Abstract
Pediatric pulmonary tuberculosis diagnosis is difficult because young children are unable to expectorate sputum samples. Testing stool for tuberculosis DNA from swallowed sputum may diagnose pulmonary tuberculosis. Hospitalized children with suspected tuberculosis had stool, nasopharyngeal, and gastric aspirates cultured that confirmed pulmonary tuberculosis in 16/236 patients. Twenty-eight stored stools from these 16 children were used to evaluate stool polymerase chain reaction (PCR) for tuberculosis diagnosis compared with 28 stool samples from 23 healthy control children. Two DNA extraction techniques were used: fast-DNA mechanical homogenization and Chelex-resin chemical extraction. DNA was tested for tuberculosis DNA with a hemi-nested IS6110 PCR. PCR after Fast-DNA processing was positive for 6/16 culture-proven tuberculosis patients versus 5/16 after Chelex extraction (sensitivity 38% and 31%, respectively). All controls were negative (specificity 100%). If sensitivity can be increased, stool PCR would be a rapid, non-invasive, and relatively bio-secure initial test for children with suspected pulmonary tuberculosis.
INTRODUCTION
Tuberculosis kills ~2 million people each year, and in 1989, the World Health Organization estimated that ~300,000 children < 15 years of age die of tuberculosis per year worldwide.1,2 Pediatric tuberculosis diagnosis is impeded by difficulty obtaining sputum samples in children and the paucibacillary nature of their disease that often necessitates invasive procedures such as gastric aspiration or bronchoscopy3 or reliance on unreliable clinical scoring systems.4 In South Africa, it has been shown that induced sputum has comparable diagnostic sensitivity to gastric aspirate in HIV-positive and -negative children,5 whereas some other studies have shown less promising results for induced sputum.6 Although less invasive than gastric aspirates, induced sputum is still unpleasant and requires precautions to prevent airborne tuberculosis transmission to staff and other patients.3 If tuberculosis could be diagnosed from stool, collection could easily take place in the field or in clinics.
The important issues and challenges in pediatric tuberculosis differ markedly between developed and developing countries. In more developed countries such as the United States, rates of tuberculin skin test reactivity in the general population are low, so the tuberculin skin test is a useful diagnostic test for tuberculosis. In contrast, in many developing countries such as Peru, interpretation of the tuberculin skin test for diagnosis of pulmonary tuberculosis is less reliable.7,8 The diagnostic yield of gastric aspiration ranges from 20% to 40%.3,9 Although children and infants may be unable to expectorate sputum samples, most sputum is swallowed,9 and tuberculosis DNA may remain intact after intestinal transit. We and others have shown that the IS6110 polymerase chain reaction (PCR) test is both sensitive and specific for detecting Mycobacterium tuberculosis in respiratory samples in adults and gastric aspirates in children.10-14 We have also shown that M. tuberculosis DNA can be detected by the above method in stool samples from adults with pulmonary tuberculosis (J Cordova and others, unpublished data). However, sensitivity was affected by DNA extraction methods. We therefore did this research to test two hypotheses: first, that stool PCR could be used to diagnose pediatric pulmonary tuberculosis as an alternative to invasive procedures in pediatric patients with suspected pulmonary tuberculosis; second, that the sensitivity of stool PCR could be improved by optimizing the DNA extraction technique.
MATERIALS AND METHODS
Subjects
A long-term clinical trial in Lima, Peru, has been evaluating pediatric tuberculosis diagnostic strategies, as reported.4 Children < 12 years of age were enrolled if there was a high clinical suspicion of tuberculosis indicated by a Stegen and Toledo15 score ≥ 5, provided that they were HIV negative and had not received tuberculosis treatment. Gastric aspirates, nasopharyngeal aspirates,16 and stool samples were collected on 2 consecutive days. If stool samples were not available on consecutive days, the first subsequent stool sample was substituted. Samples were tested for tuberculosis by auramine microscopy17 and culture using the microscopic-observation drug-susceptibility (MODS) technique.18,19 All patients with a clinical or culture proven diagnosis of tuberculosis were provided with standard anti-tubercular chemo-therapy by the national tuberculosis control program.1
As negative controls, stool samples and nasopharyngeal aspirates were also tested from asymptomatic children living in a well-described Peruvian peri-urban shantytown.8,20,21 These age-matched control children all had normal clinical examinations, no tuberculosis risk factors, and negative tuberculin skin tests (< 10-mm induration after 5 IU intradermal 0.1-mL injection of Tubersol; Aventis Pasteur, Toronto, Canada). Stool samples without additives from all patients and controls were stored for up to 2 years at −20°C.
The study protocol and consents were approved by the institutional review boards at Tulane Medical Center, Johns Hopkins Bloomberg School of Public Health, Association Benefica PRISMA, and the Instituto de Salud del Niño of Lima, Peru. Written informed consent was obtained from all patients or their parents or guardians, and the human experimentation guidelines of the US Department of Health and Human Services were followed.
Two years after the beginning of the study, we found that a recently developed Fast-DNA extraction method increased tuberculosis PCR detection from adult patients' stool specimens. Because of this new knowledge, a study was set up using the stored stool samples from pediatric patients who had a positive M. tuberculosis culture compared with stored stool samples from healthy controls. Laboratory personnel, blinded to the nature of the samples, extracted DNA from one half of each sample using 1) Fast-DNA homogenization and from the other half using 2) Chelex resin extraction. IS6100 PCR was performed on the DNA samples for tuberculosis detection.
Fast DNA extraction method
Stool (200 mg) was homogenized with 1 mL phosphate buffer, transferred to a lysing matrix tube with 125 μL of homogenization buffer, and processed in the FastPrep instrument (Qbiogene, Irvine, CA) for 30 seconds at speed 5.5. The product was centrifuged at 14,000g for 1 minute, and 250 μL of protein precipitation solution was added to the supernatant. The product was centrifuged at 14,000g for 15 minutes, and the supernatant was processed according to the manufacturer's instructions with the DNA product eluted in 60 μL of water.
Chelex extraction and stool decontamination
A stool sample (200 mg) was homogenized in 6 mL of water, and 2 mL of the supernatant was decontaminated with n-acetyl-L-cysteine and sodium hydroxide for 15 minutes as described.11,14,17 Ethanol was added to a final concentration of 60% to kill the mycobacteria. The decontaminated pellet was washed with TE-Triton X-100 buffer and resuspended in 100 μL of Chelex 10%/Triton X-100 1% TE. The samples were boiled for 30 minutes, cooled at −20°C for 10 minutes, and centrifuged at 6,000g for 5 minutes, and the 3-μL supernatant was used for PCR.11
Dilution
The DNA products were tested using PCR both directly and after 1:5 dilution to decrease inhibitors. Dilutions were made in DNAse and pyrogen-free water for Fast-DNA and in Triton-Tris-EDTA for Chelex.17
IS6110-PCR procedure
Hemi-nested PCR reactions were performed on each sample, using 0.22 mmol/L outer and 0.52 mmol/L inner primers that have been shown to be specific to M. tuberculosis, as described.11,13,22 Duplicate PCR reactions were performed for each sample with Taq polymerase (Promega, Madison, WI) and TaqStart (Clontech, Mountain View, CA). Negative PCR controls consisted of the PCR mixture without sample. Positive PCR controls were auramine-negative gastric aspirate culture-positive pediatric samples. The IS6110-PCR (Genbank accession M29899) protocol took < 6 hours to perform.
Statistical analysis
The gold standard for a diagnosis of proven tuberculosis was M. tuberculosis isolation in laboratory culture. Sensitivity and specificity with 95% confidence intervals were estimated for each test. Proportions were compared using Z-tests for proportions. All tests were two-tailed and were performed under a 95% confidence level with the statistical software STATA 9.0 (Statacorp, College Station, TX).
RESULTS
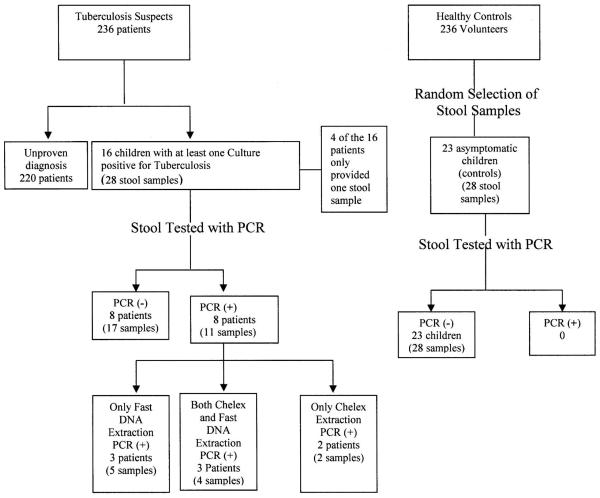
The study standards for reporting of diagnostic accuracy flow chart is shown in Figure 1, and the patient characteristics are shown in Table 1. Four patients were female, and there were no significant sex differences for patients and controls (P = 0.2). Tuberculin skin tests were positive for 14/16 (87%) of tuberculosis cases. The mean Stegen and Toledo score for tuberculosis cases was 7.4%, and 69% had a score ≥ 7, indicating highly probable tuberculosis.
Figure 1.
Standards for reporting of diagnostic accuracy flow chart.
Table 1.
Patient characteristics
| Characteristic | Patients (N = 16) |
|---|---|
| Patient age (years) | |
| Median age (IQR) | 5.05 |
| Mean years ± SD | 4.42 ± 3.04 |
| Range | 3 weeks to 11 years |
| Ratio of males to females | 12:4 |
| Stegen and Toledo score | |
| Mean Stegen Toledo score ± SD | 7.44 ± 2.28 |
| Score of ≥ 7 (highly probable tuberculosis) | 11 (69%) |
| Range | 5–13 (probable to highly- probable tuberculosis) |
| Tuberculin skin test positive (≥ 10 mm) | 14 (88%) |
| Cough > 2 weeks | 6 (38%) |
| Suspect radiograph | 11 (69%) |
| Pulmonary infiltrate on radiograph | 2 (13%) |
| Contact with adult tuberculosis patient | 14 (88%) |
The distribution of the patient population is shown, characterizing demographics and clinical Stegen and Toledo score and its respective components.
The mean age of the controls was 7.4 years, which was significantly greater than the patients (4.4 years; P = 0.01). All of the controls had a negative tuberculin skin test and a Stegen and Toledo score of zero.
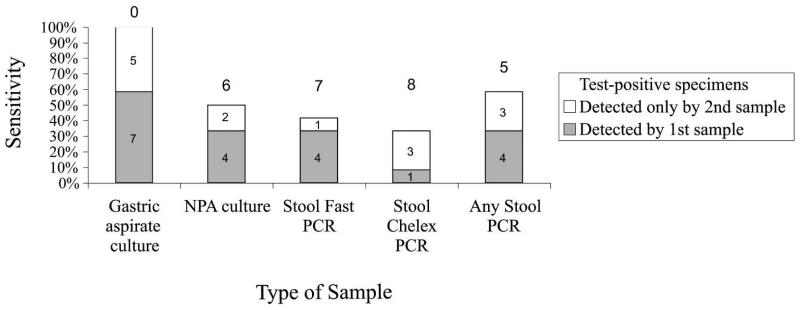
Table 2 shows that stool specimens with DNA extracted by the Fast-DNA method were PCR positive in 6/16 culture-positive patients versus 5/16 patients when stool was extracted by the Chelex method (sensitivity 38% versus 31%, respectively, P = 0.7). Figure 2 shows the contribution of second samples to diagnostic sensitivity.
Table 2.
Stool PCR sensitivity
| (A) Analysis by subject | ||||
| Fast DNA stool PCR | Tuberculosis cases (N = 16) | Healthy controls (N = 23) | Sensitivity | Specificity |
|
| ||||
| Fast PCR (+) | 6 | 0 | 38% | 100% |
| Fast PCR (−) | 10 | 23 | ||
|
| ||||
| Chelex stool PCR | Tuberculosis cases (N = 16) | Healthy controls (N = 23) | ||
|
| ||||
| Chelex PCR (+) | 5 | 0 | 31% | 100% |
| Chelex PCR (−) | 11 | 23 | ||
|
(B) Analysis by specimen | ||||
| Stool and culture samples from cases (N = 28) |
Healthy control samples (N = 28) |
Sensitivity | Specificity | |
|
| ||||
| Fast (+) stool PCR | 9 | 0 | 31% | 100% |
| Fast (−) stool PCR | 19 | 28 | ||
| Chelex (+) stool PCR | 6 | 0 | 21% | 100% |
| Chelex (−) stool PCR | 22 | 28 | ||
| Gastric aspirate culture (+) | 20 | Not done | 71% | NA |
| Gastric aspirate culture (−) | 8 | Not done | ||
| Nasopharyngeal aspirate culture (+) | 9 (missing 1) | 28 | 31% | 100% |
| Nasopharyngeal aspirate culture (−) | 18 | 28 | ||
The sensitivity and specificity of stool PCR for the detection of M. tuberculosis is shown, analyzed by DNA extraction diagnostic method amongst children with at least one positive TB culture and from healthy control children. (A) Analysis by subject and (B) analysis by specimen: sensitivity and specificity of stool PCR of gastric aspirate culture and nasopharyngeal culture for detection of M. tuberculosis.
Figure 2.
Contribution of second samples to diagnostic sensitivity. The gray bars represent tuberculosis detected by the first sample, whereas the white bars depict the added diagnostic yield of the second sample. The number above each set of bars represents the number of false negatives. This graph only includes the 12 patients who had two samples taken. NPA = nasopharyngeal aspirate.
Table 2B shows that, among samples from the 16 culture positive patients, 9/28 (32%) were PCR positive in samples extracted by Fast DNA compared with 6/28 (21%) extracted by Chelex. Although Fast DNA tended to be more sensitive than Chelex, the difference was not statistically significant (P = 0.3). The results from Fast-DNA and Chelex tests were highly concordant with 75% agreement (κ = 0.37, P = 0.02). None of the 28 samples from 23 control participants were positive by PCR with stool DNA extracted by either Fast DNA or Chelex (100% specificity).
All patients with culture-proven tuberculosis had at least one gastric culture–positive specimen (diagnostic sensitivity by patient, 100%), whereas 20/28 gastric aspirate samples from these patients were culture positive versus 9/27 nasopharyngeal aspirate cultures (sensitivity by diagnostic sample 71% versus 31%, respectively, P = 0.01). Gastric aspirates were not collected from controls, but all of the 28 nasopharyngeal cultures from the 23 controls were culture negative (100% specificity).
PCR assays were performed on both undiluted stools and stools diluted 1:5 in aqueous solutions, as described above. For both the Chelex and Fast-DNA extraction sample products, the 1:5 diluted samples had greater sensitivity than the undiluted specimens, yielding all but one of the positive PCR results (5/28 [18%] by Chelex and 8/28 [29%] by Fast DNA). The DNA extracts from the Chelex and Fast-DNA undiluted samples were less adequate for PCR than the diluted DNA extracts, yielding positive results in 4/28 (14%) and 5/28 (18%) specimens, respectively.
Table 3 shows clinical risk factors associated with a positive stool PCR result. Stool PCR was significantly more likely to be positive in samples from tuberculosis patients with cough for > 2 weeks, patients with a culture-positive paired specimen (stool, gastric aspirate, or nasopharyngeal aspirate), and patients with an auramine stain–positive paired stool sample.
Table 3.
Logistic regression of factors associated with a positive PCR result
| Stool sample PCR (+) (N = 11) |
Stool sample PCR (−) (N = 17) |
Odds ratio | P value | |
|---|---|---|---|---|
| Samples from children with positive tuberculin skin test | 8 | 17 | 0 95% confidence interval [0–0.71] |
0.03 |
| Sample from children with cough for > 2 weeks | 8 | 4 | 8.6 | 0.01 |
| Paired gastric aspirate culture positive | 10 | 10 | 7 | 0.066 |
| Stool sample auramine microscopy positive | 3 | 0 | > 1,000* | 0.02 |
| At least one paired culture-positive specimen (gastric aspirate, nasopharyngeal aspirate, or stool) | 11 | 10 | > 1,000* | 0.01 |
Logistic regression of factors associated with a positive PCR result among 28 stool samples from 16 children with culture-proven pulmonary tuberculosis.
The following factors were also studied but did not approach statistical significance: age (older than 5 years), OR = 1.2, P = 0.8; male sex, Odds ratio (OR) = 1.9, P = 0.5; Stegen Toledo score > 7, OR = 1.45, P = 0.7; suspect radiograph, OR = 0.31, P = 0.3; pulmonary infiltrate on radiograph, OR = 6, P = 0.1; contact with adult tuberculosis patient, OR = 0.6, P = 0.6; nasopharyngeal aspirate auramine microscopy positive, OR = 2.5, P = 0.3; gastric aspirate auramine microscopy positive, OR = 6, P = 0.1; stool culture positive, OR = 3.5, P = 0.3.
Because all stool samples with paired culture-positive gastric aspirate samples were PCR positive and no stool sample that was auramine microscopy positive was stool PCR negative, the odds ratios for these factors approached infinity and therefore cannot be exactly calculated.
DISCUSSION
This study showed that IS6110-PCR with Fast-DNA stool sample processing diagnosed pediatric pulmonary tuberculosis with 38% sensitivity and 100% specificity compared with cultures of multiple samples as the gold standard. Consequently, stool PCR had the same sensitivity as nasopharyngeal aspirate cultures. We compared the Fast-DNA extraction method to Chelex extraction and found no significant difference in sensitivity, although the Fast-DNA tended to perform better. Even though all the patients in this study had at least one culture positive for tuberculosis, not every sample tested positive by culture. Culture and PCR testing of duplicate clinical specimens with multiple laboratory methodologies increased diagnostic sensitivity, suggesting that specimens were close to the threshold of detection sensitivity in this paucibacillary childhood infection.
Diagnosing tuberculosis in stool using a DNA extraction method and hemi-nested IS6110 PCR assay is potentially a useful initial test for tuberculosis in children, especially if sensitivity can be increased. Patients who test positive may avoid multiple invasive procedures. Furthermore, stool PCR took 1 day, implying that some pediatric tuberculosis cases could be identified much more quickly than the several weeks required for tuberculosis culture. It would also be possible to detect rifampicin resistance using the heteroduplex technique that we have reported,11,23 although culture-based testing showed that none of these strains were rifampicin resistant and heteroduplex-PCR was therefore not performed in this study.
This proof of principle study showed that stool PCR has the capacity to diagnose pediatric tuberculosis. Ongoing research aims to increase sensitivity to make stool PCR a valuable initial test for children with suspected pulmonary tuberculosis. The greater sensitivity of diluted compared with undiluted stool samples implies the presence of PCR inhibitors in these clinical samples. The dilution step reduced the adverse effect of inhibitors on PCR sensitivity but will also have decreased the concentration of M. tuberculosis DNA, inevitably reducing diagnostic sensitivity. We are evaluating substituting DNA concentration and purification columns in place of dilution for removing inhibitors while increasing DNA concentration and hence sensitivity.
Studies evaluating pediatric tuberculosis diagnostics are confounded by the inadequacy of gold standard tests and the rarity of proven tuberculosis. We focused on children with culture-proven tuberculosis compared with healthy controls to evaluate stool PCR compared with a definite gold standard. However, 472 children had to undergo extensive studies to define the relatively modest sample sizes involved in the current research. Although this was probably the optimal strategy for defining with certainty the sensitivity and specificity of stool PCR, it should be noted that most children receiving treatment for pulmonary tuberculosis are culture negative and only have an uncertain, presumed diagnosis. Stool PCR positivity would likely be less frequent in clinical practice involving culture-negative and culture-proven tuberculosis, but tuberculosis diagnosis is generally uncertain in these cases, so implications for the true sensitivity of the test are difficult to define for culture-negative patients.
When deciding which DNA extraction method to use, various factors need to be considered. Chelex ($1/sample materials) is considerably less expensive than the Fast-DNA kits ($14/sample kit cost, plus single machine purchase), so if the sensitivity of Chelex extraction could be improved, it would be ideal. Chelex requires a decontamination process, whereas Fast-DNA does not. Chelex extraction was technically simple and rapid, requiring < 2 hours to process 12 samples. The Fast-DNA extraction process required commercial kits and approximately twice as much laboratory time per sample. Fast-DNA extraction tended to provide greater PCR sensitivity, but this difference was not statistically significant, and the relative efficiency of these DNA extraction techniques is the subject of our ongoing research.
Significant predictors that a pediatric tuberculosis patient's stool sample would be PCR positive included the presence of a paired gastric aspirate or stool sample that was auramine stain positive for mycobacteria. Therefore, stool PCR is more likely to be positive in patients with microbiological evidence of increased bacillary load. In many pediatric patients, M. tuberculosis may never enter the bronchi but instead remain in the lymph node.24 Children with prolonged cough were significantly more likely to have a positive stool PCR sample, probably because they are more likely to have infection in their bronchus and therefore are more likely to produce sputum that will then be swallowed. Children with a negative tuberculin skin test were more likely to have a positive stool PCR sample, probably because an anergic tuberculin skin test response may indicate more severe tuberculosis with higher bacillary load.25
A limitation of this study is the small sample population because of the small proportion of children with suspected tuberculosis who had a culture-proven diagnosis. However, this study involved 472 children, one half of whom had suspected pulmonary tuberculosis and underwent extensive invasive testing with > 2,000 laboratory cultures to identify this number of culture-proven cases. This indicates the low yield of culture confirmation of tuberculosis in children and emphasizes the urgent need for new, more sensitive diagnostic tests despite the large trials required for their evaluation. Other limitations include the fact that two stool samples were not collected from every child with a positive upper gastric-intestinal aspirate and that it was not always possible to collect the stool sample on the same day as the upper gastric-intestinal aspirate was performed. For diagnosing pediatric tuberculosis, two gastric aspirates are usually taken on consecutive days to increase sensitivity. This practice was supported by our study, which also suggested that if two stool samples were collected from every patient, the sensitivity of the stool PCR test would improve. The stool samples used in this study had been stored frozen at −20°C for ~2 years, which may have reduced the quantity or integrity of mycobacterial DNA, potentially reducing the sensitivity of the PCR tests. Although the specificity was 100%, the number of control samples was relatively limited, and a larger sample size would be required to confidently determine true specificity.
In conclusion, stool PCR is a specific, moderately sensitive technique for the diagnosis of pulmonary tuberculosis in culture-positive pediatric patients and has potential use as a rapid, non-invasive preliminary test for pediatric tuberculosis. The DNA extraction and PCR techniques are being refined to increase diagnostic sensitivity. Because of the high specificity, a positive stool PCR may allow clinicians to rapidly identify some children highly likely to have culture-positive pulmonary tuberculosis and initiate treatment at an earlier stage.
Acknowledgments
We thank the numerous professional colleagues whose generous collaboration made this work possible and the patients who kindly agreed to give informed consent to provide gastric aspirates, nasopharyngeal aspirates, and stool samples in the interests of medical progress.
Financial support: This study was funded by NIH Project 1 RO1 AI49139-01A1, The National Institute of Allergy and Infectious Diseases; Wellcome Trust Clinical Fellowships in Tropical Medicine; the Global Research Training grant; and the charity Innovation For Health And Development (IFHAD).
Footnotes
Disclosure: The authors declare that they have no conflict of interests in relation to this work.
REFERENCES
- 1.WHO-WHO/TUB . Guidelines for Tuberculosis Treatment in Adults and Children in National Tuberculosis Programs. Tuberculosis Unit Division of Communicable Diseases. Third edition World Health Organization; Geneva: 2003. [Google Scholar]
- 2.WHO . Tuberculosis. World Health Organization; Geneva: 1995. Fact sheet No. 93. [Google Scholar]
- 3.Khan EA, Starke JR. Diagnosing tuberculosis in children: increased need for better methods. Emerg Infect Dis. 1995;1:115–123. doi: 10.3201/eid0104.950402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberhelman RA, Soto-Castellares G, Caviedes L, Castillo ME, Kissinger P, Moore DA, Evans C, Gilman RH. Improved recovery of Mycobacterium tuberculosis from children using the microscopic observation drug susceptibility method. Pediatrics. 2006;118:e100–e106. doi: 10.1542/peds.2005-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zar HJ, Hanslo D, Apolles P, Swingler G, Hussey G. Induced sputum versus gastric lavage for microbiological confirmation of pulmonary tuberculosis in infants and young children: a prospective study. Lancet. 2005;365:130–134. doi: 10.1016/S0140-6736(05)17702-2. [DOI] [PubMed] [Google Scholar]
- 6.Schoch OD, Rieder P, Tueller C, Altpeter E, Zellweger JP, Rieder HL, Krause M, Thurnheer R. Diagnostic yield of sputum, induced sputum, and bronchoscopy after radiologic tuberculosis screening. Am J Respir Crit Care Med. 2007;175:80–86. doi: 10.1164/rccm.200608-1092OC. [DOI] [PubMed] [Google Scholar]
- 7.Getchell WS, Davis CE, Gilman J, Urueta G, Ruiz-Huidrobo E, Gilman RH. Basic epidemiology of tuberculosis in Peru: a prevalence study of tuberculin sensitivity in a pueblo joven. Am J Trop Med Hyg. 1992;47:721–729. doi: 10.4269/ajtmh.1992.47.721. [DOI] [PubMed] [Google Scholar]
- 8.Sanghavi DM, Gilman RH, Lescano-Guevara AG, Checkley W, Cabrerea L, Cardenas V. Hyperendemic pulmonary tuberculosis in a Peruvian shantytown. Am J Epidemiol. 1998;148:384–389. doi: 10.1093/oxfordjournals.aje.a009657. [DOI] [PubMed] [Google Scholar]
- 9.Ampofo KK, Saiman L. Pediatric tuberculosis. Pediatr Ann. 2002;31:98–108. doi: 10.3928/0090-4481-20020201-07. [DOI] [PubMed] [Google Scholar]
- 10.Smith KC, Starke JR, Eisenach K, Ong LT, Denby M. Detection of Mycobacterium tuberculosis in clinical specimens from children using a polymerase chain reaction. An Pediatr (Barc) 1996;97:155–160. [PubMed] [Google Scholar]
- 11.Montenegro SH, Gilman RH, Sheen P, Cama R, Caviedes L, Hopper T, Chambers R, Oberhelman RA. Improved detection of Mycobacterium tuberculosis in Peruvian children by use of a heminested IS6110 polymerase chain reaction assay. Clin Infect Dis. 2003;36:16–23. doi: 10.1086/344900. [DOI] [PubMed] [Google Scholar]
- 12.Neu N, Saiman L, San Gabriel P, Whittier S, Knirsch C, Ruzal-Shapiro C, Della-Latta P. Diagnosis of pediatric tuberculosis in the modern era. Pediatr Infect Dis. 1999;18:122–126. doi: 10.1097/00006454-199902000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Dalovisio JR, Montenegro-James S, Kemmerly SA, Genre CF, Chambers R, Greer D, Pankey GA, Failla DM, Haydel KG, Hutchinson L, Lindley MF, Nunez BM, Praba A, Eisenach KD, Cooper ES. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens. Clin Infect Dis. 1996;23:1099–1106. doi: 10.1093/clinids/23.5.1099. [DOI] [PubMed] [Google Scholar]
- 14.Delacourt C, Poveda JD, Chureau C, Beydon N, Mahut B, de Blic J, Scheinmann P, Garrigue G. Use of polymerase chain reaction for improved diagnosis of tuberculosis in children. J Pediatr. 1995;126:703–709. doi: 10.1016/s0022-3476(95)70396-9. [DOI] [PubMed] [Google Scholar]
- 15.Stegen G, Jones K, Kaplan P. Criteria for guidance in the diagnosis of tuberculosis. Pediatrics. 1969;43:260–263. [PubMed] [Google Scholar]
- 16.Franchi LM, Cama RI, Gilman RH, Montenegro-James S, Sheen P. Detection of Mycobacterium tuberculosis in nasopharyngeal aspirate samples in children. Lancet. 1998;352:1681–1682. doi: 10.1016/s0140-6736(05)61454-7. [DOI] [PubMed] [Google Scholar]
- 17.Kent BD, Kubica GP. Public Health Mycobacteriology: A Guide for the Level III Laboratory. US Dept. Health and Human Services, Centers for Disease Control; Atlanta: 1985. pp. 36–39. 47–69, 185–187. [Google Scholar]
- 18.Caviedes L, Lee TS, Gilman RH, Sheen P, Spellman E, Lee EH, Berg DE, Montenegro-James S. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. J Clin Microbiol. 2000;38:1203–1208. doi: 10.1128/jcm.38.3.1203-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore DA, Evans CA, Gilman RH, Caviedes L, Coronel J, Vivar A, Sanchez E, Piñedo Y, Saravia JC, Salazar C, Oberhelman R, Hollm-Delgado MG, LaChira D, Escombe AR, Friedland JS. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355:1539–1550. doi: 10.1056/NEJMoa055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Getchell WS, Davis CE, Gilman J, Urueta G, Ruiz-Huidobro E, Gilman RH. Basic epidemiology of tuberculosis in Peru: a prevalence study of tuberculin sensitivity in a pueblo joven. Am J Trop Med Hyg. 1992;47:721–729. doi: 10.4269/ajtmh.1992.47.721. [DOI] [PubMed] [Google Scholar]
- 21.Salazar GE, Schmitz TL, Cama R, Sheen P, Franchi LM, Centeno G, Valera C, Leyva M, Montenegro-James S, Oberhelman R, Gilman RH, Thompson MJ, Working Group on TB in Peru Pulmonary tuberculosis in children in a developing country. Pediatrics. 2001;108:448–453. doi: 10.1542/peds.108.2.448. [DOI] [PubMed] [Google Scholar]
- 22.Escalante P, Ramaswamy S, Sanabria H, Soini H, Pan X, Valiente-Castillo O, Musser JM. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tubercle Lung Dis. 1998;79:111–118. doi: 10.1054/tuld.1998.0013. [DOI] [PubMed] [Google Scholar]
- 23.Williams DL, Spring L, Gillis TP, Salfinger M, Persing DH. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin Infect Dis. 1998;26:446–450. doi: 10.1086/516313. [DOI] [PubMed] [Google Scholar]
- 24.Leung AN, Muller NL, Pineda PR, FitzGerald JM. Primary tuberculosis in childhood: radiographic manifestations. Radiology. 1992;182:87–91. doi: 10.1148/radiology.182.1.1727316. [DOI] [PubMed] [Google Scholar]
- 25.American Thoracic Society Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med. 2000;161:1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]