IQGAP1 Rac1/Cdc42 binding site, suggesting that this binding site is also crucial for Akt and mTOR association. It provided possibility of interaction between mTOR and Akt.
It is demonstrated that IQGAP1 may be recruited to a particular subcellular domain by the EGF receptor (Roy et al., 2005). IQGAP1 translocolizes between different subcellular regions. For example, activation of E-cadherin (Kuroda et al., 1998) induce IQGAP1 accumulation at the plasma membrane, on the other hand, increasing intracellular Ca2+ concentrations can remove IQGAP1 from the cell membrane (Mateer et al., 2002). Our results showed that loss of Akt phosphorylation in the presence of the ΔGRD, the dominant negative mutation, suggests that Akt phosphorylation state is IQGAP1 activity dependent. As a consequence, dominant negative IQGAP1 reduced liver cancer cells proliferation significantly. This might because translocation of IQGAP1 to cells membrane was abolished when its activation was inhibited.
In the present study, we gain further insight into the participation of IQGAP1 in the PI3K/Akt signaling cascade. Among other functions, a novel association between IQGAP1 and mTOR or Akt was identified. Evidence strongly suggests that IQGAP1 is a scaffold molecule that facilitates interaction of mTOR and Akt. Our model for how IQGAP1, PI3K, Ak and mTOR interaction promotes cancer progression is a clue for further study liver cancer development. We discovered this interaction in hepatocellular carcinoma cells, but it may also occur in other cancerous cell types. IQGAP might be a target for individualized therapy.
Methods
Patients and biopsies
Biopsies from hepatocellular carcinoma tumor tissue and adjacent normal tissue were collected from 56 liver cancer patients and kept frozen at -80C. Protein was extracted from biopsies with lysis buffer (50 mM Tris-HCl pH 7.4, 1% (v/v) Triton X-100, 1 mM EDTA, 1 mM leupeptin, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 1mM Na3VO4).
Plasmids
Plasmids containing genes for Myc-tagged wild type human IQGAP1 in pcDNA3, Vectors Myc-IQGAP1ΔMK24 or IQGAP1ΔGRD was used. A gene for small hairpin (sh)RNA targeting IQGAP1 were constructed as described previously (Jadeski et al., 2008).
Cell culture and transfection
HEK293 cells and Human hepatocellular carcinoma cell line HepG2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS). Where indicated, the cells were transiently transfected with 10 µg of pcDNA3 (empty vector), Myc-IQGAP1, or Myc-IQGAP1ΔMK24 using FuGENE 6 (Roche). HepG2 cells were also stably transfected with empty pcDNA3 vector (HepG2/V), Myc-IQGAP1ΔGRD (ΔGRD) or pcDNA3-Myc-IQGAP1 (HepG2/I). IQGAP1 protein expression was three-fold higher in HepG2/I cells than in HepG2/V cells (data not shown). Stable knockdown of IQGAP1 was obtained by integrating a specific shRNA gene targeting IQGAP1 into the HepG2 genome. IQGAP1 protein expression in these cells (termed HepG2-shI cells) was reduced by 85% (data not shown).
Western blotting
Cultured cells were washed three times in serum-free medium and lysed with RIPA buffer (50 mM Tris-HCl pH 7.4, 1% (v/v) Triton X-100, 1 mM EDTA, 1 mM leupeptin, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 1mM Na3VO4), or frozen samples of human hepatocellular carcinoma and normal liver tissue were thawed and homogenized. Equal amounts of protein were separated by 12% SDS-PAGE and transferred to PVDF membranes. Immunoblots were probed with anti-IQGAP1, anti-Akt (ser473) monoclonal antibody or anti-tubulin antibody (Sigma). Complexes were visualized with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) and developed by ECL (Amersham Pharmacia Biotech). Results were quantified using Totallab TL100 software.
Cell proliferation assay
Incorporation of [3H] thymidine was used to evaluate cell proliferation as described previously (Jadeski et al. 2008). Cells were made quiescent by culturing for 24 h in DMEM with 0.5% FBS, and DNA synthesis was measured by labeling 2 × 105 cells with 1 µCi/ml [3H] thymidine for 18 h, washing with ice-cold 0.5% trichloroacetic acid and lysing with 0.25 M NaOH. Radioactivity of 600 µl of lysate was evaluated by liquid scintillation spectrometry. Each experiment was performed in triplicate.
Soft agar assays
Soft agar assays were performed as described (Jadeski et al., 2008). Colony growth was determined by plating 1 × 104 HepG2/V, HepG2/I or HepG2-shI cells in 0.3% agarose with a 0.5% agarose underlay of 1 × 104 cells/well, in 6-well plates. Colonies >0.2 mm were counted after 14 days at 37℃ in 5% CO2. Each experiment was performed in triplicate.
Animals and in vivo primary tumor growth assay
Nude BABL/c mice, 5-to-6-weeks-old, were maintained in a 12 h day/night cycle with free access to food and water. All facilities, including food and water, were autoclaved. All operations were performed in sterile conditions.
To establish xenografts of liver cancer cells (HepG2/V, HepG2/I and HepG2-shI), cells were injected subcutaneously into the flank, at 1 × 105 cells/site, with 8 mice/group. Mice were sacrificed after 1-5 weeks after implantation, tumor numbers were counted, and tumor xenografts were removed and weighed.
Co-immunoprecipitation
To investigate interaction of IQGAP1 with mTOR and Akt, immunoprecipitation was performed in HEK293 cells with anti-myc antibody, anti-mTOR or anti-Akt antibody, respectively (Cell Signaling). Immunoprecipitates were analyzed by Western blotting as above, using anti-mTOR or anti-Akt antibody (Cell Signaling).
Statistical analysis
Data are shown as averages with standard deviation (SD). Significance was analyzed by Student's t-test, using Statistical SPSS software package (SPSS Inc, Chicago). Differences were considered statistically different at P < 0.05.
Supplemental data
Supplemental Data include a table and can be found with this article online at http://e-emm.or.kr/article/article_files/SP-42-7-01.pdf.
Acknowledgement
The work was supported by The National Basic Research Program (973 Program)(2007CB512905), The State S&T Projects (11th Five Year) (2008ZX10002-007).
Abbreviations
- GAP
GTP-activating protein
- GRD
RasGAP-related domain
- IQ
isoleucine-glutamine
Supplementary Material
Supplemental Data
References
- 1.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong P, Nabeshima K, Nishimura N, Kawakami T, Hachisuga T, Kawarabayashi T, Iwasaki H. Overexpression and diffuse expression pattern of IQGAP1 at invasion fronts are independent prognostic parameters in ovarian carcinomas. Cancer Lett. 2006;243:120–127. doi: 10.1016/j.canlet.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Dubrovska A, Kim S, Salamone RJ, Walker JR, Maria SM, García-Echeverría C, Schultz PG, Reddy VA. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci USA. 2009;106:268–273. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franke TF. PI3K/Akt: getting it right matters Akt signaling in animal physiology and human disease. Oncogene. 2008;27:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 5.Jadeski L, Mataraza JM, Jeong HW, Li Z, Sacks DB. IQGAP1 Stimulates Proliferation and Enhances Tumorigenesis of Human Breast Epithelial Cells. J Biol Chem. 2008;283:1008–1017. doi: 10.1074/jbc.M708466200. [DOI] [PubMed] [Google Scholar]
- 6.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, Shoji I, Matsuura Y, Yonehara S, Kaibuchi K. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell-cell adhesion. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 7.Mateer SC, McDaniel AE, Nicolas V, Habermacher GM, Lin MJ, Cromer DA, King ME, Bloom GS. The mechanism for regulation of the F-actin binding activity of IQGAP1 by calcium/calmodulin. J Biol Chem. 2002;277:12324–12333. doi: 10.1074/jbc.M109535200. [DOI] [PubMed] [Google Scholar]
- 8.Mataraza JM, Li Z, Jeong HW, Brown MD, Sacks DB. Multiple proteins mediate IQGAP1-stimulated cell migration. Cell Signal. 2007;19:1857–1865. doi: 10.1016/j.cellsig.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nabeshima K, Shimao Y, Inoue T, Koono M. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 2002;176:101–109. doi: 10.1016/s0304-3835(01)00742-x. [DOI] [PubMed] [Google Scholar]
- 10.Owen D, Campbell LJ, Littlefield K, Evetts KA, Li Z, Sacks DB, Lowe PN, Mott HR. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: interfaces differ between the complexes. J Biol Chem. 2008;283:1692–1704. doi: 10.1074/jbc.M707257200. [DOI] [PubMed] [Google Scholar]
- 11.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem. 2004;279:17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- 12.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25:7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 14.Wang JB, Sonn R, Tekletsadik YK, Samorodnitsky D, Osman MA. IQGAP1 regulates cell proliferation through a novel CDC42-mTOR pathway. J Cell Sci. 2009;122:2024–2033. doi: 10.1242/jcs.044644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E, 3rd, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data