Abstract
The advent of sequencing and structural genomics projects has provided a dramatic boost in the number of protein structures and sequences. Due to the high-throughput nature of these projects, many of the molecules are uncharacterised and their functions unknown. This, in turn, has led to the need for a greater number and diversity of tools and databases providing annotation through transfer based on homology and prediction methods. Though many such tools to annotate protein sequence and structure exist, they are spread throughout the world, often with dedicated individual web pages. This situation does not provide a consensus view of the data and hinders comparison between methods. Integration of these methods is needed. So far this has not been possible since there was no common vocabulary available that could be used as a standard language. A variety of terms could be used to describe any particular feature ranging from different spellings to completely different terms. The Protein Feature Ontology (http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=BS) is a structured controlled vocabulary for features of a protein sequence or structure. It provides a common language for tools and methods to use, so that integration and comparison of their annotations is possible. The Protein Feature Ontology comprises approximately 100 positional terms (located in a particular region of the sequence), which have been integrated into the Sequence Ontology (SO). 40 non-positional terms which describe general protein properties have also been defined and, in addition, post-translational modifications are described by using an already existing ontology, the Protein Modification Ontology (MOD). The Protein Feature Ontology has been used by the BioSapiens Network of Excellence, a consortium comprising 19 partner sites in 14 European countries generating over 150 distinct annotation types for protein sequences and structures.
Introduction
New Data
Genome sequencing has elucidated the locations of many genes on more than 700 genomes (1). Understanding human variation and disease requires knowledge of the role of each amino acid in a protein and how mutations or alternative splicing events can change function and phenotype. In addition, structural genomics initiatives are providing a wealth of new structural information. Protein structure is more conserved than sequence during evolution, with about 600 experimentally determined structural models released by the PDB each month. Many of these proteins have unknown functions, but more distant evolutionary links can be identified between proteins, ultimately revealing more about the characteristics of a protein family.
Prediction and Computational Methods
The development of tools for comparisons between structure and sequence has increased rapidly, since these automatic methods are crucial in order to fill in the functional space between characterised and uncharacterised protein sequences and structures. Any knowledge we have of a protein is attached to the sequence and structure through annotation. Tools for the annotation of these data by automatic methods or the manual annotation of experimental data from the literature has become increasingly important (2). Many computational biology laboratories specialise in different aspects of proteome annotation for a range of features and processes: modifications (phosphorylation, lipidation etc), secondary structure prediction methods, fold recognition, effects of alternative splicing, single nucleotide polymorphisms (SNPs), domain and function assignment for catalytic residues, metal and ligand binding residues, protein–protein interactions and protein-DNA interfaces.
Challenges in Accessing Annotations
We now have a host of computational methods and tools available to help us learn more about sequences and structures. However these tools and databases are spread throughout the world and are numerous, often with more than one method annotating a similar feature. The potential user finds it hard to query them all and compare the results across different methods. At best, the user must traverse multiple websites, using a click and drag approach. The nature of bioinformatics also means that these tools can change rapidly as the software is developed to include cutting edge research findings and favoured web servers may even change location. More knowledge can be gained by combining and comparing annotations from these many sources. This inherently relies on the organisation and presentation of the data displayed, and consistency is crucial. An effective ‘first step’ for dealing with such a problem is to develop an ontology of protein features. An ontology is a standardised set of structured and precisely defined terms and relationships, providing a platform for both manual and automated reasoning in a dynamic environment so that changes can occur as different uses arise and new terms added. From this, annotations which adopt the ontology can be integrated into a single location, aiding comparisons between them.
The Gene Ontology resource (The Gene Ontology Consortium, 3,4) has been developed to describe gene and gene product attributes. This collaborative resource was set up to provide consistent descriptions of gene products that can be used by different databases annotating different species. There are three categories; biological process describing the biological objective of the gene or the gene product, molecular function, describing the biochemical activity and cellular component referring to the place in the cell where the gene product is active. The Sequence Ontology (5) has been developed to facilitate exchange, analysis and management of genomic annotation data. This standard has been used to underpin the features stored in the sequence databases of model organisms (6) and to standardize the annotation exchange formats (www.sequenceontology.org/gff3.shtml). It is used by many of the model organism communities to annotate their sequence features, such as FlyBase (7), WormBase (8), DictyBase (9) and SGD (10).
Ontologies have also been created for other aspects of biology such as the MOD ontology for post-translational modifications (11), PSI-MI for molecular interactions (12) and the Pathway ontology (13) as well as a number of specialist database ontologies (7,14-17). There is also an initiative to create an all-encompassing ‘protein ontology’ by the Protein Ontology (PRO) Consortium (18). This ontology will model all aspects of proteins from their evolution to their form and function. This project is still in its infancy and so far provides an ontological description (from bottom up) of protein modifications, sequence forms (including alternative splicing, mutant forms, cleaved and post-translationally modified products), the whole protein unit, detected sequence domains, structural domains, and the evolutionary unit. This top level ontology is created by linking many resources which already exist, including the Gene Ontology, the protein modification ontology PSI-MOD, and the human disease ontology.
The Protein Feature Ontology
This ontology was created in order to provide a method for the comparison of annotations of protein features. The terms that are commonly used need to be standardised so that similar terms can be identified and compared. Inconsistencies such as different spellings, casings and most importantly, a range of synonymous names are often used for the same annotation. For example, a predicted transmembrane segment of peptide is annotated as both TRANSMEM (http://phobius.binf.ku.dk/) and Membrane (http://www.cbs.dtu.dk/services/) by different servers. A domain annotation is traditionally indicated by the name of the method which provided it, for example, InterPro (19) provides domain annotations such as SMART, ProDom, SCOP rather than the annotation domain. These annotations clearly make sense in their own contexts but when annotations are brought together from a number of sources, a more uniform approach needs to be adopted to provide effective comparison.
Such a project of integration has been undertaken by the BioSapiens Network of Excellence (The BioSapiens Network of Excellence, 200520), a consortium comprising 19 participating partners from bioinformatics laboratories in 14 European countries. A main goal of their work is to bring together annotations created by their in-house methods and algorithms in order to create a European ‘virtual institute of annotations’. These annotations are derived from methods of manual annotation and informatics tools, resulting in a set of protein/nucleic acid sequence and structural annotations from some of the leading bioinformatics laboratories in the world. Access to this information has been achieved technically through the implementation of a distributed annotation system (DAS (21)). This system comprises both a central reference server (in the case of protein annotations serving UniProt Knowledgebase (UniProtKB) sequences) and individual annotation servers which provide the annotations for sequences held in the central reference server. This information is then interpreted by a DAS client which reads the sequence and the annotations and displays the information in a human readable format. This allows the annotations from each partner site to remain under the control of the partner but to be co-ordinated and instantly brought together at a central point. Three main DAS clients exist; the Ensembl genome browser (22) for both genomic and proteomic annotations, Dasty2 (Jimenez, R. in press) for protein sequence annotations and Spice (23) for both protein sequence and structural annotations. The beauty of this method is that individual sites control their own DAS sources and therefore it is open to all to participate regardless of location or agreement. At present, the BioSapiens Network of Excellence has collected 40 different distributed annotation sources for protein sequence and structure providing over 150 annotation types. This method of collecting and centralising the display of annotations from disparate laboratories across Europe is unprecedented and as such, provides a unique data source as well as a central platform for participating laboratories to display their data. However, with the control of the information lying with each partner site, a lack of consistency in the annotation terms provided by each source has evolved. The adoption of a controlled vocabulary of protein features would not only allow ‘like’ annotations from different servers to be identified and viewed together on the clients, but also enable complex manipulation of these data both manually and automatically, deriving relationships between annotation methods and enabling the creation of intra-method consensus tracks.
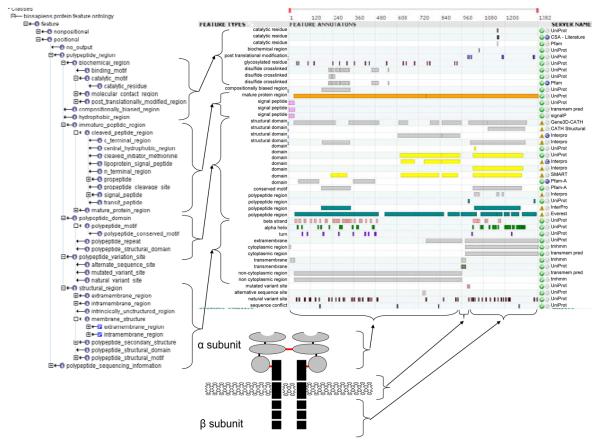
The Protein Feature Ontology is a set of terms which describe the features which make up protein function and form (Figure 1), from features describing the local structure such as residues involved in disulfide bonds, helices, strands and motifs (such as the helix-turn-helix motif) to overall tertiary structure marking a globular domain. Functional residues such as those which are important for signalling or catalysis can be annotated as well as those in contact with ligands or involved in protein interactions. The scope of this ontology is illustrated in Figure 1 which shows selected DAS tracks annotating features on the alpha and beta subunits of the insulin receptor. The protein feature ontology serves to provide a uniform description of the features and properties of a protein. In addition to this, each track is provided with an evidence code (http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=ECO) to display provenance (also shown on Figure 1).
Figure 1.
Selected DAS tracks annotating features on the alpha and beta subunits of the insulin receptor. The tracks are showed using the Dasty2 client. Evidence codes are listed as: inferred by curator (IC), inferred from sequence similarity (ISeqS), inferred from structural similarity (ISS), inferred from electronic annotation (IE), inferred from motif similarity (IM).
The ontology is divided into two parts: Non-positional terms which refer to the whole protein sequence or structure and Positional terms which refer to a specific residue or range of residues in the protein. For positional terms, features are located using sequence residue numbers and the properties of these features describe an attribute of the feature, for example, residues 5-130 are described as a domain. Figure 1 provides an illustration of these types of terms. Other terms are not associated with a particular region of the sequence (non-positional), but instead provide a description of the properties of the whole protein such as an associated publication or links to related sources of information such as GO term annotation or EC annotation. The protein feature ontology currently comprises approximately 140 terms: 100 positional terms and 40 non-positional terms.
Positional Annotations
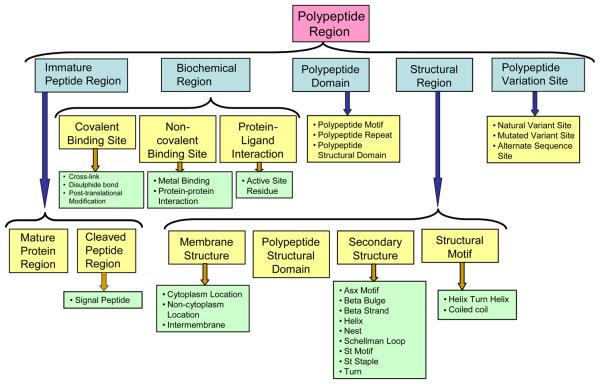
Positional terms (Figure 2) describe features that can be “associated” with a particular region in the peptide. These annotations are derived from programs or methods which detect features on the protein sequence or structure. Examples of such features include the specific role of an amino acid residue such as its catalytic activity, involvement in the binding of a metal ion, an indication of the location within the cell such as intramembrane, or the structural conformation of the residue range such as the alpha-helix. These terms fall within the scope of the Sequence Ontology and as a result, these terms have been integrated into the SO. The Protein Feature Ontology was created in collaboration with UniProtKB and the UniProtKB Feature Types were used as the starting point. All UniProtKB feature types exist in the ontology, but in order to fit in with the Sequence Ontology naming schemes, some term names have been modified. For example, the word polypeptide has been added to ontology terms to disambiguate them from more general terms in SO, which includes a wider selection of features. Motif becomes polypeptide_motif. A mapping between the ontology and the UniProtKB feature types is being maintained so that it is possible to automatically map between them. The mapping is also maintained in the SO synonyms.
Figure 2.
Representing child terms in the parent category polypeptide_region. This category contains both structural and functional feature annotations. A large number of tools and databases provide annotations which fall into this parent category and as a result will be able to be viewed and compared together.
The 3 sections in SO that are populated by positional protein feature terms are:
- Polypeptide_region: A continuous sequence or single residue in a reference/mature protein sequence. Within this category lie a number of terms:
- Biochemical_region, amino acids involved in binding, interactions, catalysis or peptide bonds (which are represented by the positions of the two flanking amino acids).
- Polypeptide_domain, describing a structurally or functionally defined protein region which has been shown to recur throughout evolution. In order to distinguish further, the term polypeptide_domain has been further categorised into three child terms. Two UniProtKB feature types have been classified here: polypeptide_motif indicating a short (up to 20 amino acids) region which is conserved in different proteins and polypeptide_repeat which indicates internal sequence repetition. In addition, the term polypeptide_structural_domain has also been created, allowing the difference between a structural domain (a structure which is self-stabilising and folds independently from the rest of the protein chain) and the parent term (polypeptide_domain) to be distinguished. This allows for the term polypeptide_structural_domain to also exist within the structural_region branch of the ontology so that annotations can be clustered and potentially viewed with secondary structural and membrane structure features.
- Also within this category is the term immature_peptide_region - the extent of the peptide after it has been translated and before any processing occurs. This is then divided into mature_protein_region, the extent of a polypeptide chain in the mature protein, and cleaved_peptide_region for regions which are cleaved during maturation (including signal_peptide and transit_peptide).
- Structural_region describes the backbone conformation of the polypeptide and includes child terms to describe both secondary structure and the structure of the protein in the membrane.
- Polypeptide_variation_site, indicates alternative sequences due to naturally occurring events such as polymorphisms and alternative splicing or experimental methods such as site-directed mutagenesis.
Polypeptide_sequencing_information: This category clusters annotations which report incompatibility in the sequence due to some experimental uncertainty.
No_output: which allows annotators to report where an analysis has been run and not produced any annotation.
All terms under the parent ‘positional’ are located primarily in SO. It is possible to view and use these terms within SO along with other SO terms or within the Protein Feature Ontology. In addition, the terms in SO can be automatically extracted to create a standalone ontology by filtering in OBO-Edit (24) for the category ‘biosapiens protein feature ontology’.
Non-positional annotations
This section classifies annotations which do not refer directly to a particular feature on the protein sequence or structure but instead refer to the full length of the protein. They typically describe those attributes which would be included in a database entry. For example, a number of methods provide a GO term as output. Each partner would classify their output with the ontology term GO_annotation, allowing the client to cluster these together in the table. Terms within this non-positional category are mainly derived from categories within a UniProt entry. Two main areas are currently covered, Uniprot comments (CC field), keyword categories (KW field) with additional entries to describe taxonomy and publication.
Format/Rules/Naming-conventions/Relationships
Where applicable, UniProtKB feature types have been used as term names. Naming conventions have been used in line with the Sequence Ontology regulations. Terms must be computer readable. Therefore, underscores are used instead of spaces, numbers spelt out and common abbreviations used. No full stops, points, slashes, hyphens or brackets are allowed but common abbreviations are used. All entries are in lower case except for common abbreviations and where there are differences, the US form of spelling is chosen. Synonyms aid ontology searching and there is no limit to the number of synonyms. Here, normal term rules do not apply so that, for example, common abbreviations are spelt out and English spellings can be stated.
The use of the controlled vocabulary allows annotations of the same feature to be viewed side by side, for example, metal binding residues identified from 3D data by PDBsum (25) will be viewed alongside metal binding residues identified by the UniProtKB curators, as both have the ontology term ‘metal_binding’. However, the structure of the ontology also provides information on how this term relates to other terms. With this information, the metal binding residues can be viewed alongside related annotations such as those which describe protein_ligand_interactions and catalytic_residues. The protein feature ontology includes two relationship types; ‘is_a’ (e.g. a helix is_a polypeptide_secondary_structure, a disulfide_bond is_acovalent_binding_site) and ‘part_of’, which indicates when a term forms only a portion of its parent: the extramembrane region is part_of the whole membrane_structure.
Additional terms to describe post-translational modifications
Also falling within the scope of the protein feature ontology are the terms and definitions describing post-translational modifications. An ontology describing this area has already been created (The Protein Modification (MOD) Ontology http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MOD) comprising approximately 1050 terms in more than 45 top-level nodes. The ontology describes alternative hierarchical paths for the classification of protein modifications. These paths describe either the molecular structure of the modification, for example, phosphorylated residue (MOD:00696, a protein modification that effectively substitutes a phosphoryl group for a hydrogen atom) or a description of the amino acid residue that is modified, for example, modified L-tyrosine residue (MOD:00919, a protein modification that modifies an L-tyrosine residue). These terms are inserted into the anchor term post_translational_modification (SO:0001089) in the composite Protein Feature Ontology.
The Open Biomedical Ontologies Community
This project has been undertaken as part of the Open Biomedical Ontologies (OBO) (26) and thus delivers the ontology in OBO format (http://www.geneontology.org/GO.format.obo-1_2.shtml). The protein feature ontology is a directed acyclic graph (DAG) which has been edited using the OBO-Edit ontology manger (24) and can be viewed using the Ontology Lookup Service at the EBI (http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=BS). As an opensource ontology, new terms are currently suggested and reviewed by BioSapiens members as well as the Sequence Ontology community. The current version, including BioSapiens terms, Sequence Ontology terms and Protein Modification terms can be downloaded from the ontology website (http://www.biosapiens.info/ontology). The BioSapiens terminology integrated into SO can also be downloaded from www.sequenceontology.org. Users should note that the BS identifier is given as an alternate ID. Suggestions and comments are welcomed on the tracker http://www.ebi.ac.uk/seqdb/jira/secure/Dashboard.jspa and the SO term tracker http://sourceforge.net/tracker/?group_id=72703. The SO community also provides a mailing list for debate of new terminology at song_devel@lists.sourceforge.net.
Conclusions
We have created an ontology for protein features in order to facilitate integration of protein feature annotations provided by a growing number of methods from around the world. An ontology is a controlled vocabulary composed of types (terms with synonyms) and the relations that hold between them. This allows two things: firstly, a standardisation of the terms that are used, allowing ‘like’ annotations to be identified and, secondly, the relationships allow automatic inferences to be drawn between annotation types. Computer programs will know that the extramembrane region and the intramembrane region are both part_ of the membrane_structure and in turn, they are all structural_regions alongside polypeptide_secondary_structures such as helices and beta_strands, allowing inferences on the exact structure and cellular location to be drawn. An initial use of the protein feature ontology is illustrated by the BioSapiens Network of Excellence. A major goal of the consortium is to provide a ‘virtual centre for annotation’ and as part of this, an ontology is needed on which to base the annotations. The implementation of this ontology will allow the annotations collected by the BioSapiens partners to be clustered and manipulated to provide greater biological meaning.
The ultimate power of this resource derives from its ability to allow combination and comparison of annotations from many sources. This inherently relies on the organisation and presentation of the data displayed, for which consistency is crucial. The creation of this resource provides biologists, biochemists and bioinformaticists with a united view of all available annotations; so that reliability of data/annotations can be better assessed. This tool is available for any integration project and in the future, will be integral to the creation of powerful distributed resources allowing small research groups to upload their data alongside large established databases adding colour and weight to all annotations provided.
Acknowledgements
With grateful thanks for the input from Eugene Kulesha, Andy Jenkinson, all participants of the ontology workshop held in February 2007 and all participants of the BioSapiens Network of Excellence. This work was funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health,’ contract number LHSG-CT-2003-503265′.
References
- 1.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic acids research. 2008;36:D475–479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves GA, Thornton JM. Integrating biological data through the genome. Human molecular genetics. 2006;15:R81–87. doi: 10.1093/hmg/ddl086. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 3.The Gene Ontology project in 2008. Nucleic acids research. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat.Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome biology. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics (Oxford, England) 2007;23:i337–346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 7.Grumbling G, Strelets V. FlyBase: anatomical data, images and queries. Nucleic acids research. 2006;34:D484–488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, Canaran P, Chan J, Chen WJ, Davis P, Fernandes J, et al. WormBase 2007. Nucleic acids research. 2008;36:D612–617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chisholm RL, Gaudet P, Just EM, Pilcher KE, Fey P, Merchant SN, Kibbe WA. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic acids research. 2006;34:D423–427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic acids research. 2004;32:D311–314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montecchi-Palazzi L, B. R, Binz P-A, Chalkley RJ, Cottrell J, Creasy D, Seymour SL, Garavelli JS. Nat. Biotechnol. 2008 doi: 10.1038/nbt0808-864. in press. [DOI] [PubMed] [Google Scholar]
- 12.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon--level 2.5 of the HUPO-PSI format for molecular interactions. BMC biology. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007--easing the path from disease to data and back again. Nucleic acids research. 2007;35:D658–662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avraham S, Tung CW, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, et al. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic acids research. 2008;36:D449–454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee SY, Dickerson J, Xu D. Bioinformatics and its applications in plant biology. Annual review of plant biology. 2006;57:335–360. doi: 10.1146/annurev.arplant.56.032604.144103. [DOI] [PubMed] [Google Scholar]
- 16.Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic acids research. 2008;36:D768–772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic acids research. 2005;33:D390–395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natale DA, Arighi CN, Barker WC, Blake J, Chang TC, Hu Z, Liu H, Smith B, Wu CH. Framework for a protein ontology. BMC bioinformatics. 2007;8(Suppl 9):S1. doi: 10.1186/1471-2105-8-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder N, Apweiler R. InterPro and InterProScan: Tools for Protein Sequence Classification and Comparison. Methods in molecular biology (Clifton, N.J. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 20.BiosSapiens Research networks: BioSapiens: a European network for integrated genome annotation. Eur J Hum Genet. 2005;13:994–997. doi: 10.1038/sj.ejhg.5201470. [DOI] [PubMed] [Google Scholar]
- 21.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spudich G, Fernandez-Suarez XM, Birney E. Genome browsing with Ensembl: a practical overview. Briefings in functional genomics & proteomics. 2007;6:202–219. doi: 10.1093/bfgp/elm025. [DOI] [PubMed] [Google Scholar]
- 23.Prlic A, Down TA, Hubbard TJ. Adding some SPICE to DAS. Bioinformatics (Oxford, England) 2005;21(Suppl 2):ii40–41. doi: 10.1093/bioinformatics/bti1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day-Richter J, Harris MA, Haendel M, Lewis S. OBO-Edit--an ontology editor for biologists. Bioinformatics (Oxford, England) 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA. Enhancing the functional annotation of PDB structures in PDBsum using key figures extracted from the literature. Bioinformatics. 2007;23:1824–1827. doi: 10.1093/bioinformatics/btm085. [DOI] [PubMed] [Google Scholar]
- 26.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]