Abstract
Poly(ADP-ribosyl)ation represents an important post-translational modification in higher eukaryotes. Several DNA repair/checkpoint proteins possess specific PAR-Binding Zinc finger (PBZ) modules critical for function. Here, we present solution structures of the two PBZ modules of APLF (Aprataxin and PNK-like factor), revealing a novel type of zinc finger. By combining in vivo PAR-binding data with NMR interaction data using PAR fragments, we suggest a structural basis for PBZ-PAR recognition.
Poly(ADP-ribose) polymerases (PARPs) catalyze the post-translational modification of proteins by the addition of chains of poly(ADP-ribose), or PAR. PAR is a highly negatively charged, branched-chain polymer formed from ADP-ribose units derived from NAD and linked via glycosidic ribose-ribose bonds. PAR is critical for many processes including DNA repair, regulation of chromosome structure, transcriptional regulation, mitosis and apoptosis, but its role is best understood in regulation of DNA repair.1-3 Many DNA damage response proteins have specific PAR binding modules, and to date three types have been described. The “macro domain” occurs in several PARP family members and histones4,5 and has been proposed to bind PAR chain termini.6 XRCC1, histones and p53 contact PAR through a short motif rich in hydrophobic and basic residues.7 Recently, a third class was identified, comprising a short, conserved zinc-dependent module termed PBZ for Poly(ADP-ribose)-Binding Zinc finger,8 and found exclusively in DNA repair and checkpoint proteins. These include APLF, a novel DNA damage response protein,9 and CHFR, which functions in the antephase checkpoint and is frequently mutated in human epithelial cancers.10 In both cases, ablation of the PBZ modules destroys function.
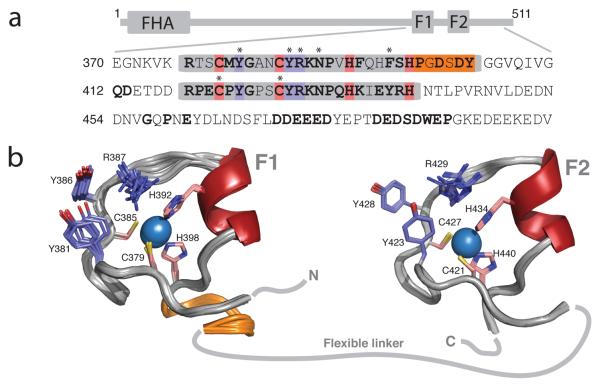
APLF contains two tandem PBZ repeats,8 here called F1 and F2 (Fig. 1). Although its function in DNA repair has yet to be defined, APLF accumulates at DNA breaks in a manner dependent both on its PBZ modules and active PAR synthesis catalyzed by PARP-1 (the most abundant PARP enzyme, which is associated with and strongly activated by DNA breaks).
Figure 1.
Structures of the PBZ modules of APLF. (a) Domain architecture and partial sequence of human APLF. The PBZ modules (F1 and F2) are boxed, metal-binding residues are pink, key PAR-binding residues violet and the additional loop of F1 orange. Highly conserved residues are bold (c.f. Supp. Fig. 11) and asterisks indicate residues mutated in our study. (b) Solution structures of APLF F1 and F2, coloured as in (a) with zinc ions as blue spheres and helices dark red; the 10 lowest energy structures are shown. Structural statistics appear in Supp. Table 1 and Supp. Figure 2.
We determined the solution structure of human APLF residues 364-451, which contains both PBZ modules and possesses comparable PAR-binding activity to wild-type APLF (Fig 1; Supp. Figs. 1-3 and Supp. Table 1). The structured regions comprise approximately residues 376-404 (F1) and 418-440 (F2). The two modules both consist of a series of loops surrounding the central zinc ion, each coordinated by two Cys and two His residues (Cys379, Cys385, His392 and His398 in F1; Cys421, Cys427, His434 and His440 in F2). The presence of two zinc ions was confirmed by mass spectroscopy (Supp. Fig. 4). Secondary structure is largely absent, comprising only a short helix near the third metal binding ligand. Aside from those of the metal-binding residues, very few sidechains are directed inwards, the only other principle contribution to the core being a conserved aromatic (Phe396/Tyr438) in the helix. The first module differs in possessing an additional structured loop at its C-terminal end, comprising residues Pro399-Tyr404, that folds back across one face of the module, stabilised mainly by contacts involving Tyr404, Ser378 and Pro399. However, within the common part of the structure both modules are very similar (backbone rmsd between F1 377-398 and F2 419-440 is 0.82Å). The two fingers were found to be structurally independent in solution, as demonstrated by the absence of any interfinger NOE interactions and the unrelated alignment tensors found for each in experiments to measure residual dipolar couplings (Supp. Fig. 5).
Since PAR itself is not conveniently available in sufficient quantity or homogeneity for structural work, we investigated its binding to the APLF PBZ modules by using fragments of PAR and related ligands. Such ligands bind much more weakly than PAR polymer, having lower local concentrations, and only target one finger at a time, but should nonetheless reveal key features of PAR recognition. We used commercially available compounds (nucleotide monophosphates and ADP ribose, here abbreviated ADPR), but none of these contain the characteristic α(1 → 2) O-glycosidic bond between ribose rings that is formed by PARP-1 and is the most unique characteristic of PAR structure. To overcome this limitation, we followed a recently published synthesis to obtain 2′-O-α-D-ribofuranosyladenosine (here abbreviated RFA), the simplest adenosine derivative to contain such a bond (Fig. 2a).11
Figure 2.
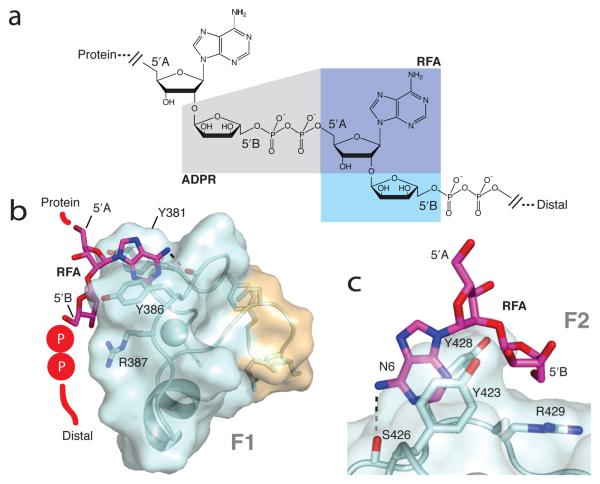
Structures of APLF PBZ modules complexed with RFA. (a) Structure of PAR and fragments; ADPR is the fragment within the grey area and RFA is that within the blue area (dark blue being common to both). Structures of RFA bound to (b) APLF F1 and (c) APLF F2. In (b) the RFA molecule is schematically extended to show how PAR may bind; (c) shows a close up of key interactions (including H-bond adenosyl NH2 – S426 O, dotted line).
In combination, chemical shift changes and NOE interactions measured with these ligands defined key features of PAR recognition (Supp. Figs. 6-7), enabling us to calculate co-structures of F1 and F2 bound to RFA (Fig. 2, Supp. Fig. 8 and Supp. Table 2). Essentially equivalent results were observed for F1 and F2 in all experiments, showing there is no large affinity difference at least for the fragments tested. The adenine ring of PAR stacks with the ring of the highly conserved surface tyrosine (Tyr381/Tyr423), and makes a weaker edge-face interaction with Tyr386 (Tyr428 in F2). The adenosyl 6-amino group probably forms a hydrogen bond to a protein backbone carbonyl (Asn384/Ser426). Contacts occur to both ribose rings of RFA, but not to ribose A of ADPR. In contrast, the C5′OH group of ribose B of RFA is positioned close to the arginine sidechain of the highly conserved CYR motif that is a defining characteristic of the PBZ module (Arg387/Arg429), strongly suggesting that the pyrophosphate linkage that would extend from this position in PAR may be recognised by this arginine. Thus, we propose that the two key elements of PAR that probably interact with a PBZ module, an adenine ring and a pyrophosphate group, are recognised only when they occur on opposite sides of an intervening α(1 → 2) O-glycosidic bond, thereby rationalising how recognition is specific to PAR (Fig. 2).
Although only monomer-level fragments of PAR were available for our structural work, limited evidence was observed for a second, weaker adenine-binding site involving Phe396 (Supp. Figs. 7 and 9). Modeling calculations of APLF F1 and a 2-unit PAR fragment showed that a second site could be occupied simultaneously with the first by successive monomers in PAR polymer, and probably both sites share common features (Supp. Fig. 9); in particular, a second adenine of PAR could stack with Phe396 while a second pyrophosphate group could be recognised by Arg376 (mutation of which abolishes binding8 and which is highly conserved), again resulting in simultaneous recognition of an adenine and pyrophosphate separated by an intervening glycosidic bond.
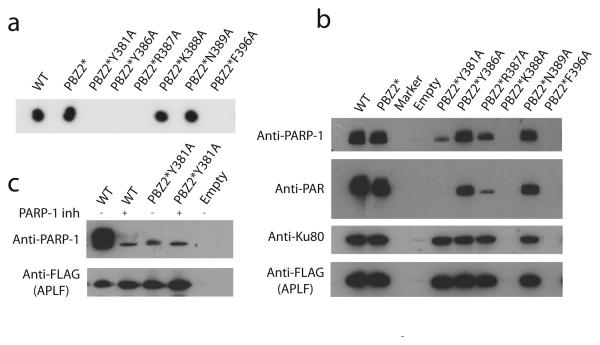
The key residues involved in PAR recognition were tested in a mutational analysis for F1 (in the context of inactivated F2), both in vitro using purified His-tagged proteins and a dot blot assay and in vivo using a FLAG-tag antibody pull-down. Consistent with our structural model, PAR binding is abolished upon mutation of Tyr381 or Arg387 (Figs. 3a,b), while for Tyr386 binding is abolished in vitro and reduced in vivo; presumably Tyr386 is somewhat more dispensable for binding than the others. In contrast, PAR binding is largely unaffected by mutation of Lys388 or Asn389 (Fig. 3a), while mutation of Phe396 probably causes misfolding, preventing assessment of its contribution to binding. Overall, both sequence conservation and mutational analysis are in full agreement with involvement of the residues we identify for PAR binding.
Figure 3.
Interactions of APLF with PAR and PARP-1 are mediated by PBZ. (a) Interactions of wild-type and PBZ-mutated APLF proteins in vitro as determined by dot-blot analysis. (b) Association of wild-type and PBZ-mutated Flag-tagged APLF with PAR in vivo. (c) PBZ mediated stabilization of APLF:PARP-1 interaction is dependent on active PAR synthesis.
Despite their similar C2H2 zinc coordination, PBZ modules are both structurally and functionally dissimilar from canonical dsDNA-binding TFIIIA-type zinc fingers. Rather, PBZ modules resemble ssRNA binding C3H1 Tandem Zinc Fingers (TZF) such as those of TIS11d and MBNL1.12,13 Both PBZ and TZF fingers largely lack secondary structure, but nonetheless have rigid backbone conformations due to the zinc coordination. For both, base recognition is achieved through mainchain-base hydrogen bonding and stacking interactions by highly conserved, solvent exposed aromatic residues; indeed, if PBZ and TZF structures are superposed using their zinc-binding atoms in a permuted order, their structures and base-interacting residues overlay closely (Supp. Fig. 10). However, while TZFs recognize specific ssRNA sequences, the PBZs of APLF recognize unique structural characteristics of PAR, which has features both of an oligonucleotide and an oligosaccharide.
Unlike ssRNA, PAR is linked covalently to proteins (e.g. PARP-1, histone tails and p53), and recognition can involve both PAR-mediated and PAR-independent components. Interestingly, our data show also that the previously described APLF:PARP-1 interaction8,14 probably comprises both, since abrogation of PAR binding either by mutation of Tyr381 and F2 or addition of PARP inhibitors reduces PARP-1 binding to a basal level, but not to zero (Fig 3c). These results also demonstrate that PAR-dependent stabilization of the PARP-1 interaction is fully mediated by PBZs. Others have reported that the PARP-1 interaction is specific to F1 of APLF;14 as the additional loop in F1 is the main structural difference between the fingers, it might account for this. As well as binding PAR, the zinc-finger region of APLF may bind proteins. The additional loop in F1 is very highly conserved, distant from the proposed PAR-binding site, and includes two aspartates, and there is also a very highly conserved DE-rich region near the C-terminus (residues 470-487; Supp. Fig. 11) reportedly implicated in aspects of APLF function;15 NMR shows the latter is largely or wholly unfolded (Supp. Fig 5). Since both regions are highly conserved but unlikely to be involved in PAR binding, they could represent protein-protein interaction sites.
In conclusion, we have determined structures for both PBZ motifs of APLF and suggested how specific recognition of PAR is achieved; this represents the first structural insight into how PBZ motifs function in nuclear signalling. Given the unusual chemical composition of PAR and its role in the cellular stress response, inhibition of PAR recognition might provide an interesting alternative to PARP-1 inhibition for future approaches to cancer therapy.
Accession codes
Atomic co-ordinates are deposited at Protein Data Bank for free F1 and F2 (codes 2kqb and 2kqc respectively), and the F1-RFA and F2-RFA complexes (codes 2kqd and 2kqe respectively). Assignments of 1H, 13C, 15N chemical shifts for free APLF 368-451 are deposited at BioMagResBank (code 16596).
Supplementary Material
Acknowledgments
We thank A. Murzin, A. Andreeva and C. Rademacher for helpful discussions, G. Smith (AstraZeneca) for the PARP inhibitor KU-0058948, W. Vranken for help with data deposition and C. Oubridge for help with mass spectroscopy. S.E. was funded by a studentship from Boehringer Ingelheim Fonds, C.B. by a fellowship from F.E.B.S., and I.A. by fellowships from Cancer Research UK and the Louis-Jeantet Foundation.
REFERENCES
- 1.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Biochem J. 1999;342(Pt 2):249–68. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MY, Zhang T, Kraus WL. Genes Dev. 2005;19:1951–67. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber V, Dantzer F, Ame JC, de Murcia G. Nat Rev Mol Cell Biol. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 4.Karras GI, et al. E.M.B.O. J. 2005;24:1911–20. [Google Scholar]
- 5.Ahel D, et al. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timinszky G, et al. Nat Struct Mol Biol. 2009;16:923–9. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 7.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. J Biol Chem. 2000;275:40974–80. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 8.Ahel I, et al. Nature. 2008;451:81–5. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 9.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. Mol Cell Biol. 2007;27:3793–803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scolnick DM, Halazonetis TD. Nature. 2000;406:430–5. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 11.Mikhailov SN, Kulikova IV, Nauwelaerts K, Herdwijn P. Tetrahedron. 2008;64:2871–2876. [Google Scholar]
- 12.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Nat Struct Mol Biol. 2004;11:257–64. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 13.Teplova M, Patel DJ. Nat Struct Mol Biol. 2008;15:1343–51. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. DNA Repair (Amst) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW. Mol Cell Biol. 2008;28:4620–8. doi: 10.1128/MCB.02243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.