Abstract
Germ-line mutations in BRCA1 predispose to breast and ovarian cancer. BRCA1-mutated tumors show genomic instability, mainly as a consequence of impaired recombinatorial DNA repair. Here we identify 53BP1 as an essential factor for sustaining the growth arrest induced by Brca1 deletion. Depletion of 53BP1 abrogates the ATM-dependent checkpoint response and G2 cell cycle arrest triggered by the accumulation of DNA breaks in Brca1-deleted cells. This effect of 53BP1 is specific to BRCA1 function, as 53BP1 depletion did not alleviate proliferation arrest or checkpoint responses in Brca2-deleted cells. Importantly, loss of 53BP1 partially restores the homologous recombination defect of Brca1-deleted cells and reverts their hypersensitivity to DNA-damaging agents. We find reduced 53BP1 expression in subsets of sporadic triple-negative and BRCA-associated breast cancers, indicating the potential clinical implications of our findings.
Introduction
BRCA1 and BRCA2 are large phosphoproteins involved in DNA damage repair through homologous recombination (HR)1. Although BRCA1 and BRCA2 share many interacting proteins, they show little homology and are thought to have different roles in HR and other processes2. BRCA1 is thought to be mainly a scaffold protein enabling interactions between different components of the HR machinery whereas BRCA2 is directly involved in loading RAD51 to sites of damage or stalled replication forks3.
Heterozygous BRCA1 and BRCA2 inactivation mutations are associated with an increased risk to develop breast and ovarian cancers. These tumors often show loss of heterozygosity of the wild type allele and mutation of the p53 tumor suppressor4, 5. Breast cancers that arise in BRCA1 mutation carriers are mostly high grade tumors with the so-called triple-negative phenotype, i.e., lacking expression of estrogen receptor (ER) and progesterone receptor (PR) and without amplification of human epidermal growth factor receptor (ERBB2/HER2)6. BRCA1 associated tumors also express basal epithelial cell markers, such as cytokeratin 5/67, and cluster with basal-like breast cancers by gene expression profiling8. There is increasing evidence that a subset of sporadic tumors with a basal-like/triple-negative phenotype may have alterations in BRCA1-related pathways9. In contrast, BRCA2 mutation carriers develop mostly ER-positive breast cancers.
Whereas BRCA1 and BRCA2 function as tumor suppressors in breast and ovarian epithelium, homozygous deletion of BRCA1 or BRCA2 appears not to be tolerated during human or mouse development and in cultured primary cells such as mouse embryonic fibroblasts (MEFs) or stem cells10. Although concomitant deletion of p53 partially alleviates these phenotypes11, the incomplete rescue suggests the involvement of other factors in BRCA1/2 associated cancers. In search for such factors, using a candidate gene approach, knockout of 53BP1 was recently shown by Cao et al to rescue Brca1 hypomorphic MEFs and mice from premature senescence12. 53BP1, a DNA damage response (DDR) factor involved in both HR and non-homologous end joining (NHEJ), is known to be an activator of p5313. However, 53BP1 also has p53 independent functions, and deletion of both 53BP1 and p53 has a synergistic effect on tumor development14, 15.
The findings of Cao et al raise some intriguing questions. First, will 53BP1 ablation also rescue cells completely deficient for BRCA1, a situation that is common in BRCA1 associated tumors? In contrast to Brca1 null mice, the Brca1Δ11/Δ11 hypomorphic mice still express the natural BRCA1-Δ11 splice variant, which contains the conserved RING and BRCT domains10. The Brca1Δ11 allele is functionally active, as demonstrated by the fact that homozygous Brca1Δ11/Δ11 mutants are viable on a p53 heterozygous background16. Other questions concern the mechanism by which deletion of 53BP1 rescues BRCA1-deficient cells and the potential relevance of 53BP1 status for BRCA1 associated cancers.
In this work, we set out to explore these questions. We performed an unbiased transposon mutagenesis screen for factors that could restore normal growth of Brca1 null cells. Similar to the observations with Brca1Δ11 hypomorphic mutants, clonal outgrowth of Brca1 null cells was rescued by a loss of function mutation of 53BP1. We show that cells lacking both BRCA1 and 53BP1 have a partially restored HR pathway. The clinical relevance of these findings is indicated by our data showing that 53BP1 expression is reduced in a subset of basal-like/triple-negative breast cancers and in BRCA1/2 associated breast tumors. These observations suggest a selection for loss of 53BP1 function in a subset of sporadic triple-negative breast cancers and cancers arising in BRCA mutation carriers.
Results
53BP1 loss rescues proliferation defects of Brca1 null cells
Brca1 deletion in p53 proficient normal cells leads to a severe proliferation defect17. Cre/loxP-based conditional Brca1 knockout models would not be useful to screen for factors that enhance growth of BRCA1-deficient cells, since Brca1 deleted cells are rapidly eliminated and the culture is rapidly overtaken by BRCA1-proficient cells. To overcome this problem, we generated R26CreERT2;Brca1SCo/Δ mouse embryonic stem (ES) cells, which contain, in addition to a Brca1Δ5-13 null allele18, a Brca1SCo Selectable Conditional knockout allele in which exons 5 and 6 are flanked by loxP recombination sites and a split puromycin resistance marker (Fig. 1a; Supplementary Fig. 1a). Furthermore, these cells contain the CreERT2 gene targeted to the Rosa26 locus, leading to expression of a tamoxifen-inducible CreERT2 recombinase fusion protein19. Incubation of these cells with 4-hydroxytamoxifen (4OHT) results in nearly complete switching of the Brca1SCo allele and consequent loss of BRCA1 protein expression (Supplementary Fig. 1b–c). Non-switched R26CreERT2;Brca1SCo/Δ cells are effectively removed by puromycin selection (Supplementary Fig. 1e).
Figure 1.
Inactivation of 53BP1 rescues proliferation defects and drug sensitivity of Brca1 null ES cells. (a) Schematic overview of mutant alleles in R26CreERT2;Brca1SCo/Δ and R26CreERT2;Brca1Δ/Δ ES cells. Before 4-hydroxytamoxifen (4OHT) mediated induction of the CreERT2 recombinase, R26CreERT2;Brca1SCo/Δ cells are BRCA1 proficient and puromycin sensitive. Addition of 4OHT leads to CreERT2-mediated deletion of Brca1 exons 5–6, resulting in BRCA1 inactivation and concomitant expression of puromycin from the PGK promoter, thereby enabling selection of BRCA1-deficient R26CreERT2;Brca1Δ/Δ ES cells. (b) Western blot analysis of 53BP1 expression in R26CreERT2;Brca1SCo/Δ ES cells non transduced or transduced with two independent lentiviral shRNA vectors against 53bp1, after treatment with 4OHT to delete the Brca1SCo allele. (c) Crystal violet staining of untransduced R26CreERT2;Brca1SCo/Δ ES cells treated with 4OHT and stably transduced with lentiviral vectors expressing a control non-targeting shRNA (NT) or two independent shRNAs against 53bp1. (d–e) Susceptibility of R26CreERT2;Brca1SCo/Δ ES cells untreated or treated with 4OHT to DNA cross-linking agents cisplatin (d) or mitomycin C (e). Cell viability was measured after 4 days. Mean ± s.d. is shown from three independent measurements.
We used the piggyBac transposon system20 to perform an insertional mutagenesis screen for factors that rescue the proliferation defect of Brca1 deleted cells (Supplementary Fig. 2). We transfected R26CreERT2;Brca1SCo/Δ ES cells with plasmids containing an engineered piggyBac transposon and mouse codon optimized piggyBac transposase. After induction of CreERT2-mediated deletion of the Brca1SCo allele with 4OHT, we assayed for clonal survival of BRCA1-deficient ES cells under puromycin selection (Supplementary Fig. 2a). The majority (294/296) of surviving colonies analyzed contained both a switched and a non-switched Brca1SCo allele, indicating strong selection for allele duplication events (data not shown). Two clones that were completely Brca1 deleted showed identical patterns of piggyBac transposon integrations (Supplementary Fig. 2b); one of which mapped to intron 16 of the 53bp1 gene (Supplementary Fig. 2c) and correlated with abrogation of 53BP1 expression (Supplementary Fig. 2d), consistent with the reported ability of 53BP1 deletion to abrogate senescence and cell death in Brca1Δ11/Δ11 hypomorphic cells12. To validate loss of 53BP1 expression as a survival factor in Brca1 null cells, we tested the effects of shRNA-mediated depletion of 53BP1 in R26CreERT2;Brca1SCo/Δ ES cells, with two different shRNAs that efficiently suppressed 53BP1 expression, as determined by western blot analysis (Fig. 1b). The robust clonal growth arrest of R26CreERT2;Brca1SCo/Δ ES cells induced by 4OHT treatment was abolished when 53BP1 was depleted with either shRNA (Fig. 1c).
53BP1 loss rescues drug hypersensitivity of Brca1 null cells
A hallmark of BRCA1-deficient tumors is their cisplatin sensitivity21. Consistent with this, we observed enhanced cytotoxicity of cisplatin in our Brca1-deleted ES cells (Fig. 1d). shRNA-mediated loss of 53BP1 fully abolished the cisplatin sensitivity induced by Brca1 inactivation (Fig. 1d). A similar reversal of drug sensitivity by 53BP1 depletion was observed for mitomycin C (Fig. 1e). Although shRNA-mediated inhibition of p53 suppressed the growth defects of Brca1-deleted cells (data not shown), it did not suppress cisplatin sensitivity, suggesting that p53 and 53BP1 provide distinct pathways for sustaining growth arrest in Brca1-deleted cells.
53BP1 loss blocks DNA damage responses in Brca1 null cells
To investigate how suppression of 53BP1 or p53 alleviates the impaired proliferation of Brca1 knockout ES cells, we analyzed cell cycle profiles of 4OHT-treated R26CreERT2;Brca1SCo/Δ cells treated with control shRNA or shRNA causing depletion of 53BP1 or p53, monitored by western blotting (Fig. 2a). In the absence of BRCA1, ES cells accumulate in G2 (Fig. 2b), which could reflect a checkpoint response induced by accumulation of unrepaired DNA damage. This G2 arrest is abrogated in 53BP1-, but not in p53-depleted cells, suggestive of an ATM-dependent checkpoint activation for which 53BP1 is essential. Consistent with this, we observed an increase in 53BP1 expression levels in Brca1-deleted cells (Fig. 2a). The less pronounced effect of p53 depletion on the G2 arrest is mirrored by the lower induction of p53 expression in response to Brca1 deletion, as detected by western blotting. p53 plays a major role in the surveillance of chromosome integrity at the G1/S transition, although evidence for a relatively weaker p53-dependent checkpoint at the G2/M transition has also been reported22, 23.
Figure 2.
53BP1 depletion rescues cell cycle defects of Brca1 null ES cells. (a) Western blot analysis of 53BP1 and p53 expression in R26CreERT2;Brca1SCo/Δ ES cells stably tranduced with lentiviral shRNA vectors against 53bp1 or p53. Samples were taken before or at 4 and 9 days after 4-hydroxytamoxifen (4OHT) induced deletion of Brca1. (b) Flow cytometry profiles of R26CreERT2;Brca1SCo/Δ ES cells stably transduced with non-targeting shRNA lentiviruses or shRNA vectors against 53bp1 and p53. Shown are percentages of cells in G1 and G2 before or nine days after 4OHT-induced Brca1 deletion by CreERT2. Mean ± s.d. is shown from two experiments.
To address the possibility that loss of 53BP1 impacts on the DNA damage response (DDR) induced by Brca1 deletion, we used MEFs, established from mice carrying a Brca1SCo allele and a Brca1Δ5–13 null allele and immortalized by TBX2 overexpression. Transient expression of Cre recombinase from a self-deleting “hit & run” (H&R) Cre retrovirus24 resulted in efficient deletion of Brca1 exons 5 and 6 and loss of BRCA1 expression as monitored by western blotting (Fig. 3a). Concomitant loss of 53BP1 expression was detected when MEFs were co-transduced with the H&R Cre and 53BP1 shRNA-encoding viruses. Upon Cre-mediated deletion of the Brca1 gene, we observed robust phosphorylation of the DNA damage checkpoint kinase CHK2, as well as p53 accumulation, indicative of an ATM-dependent DDR in MEFs (Fig. 3a; +Cre; GFPsh). Similar to the G2 arrest, this ATM-dependent checkpoint response was markedly attenuated upon 53BP1 inhibition (Fig. 3a; +Cre; 53BP1sh1 and 2).
Figure 3.
53BP1 depletion abrogates CHK2-mediated DNA damage responses and rescues proliferation defects in Brca1 null but not Brca2 null MEFs. (a) Western blot analysis of cell extracts from Brca1SCo/Δ MEFs infected with retroviruses expressing self-deleting Cre recombinase (+Cre) or empty vector (−Cre), together with retroviruses expressing 53BP1 or GFP control shRNAs. SMC1 and tubulin were used as loading controls. NSB: non-specific band. (b) Quantification of chromatid and chromosome break frequency in metaphase spreads prepared from cells treated as in (a). At least 100 metaphases were scored for each sample. Shown are average number of events per metaphase ± s.d. (c) Proliferation curves of Brca1SCo/Δ or Brca2F/Δ MEFs infected with retroviruses expressing self-deleting Cre recombinase (+Cre) or empty vector (−Cre), together with retroviruses expressing p53, 53BP1 or GFP control shRNAs. (d) Western blot analysis of cell extracts from Brca2F/Δ MEFs treated as in (c). Tubulin was used as a loading control.
Consistent with unrepaired DNA damage being the cause of the observed G2/M arrest and checkpoint activation triggered by Brca1 deletion, we observed a marked increase in chromatid and chromosome breaks in BRCA1-deficient MEFs (Fig. 3b). shRNA-mediated 53BP1 depletion in these cells led to a decrease in the occurrence of DNA breaks, reflected in diminished checkpoint responses.
In contrast to Brca1 knockout MEFs, proliferation of Brca2 knockout MEFs25 is not rescued by shRNA-mediated depletion of 53BP1 (Fig. 3c). In contrast, p53 inhibition efficiently restored the proliferative capacity of Brca2-deleted cells. Consistent with the role of 53BP1 in mediating checkpoint responses specifically in cells lacking Brca1, 53BP1 abrogation did not affect CHK2 phosphorylation in BRCA2-deficient MEFs (Fig. 3d). This observation reflects the fundamentally distinct roles played by the BRCA1 and BRCA2 tumor suppressor proteins in the DDR: whilst BRCA1 is required for the initial steps of the DDR response and signal amplification, BRCA2 functions downstream of the checkpoint signaling, promoting the homologous recombination pathway of DNA repair. Since 53BP1 acts during the early chromatin remodeling events at the break, it is more likely to affect BRCA1-dependent signaling. In contrast, BRCA2 activation requires the damage signal generated at the break to be transduced downstream through at least two parallel pathways involving several response factors26. Thus, the loss of 53BP1 can be compensated by other regulatory mechanisms acting between the early events at the chromatin surrounding the break and the initiation of repair reactions. Although shRNA-mediated depletion of p53 rescues the proliferation defect of BRCA2-deficient cells, DDR activation illustrated by CHK2 phosphorylation persists. This suggests that CHK2-mediated p53 activation contributes to the senescence response induced by loss of BRCA2. Consistent with the role of BRCA2 as the loader of RAD51 onto DSBs, the initiating step of recombinational DNA repair, we did not detect any rescue of RAD51 foci in BRCA2-deficient cells when either p53 or 53BP1 were depleted (data not shown).
Together, these results suggest that 53BP1 is required for efficient ATM-dependent checkpoint signaling to arrest cell cycle progression in response to DNA damage accumulation in Brca1-deleted cells. Alternatively, 53BP1 loss might lead to more efficient DNA double-strand break (DSB) repair and thereby reduce DDR activation.
53BP1 loss partially restores HR in Brca1 null cells
The observed abrogation of G2 accumulation, sensitivity to DNA cross-linkers, and DDR may be due to a stimulation of BRCA1 independent DNA repair. Alternatively, checkpoint release might prevent the formation of DNA breaks by collapsing replication forks or allow the cells to continue cycling without actual repair of the damage. To investigate the effects of 53BP1 depletion on DNA repair we analyzed RAD51 foci formation in Brca1SCo/Δ and Brca1Δ/Δ ES cells following treatment with ionizing irradiation (Fig. 4a and 4c). Analysis of these IR induced RAD51 foci revealed a diminished response upon deletion of Brca1, in line with previous observations27. shRNA-mediated depletion of 53BP1 enhanced RAD51 foci formation in the absence of BRCA1, suggesting up-regulation of HR in a BRCA1 independent manner. This process is not specific for ES cells, as we observed the same phenomenon in MEFs (Fig. 4b and 4d).
Figure 4.
53BP1 depletion rescues RAD51 foci in Brca1 null cells. (a) RAD51 foci formation in R26CreERT2;Brca1SCo/Δ ES cells untreated or treated with 4OHT and transduced with lentiviruses expressing non-targeting shRNAs (NTsh) or shRNAs targeting 53bp1(53BP1sh). Cells were irradiated with 10 Gy, fixed after 6 hours, and stained with anti-RAD51 antibody (red). Nuclei were visualized with DAPI (blue). (b) RAD51 foci formation in Brca1SCo/Δ MEFs infected with retroviruses expressing self-deleting Cre recombinase (+Cre) or empty vector (−Cre), together with retroviruses expressing 53BP1 or GFP control shRNAs. Cells were irradiated with 10 Gy, fixed after 2 hours, and stained with anti-γH2AX (red) and anti-RAD51 antibodies (green). Nuclei were visualized with DAPI (blue). (c) Quantification of RAD51 foci in R26CreERT2;Brca1SCo/Δ ES cells treated as in (a). The percentage of cells with ≥10 Rad51 foci per cell are plotted for each condition. At least 20 nuclei were analyzed for each treatment. (d) Quantification of RAD51 foci in Brca1SCo/Δ MEFs treated as in (b). The percentage of cells with ≥10 RAD51 foci per cell are plotted for each condition. At least 50 nuclei were analyzed for each treatment.
To determine whether 53BP1 depletion rescues homology-directed repair (HDR) in BRCA1-deficient cells we measured gene targeting efficiencies in R26CreERT2;Brca1SCo/Δ ES cells with or without shRNA-mediated depletion of 53BP1 or p53, using an isogenic Rb targeting construct with a PGK-neo selection marker28. While CreERT2-mediated deletion of Brca1 by addition of 4OHT abolished Rb gene targeting in R26CreERT2;Brca1SCo/Δ ES cells, this effect could not be reversed by shRNA-mediated depletion of p53 (Table 1). In contrast, correct integration of the targeting vector at the Rb locus was observed in 2/160 53BP1-depleted R26CreERT2;Brca1Δ/Δ ES cell colonies, compared to 8/78 R26CreERT2;Brca1SCo/Δ colonies, suggesting that 53BP1 loss leads to partial restoration of HDR in Brca1 null cells. This notion was supported by gene targeting experiments with a promoterless Pim1-neo targeting construct29, showing correct integration in 4/47 53BP1-depleted R26CreERT2;Brca1Δ/Δ ES colonies, compared to 9/56 R26CreERT2;Brca1SCo/Δ colonies (Table 1). We conclude that depletion of 53BP1 restores HR activity in Brca1 null cells to 10–50% of BRCA1-proficient cells.
Table 1.
Gene targeting frequencies in R26CreERT2;Brca1SCo/Δ ES cells
| G418-resistant colonies |
|||||
|---|---|---|---|---|---|
| 4OHT* | shRNA | Total analyzed | Non-targeted (%) | Targeted (%) | |
| − | NT | 78 | 70 (90) | 8 (10) | |
| Rb | + | NT | 78 | 78 (100) | 0 (0) |
| + | 53BP1 | 160 | 158 (99) | 2 (1) | |
| + | p53 | 124 | 124 (100) | 0 (0) | |
| − | NT | 56 | 47 (84) | 9 (16) | |
| Pim1 | + | NT | 11 | 11 (100) | 0 (0) |
| + | 53BP1 | 47 | 43 (91.5) | 4 (8.5) | |
4-hydroxytamoxifen (4OHT) was added to induce CreERT2-mediated deletion of the Brca1SCo allele
Basal-like breast cancers have low levels of 53BP1
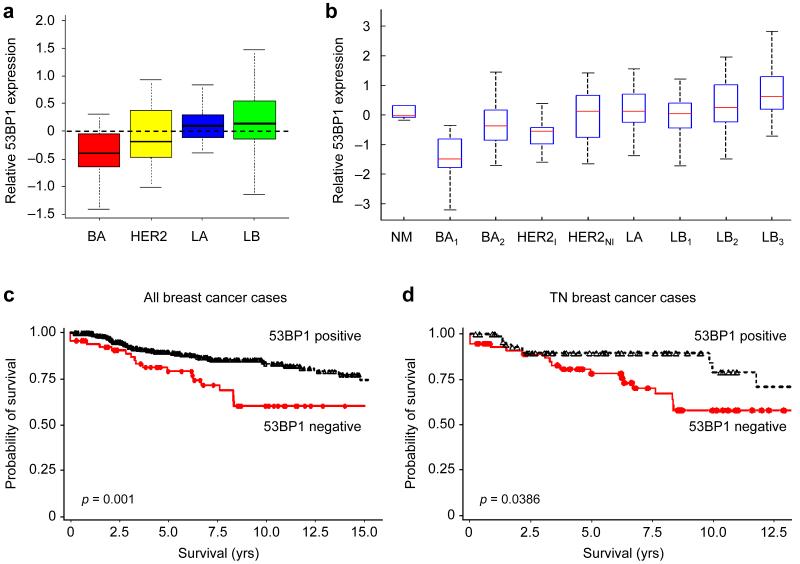
The majority of BRCA1 associated tumors carry p53 mutations30, 31. It is known that p53 loss can at least partially rescue BRCA1-deficient cells11, 32. However, there may still be additional selection for aberrant expression of 53BP1, as suggested by our in vitro results and by the synergism in tumorigenesis observed in 53bp1−/−;p53−/− knockout mice14, 15. Therefore we analyzed levels of 53BP1 mRNA in a publicly available database of gene expression array data from 286 breast cancer specimens33, all early stage (lymph node negative) and treated with surgery and radiation therapy alone. We used previously described unsupervised clustering methods to identify basal-like breast cancers (BLC), HER2-positive breast cancers, and Luminal A and Luminal B subclasses of breast cancer34. BLC tumors are characterized clinically as high grade, invasive breast cancers that lack expression of ER, PR, and HER2 (“triple-negative” phenotype) and are associated with younger age of onset. Luminal tumors are characterized by expression of the estrogen receptor, with Luminal A tumors being mostly low grade ER+ tumors, and Luminal B tumors being mostly high grade ER+ tumors. We next calculated mean levels of 53BP1 expression for each breast cancer subtype (Fig. 5a). Lowest 53BP1 expression levels were found in the BLC subclass.
Figure 5.
53BP1 expression is reduced in a subset of human BLBC. (a) Boxplots showing 53BP1 expression levels among breast cancer subtypes. Gene expression array data from 286 early stage breast cancers published by Wang et al.33 were clustered to classify tumors into basal (BA), HER2-positive (HER2), Luminal A (LA) and Luminal B (LB). The mean expression values of 53BP1 are shown for each subgroup. (b) Boxplots showing 53BP1 expression levels among different breast cancer subtypes defined by robust consensus clustering: normal (NM), basal (BA1, BA2), HER2-positive (HER2I, HER2NI), and luminal (LA, LB1, LB2, LB3). The data are normalized with mean expression of the combined data being set to 0. Expression of 53BP1 is significantly lower in the BA1 subtype (p<0.0001 vs. Normal; p<0.0002 vs. BA2). (c-d) Distant relapse-free survival stratified by 53BP1 protein expression in all breast cancers (c) and in triple-negative (TN) breast cancers (d). Kaplan-Meyer survival curves for distant relapse-free survival are shown for breast cancers that scored positive for 53BP1 staining (black lines) and those that scored negative for 53BP1 staining (red lines).
We have previously shown that a robust consensus clustering approach of breast cancer gene expression data reveals two subclasses of BLC, which we labeled BA1 and BA234. This approach also identifies 2 subclasses of HER2-positive cancers and 3 subclasses of Luminal B tumors. 53BP1 mRNA expression in these different subclasses was examined in a combined data set that includes the published data set of Wang et al.33 and Richardson et al.35. In this combined dataset, we found that 53BP1 expression was clearly lowest in the BA1 subtype of BLC (p<0.0001 vs. Normal, p<0.0002 vs. BA2) (Fig. 5b). These data suggest that a biologically relevant subset of BLC have low 53BP1 expression. To validate this finding at the protein level, we assayed a set of 504 breast cancer specimens from the Yale cohort by immunohistochemistry on tissue microarrays. The clinical characteristics of the tumors in this collection are shown in Supplementary Table 2. Four hundred forty-four cases were evaluable for 53BP1 status. A specimen was scored as lacking 53BP1 staining if fewer than 10% of tumor cells showed nuclear staining with this antibody (Supplementary Fig. 3). Out of 444 evaluable breast cancer specimens, 65 (14.6%) were scored as being negative for 53BP1.
53BP1 loss is associated with triple-negative phenotype
In the Yale cohort of 444 tumors, lack of 53BP1 staining correlated independently with lack of ER expression, lack of PR expression, and lack of HER2 overexpression as assessed by immunohistochemistry (Table 2). There was striking correlation between absence of 53BP1 staining and the triple-negative phenotype, defined as being ER−, PR− and HER2-negative. Of the 63 tumors in the Yale cohort that lacked 53BP1 staining and for which ER, PR and HER2 status were available, 57 tumors (90.5%) of these were triple-negative tumors. Of the 132 triple-negative tumors assayed, 57 (43%) were 53BP1 negative, while of the 311 non-triple-negative tumors only 6 (2%) had reduced 53BP1 staining. This correlation was highly statistically significant (P<0.0001). The IHC data are consistent with the gene expression data and confirm that a subset of BLCs and triple-negative tumors have a profoundly decreased 53BP1 expression. Loss of 53BP1 was associated with age<50, with most (67%) of 53BP1 negative tumors occurring in women less than 50 years old. There was no correlation with tumor size, lymph node status, or race.
Table 2.
Low 53BP1 expression correlates with triple-negative status (Yale cohort)
| 53BP1 expression |
||||
|---|---|---|---|---|
| Features | Total (%) | Positive (%) | Negative (%) | p * |
| ER | ||||
| positive | 243 (55) | 239 (63) | 4 (6) | |
| negative | 200 (45) | 140 (37) | 60 (94) | <0.0001 |
| PR | ||||
| positive | 226 (51) | 222 (59) | 4 (6) | |
| negative | 217 (49) | 157 (41) | 60 (94) | <0.0001 |
| ERBB2/HER2 | ||||
| positive | 78 (18) | 77 (20) | 1 (2) | |
| negative | 366 (82) | 303 (80) | 63 (98) | <0.0001 |
| Triple-negative (TN) | ||||
| Not TN | 311 (70) | 305 (80) | 6 (10) | |
| TN | 132 (30) | 75 (20) | 57 (90) | <0.0001 |
Fisher’s exact test.
53BP1 loss is associated with BRCA1/2 mutation status
To validate and extend the data from the Yale cohort, including assessment of 53BP1 protein expression in BRCA1/2-mutated familial breast carcinomas, an independent larger tissue microarray analysis was performed on the Helsinki cohort of 1187 patients (Table 3). These data were obtained using different 53BP1 antibodies, and analyzed independently by separate pathologists (see Supplementary Fig. 4 for examples of 53BP1 staining patterns in the Helsinki cohort). Confirming the results obtained from the Yale cohort, lack of 53BP1 staining in the Helsinki cohort correlated independently with lack of ER expression (p=0.000004), lack of PR expression (p=0.003), and the triple-negative phenotype (p=0.000004). In addition, the familial tumors from BRCA1/2 mutation carriers (79 tumors) showed the highest degree of aberrant 53BP1 reduction or loss, as compared to sporadic tumors (n=374, p=0.000003), familial carcinomas not attributable to BRCA1/2 mutations (n=734, p=0.001), or all non-BRCA1/2 tumors (n=1108, p=0.0001). Both BRCA1 and BRCA2 associated tumors showed a significantly increased incidence of reduced 53BBP1 staining when compared with non-BRCA1/2 tumors (p=0.003 for BRCA1 and p=0.008 for BRCA2). Overall, these results demonstrate that loss of 53BP1 is more frequent among the most aggressive and difficult-to-treat triple-negative tumors, and in tumors with BRCA1/2 mutations.
Table 3.
Loss or reduction of 53BP1 expression correlates with triple-negative status and BRCA1/2 mutation status (Helsinki cohort)
| 53BP1 |
||||
|---|---|---|---|---|
| Features | Total (%) | Normal (%) | Aberrant (%) | p * |
| ER | 1053 | |||
| positive | 834 (79.2) | 823 (80.3) | 11 (39.3) | |
| negative | 219 (20.8) | 202 (19.7) | 17 (60.7) | 0.000004 |
| PR | 1050 | |||
| positive | 703 (67.0) | 692 (67.7) | 11 (39.3) | |
| negative | 347 (33.0) | 330 (32.3) | 17 (60.7) | 0.003 |
| ERBB2/HER2 | 1075 | |||
| positive | 145 (13.5) | 142 (13.6) | 3 (10.3) | |
| negative | 930 (86.5) | 904 (86.4) | 26 (89.7) | 0.8 |
| Triple-negative (TN) | 1018 | |||
| Not TN | 875 (86.0) | 861 (87.0) | 14 (50.0) | |
| TN | 143 (14.0) | 129 (13.0) | 14 (50.0) | 0.000004 |
| BRCA1/2 mutation | 1187 | |||
| non-BRCA1/2 | 1108 (93.4) | 1079 (94.0) | 29 (74.4) | |
| sporadic | 374 | 371 | 3 | 0.000003 † |
| familial non-BRCA1/2 | 734 | 708 | 26 | 0.001 & |
| BRCA1/2 | 79 (6.6) | 69 (6.0) | 10 (25.6) | 0.0001 ‡ |
| BRCA1 | 35 | 30 | 5 | 0.003 § |
| BRCA2 | 44 | 39 | 5 | 0.008 ¶ |
Fisher’s exact test
BRCA1/2 vs. sporadic
BRCA1/2 vs. familial non-BRCA1/2 tumors
BRCA1/2 vs. all non-BRCA1/2 tumors (sporadic + familial non-BRCA1/2)
BRCA1 vs. all non-BRCA1/2 tumors
BRCA2 vs. all non-BRCA1/2 tumors.
53BP1 expression and distant metastasis-free survival
Survival data were available for analysis in the Yale cohort. There was significant association between 53BP1status and distant metastasis-free survival, with 53BP1 negative tumors having significantly lower metastasis-free survival (Fig. 5c) (p=0.001). As most tumors that lack 53BP1 have a “triple-negative” phenotype, we analyzed distant metastasis-free survival in triple-negative breast cancers stratified by 53BP1 status. Amongst the triple-negative tumors, those that lack normal 53BP1 staining have worse metastasis-free survival, (p=0.039) (Fig. 5d). As cancers in this data set were mostly early stage, lymph node-negative cancers that did not receive any adjuvant treatment, these data suggest that early stage triple-negative tumors with reduced 53BP1 may have a greater likelihood of metastasis in the absence of systemic treatment, when compared with triple-negative tumors with intact 53BP1.
Discussion
BRCA1 is a large ubiquitously expressed protein that has a major role in DNA damage repair by HR. Its activity is extensively regulated by phosphorylation36, sumoylation37-39 and interactions with many other proteins. Perhaps not surprisingly, BRCA1 is not only involved in HR but also in many other processes like cell cycle control and transcriptional regulation39, 40. Despite its widespread expression and non-cell type specific functions, mutations in BRCA1 are mainly associated with increased risk of breast and ovarian tumorigenesis. Loss of BRCA1 leads to severe proliferation defects in normal, non-cancerous cells, for instance leading to lethality during embryonic development. Therefore, it seems likely that there are survival factors that allow BRCA1-deficient tumor cells to expand. In an unbiased genetic screen we found that loss of 53BP1 rescues clonal outgrowth of Brca1 null ES cells. This result confirms the recently described rescue of Brca1Δ11/Δ11 hypomorphic mice by 53BP1 knockout12. In addition, it shows that expression of the BRCA1-Δ11 splice variant is not required for rescue of BRCA1 deficiency by 53BP1 loss. This is important since many human BRCA1 associated cancers are characterized by complete loss of BRCA1 expression.
Further characterization indicated that BRCA1 and 53BP1 double-deficient cells are no longer hypersensitive to DNA cross-linking agents and do not spontaneously form DSBs or activate DDR. Suppression of IR-induced CHK2 phosphorylation has been previously observed upon RNAi-mediated 53BP1 depletion in U2OS cells13 and in 53bp1−/− MEFs41. Here we show that 53BP1 status specifically affects spontaneous induction of CHK2 phosphorylation when BRCA1 is lost. Our results are consistent with the data reported by Bunting et al on 53BP1 mediated suppression of DNA resection at DSBs and accumulation of single stranded DNA ends in BRCA1 and 53BP1 double-deficient cells42. In cell-free extracts, DNA damage-induced ATM activation and CHK2 phosphorylation are inhibited by 3′ single-stranded DNA overhangs generated during DNA break processing in S/G243. The decreased CHK2 phosphorylation that we observe in 53BP1-depleted Brca1 null cells could therefore result from decreased ATM signaling due to increased DNA resection, or from increased DSB repair. The latter possibility is supported by the restoration of IR-induced RAD51 foci formation and the partial restoration of HR in 53BP1-depleted Brca1 null cells.
Although our data suggest that a certain level of HR repair can take place in the absence of both BRCA1 and 53BP1, 53BP1-depleted Brca1 null cells remain HR defective since gene targeting frequencies in these cells are reduced by a factor of 2–10 when compared to wildtype cells. In line with this, BRCA1-deficient tumors show excellent responses to therapies exploiting a HR defect, such as platinum drugs44 or PARP inhibitors45. Also Brca1-mutated mouse mammary tumors are highly sensitive to treatment with PARP inhibitors46 or platinum drugs47. While human BRCA1-mutated tumors can develop resistance to carboplatin by genetic reversion of the BRCA1 mutation48 – stressing the importance of BRCA1 function for HR – mouse mammary tumor models with large deletions in Brca1 cannot employ this mechanism and remain sensitive to cisplatin or carboplatin, even after multiple rounds of treatment47. In contrast, Brca1Δ11/Δ11 mouse mammary tumors, which only express the BRCA1-Δ11 isoform, readily become resistant to cisplatin despite the fact that Brca1 exon 11 sequences are irreversibly deleted49. This feature might be indicative of residual activity of the BRCA1-Δ11 isoform in HR.
At present it is not clear whether 53BP1 loss contributes to development, therapy response and/or acquired resistance of BRCA1-deficient tumors. To explore this, we examined 53BP1 expression in independent cohorts of breast cancer patients from USA and Finland, respectively. These two tumor sets were analyzed by different antibodies and reviewed by independent pathologists using separate criteria, yet both cohorts showed a striking correlation of low 53BP1 expression levels with triple-negative tumors. Since most BRCA1-mutated breast cancers cluster in this subgroup, it was no surprise to find that also these tumors lack 53BP1 expression more often than other subsets of breast tumors. However, also the often hormone receptor positive BRCA2-associated tumors are significantly enriched for 53BP1 aberrations. This may be indicative of a common selection for 53BP1 ablation in both types of HR deficient breast cancers. This might occur via different routes, given the persistence of DDR activation and growth impairment in 53BP1 depleted BRCA2-deficient cells.
In conclusion, we have shown that 53BP1 loss alleviates the proliferation defect and DNA-damage hypersensitivity of Brca1 null cells and leads to partial restoration of HR in these cells. Furthermore, aberrant expression of 53BP1 is more common in BRCA1/2 associated breast cancers, which may hint at a role for 53BP1 loss in these tumors. 53BP1 is also lost in a subset of sporadic triple-negative breast cancers, suggesting a broader role for abnormalities in this pathway in breast tumorigenesis. Our results suggest that loss of 53BP1 may promote survival of BRCA1-deficient tumor cells after DNA damage induced by chemotherapy or irradiation. It is possible that 53BP1 loss may have different effects in BRCA1-deficient tumors vs. sporadic triple-negative breast cancers. Regardless, 53BP1 might represent a candidate biomarker for predicting response of HR defective tumors to PARP inhibitors or platinum drugs.
Supplementary Material
Acknowledgements
We wish to thank Marcelle Treur-Mulder for her help with targeting of the Brca1SCo allele; Wei Wang and Pentao Liu (Wellcome Trust Sanger Institute, Hinxton, UK) for their kind gift of the piggyBac transposon MSCV5’ LTR transposon and the mPB transposase; Ewart de Bruin, Edwin Cuppen and Marco Koudijs for help with PCR amplification of the piggyBac insertions; Jun Kong and Daoud Sie for mapping of piggyBac insertions; Karoly Szuhai for multi-color fluorescence in situ hybridization karyotyping; Sjaak Philipsen (Erasmus University, Rotterdam, The Netherlands) for providing shRNA vectors; and Hein te Riele and Piet Borst for their comments on the manuscript. We thank Erhan Bilal and Gyan Bhanot for their help with analysis of microarray data and Päivi Heikkilä and Kristiina Aittomäki for their help with the Finnish breast cancer data and sample collection. We are most grateful to the patients who provided clinical samples that were analyzed in this study.
Work in J.J. laboratory is supported by the Dutch Cancer Society (KWF), the Netherlands Organization for Scientific Research (NWO), and the European Community 7th Framework Program (EuroSyStem project). Work in M.T. laboratory is supported by Cancer Research UK and Breast Cancer Campaign. Work in S.G. laboratory is supported by the U. S. Department of Defense (A.A.), the National Cancer Institute (S.G.), the Sidney Kimmel Foundation (S.G.) and the Breast Cancer Research Foundation (S.G.). B.G.H. is supported by the BCRF and the NCI. Work in J.B. laboratory is supported by the Danish Cancer Society, Danish National Research Foundation, Vilhelm Pedersen and Hustrus Mindelegat, Czech Ministry of Education (MSMT), and the European Community 7th Framework Program (projects GENICA and INFLA-CARE). Work in H.N. laboratory is supported by the Helsinki University Central Hospital Research Fund, Finnish Cancer Society, Academy of Finland (132473) and the Sigrid Juselius Foundation.
Appendix
Methods
R26CreERT2;Brca1SCo/Δ ES cells and mutagenesis screen
Details on the generation of R26CreERT2;Brca1SCo/Δ ES cells and the piggyBac transposon mutagenesis screen are described in the Supplementary Information.
MEF isolation and immortalization
Heads and organs of E13.5 embryos were removed and remaining tissue was minced, rinsed in PBS, and incubated for 30 min in 0.5 ml of 0.05% trypsin-EDTA (GIBCO) at 37°C. Cell aggregates were dissociated in DMEM supplemented with 10% FBS (GIBCO) and penicillin-streptomycin (GIBCO). Cells were plated on 10-cm dishes and cryopreserved after two days as passage 1 MEFs. MEFs were immortalized by infection with TBX2 retrovirus50.
Retroviral transductions
MEFs were transduced with pRetroSuper retroviruses encoding shRNAs targeting GFP51, 53BP1 (sh1: 5′-GCTATTGTGGAGATTGTGTTT-3′; sh2: 5′-GCGTAGAAGATATTTCACCTA-3′) or p5352 as described previously53. Briefly, HEK293T packaging cells were transfected with pCL-Eco helper vector together with either pRetroSuper alone or pRetroSuper plus retroviral vector encoding ‘Hit-and-run’ Cre recombinase24. The culture medium was replaced 24 hr after transfection. Recipient MEFs were plated and infected 24 hr later with retroviral supernatants. Additional infections were performed after 24 and 32 hr. Twenty-four hours after the last infection, cells were washed and grown in medium containing 2-3 mg ml−1 puromycin.
Lentiviral transductions
R26CreERT2;Brca1SCo/Δ ES cells were transduced with pLKO-puro shRNA lentiviruses obtained from Mission library clones (Sigma). In addition to the 53BP1 shRNAs mentioned above, we used shRNAs targeting p53 (sh1: 5′-CTACAAGAAGTCACAGCACAT-3′; sh2: 5′-AGAGTATTTCACCCTCAAGAT-3′) and a non-targeting (NT) shRNA (SHC202: 5′-CAACAAGATGAAGAGCACCAA-3′). After selection with 1.8 μg ml−1 puromycin, cells were switched by overnight incubation with 0.5 μg ml−1 4OHT. Four days after switching cells were seeded at 1000 cells/cm2 and assayed for clonal growth. Surviving colonies were fixed in formalin and stained with crystal violet.
Cell cycle analysis
Cells were incubated with BrdU for 1 hr, fixed with ethanol and incubated with mouse anti-BrdU (clone BU20A; Dako) and goat anti-mouse FITC conjugated secondary antibodies (DAKO). Cell cycle distribution was measured by flow cytometry and analyzed using FlowJo software (FlowJo).
Cytotoxicity assays
R26CreERT2;Brca1SCo/Δ ES cells were seeded in triplicate at 1000 or 3000 (in case of switched cells with non-targeting shRNA) cells per well in 96-well plates. One day after seeding, cells were fed with medium containing cisplatin or mitomycin C was added the next day. Five days later, cell viability was assayed in an Envision plate reader (Perkin Elmer) using resazurin (cell titer blue; Promega).
MEF proliferation assays
MEFs were seeded at 5000 cells per well in 96-well plates. Cell number was determined after 24, 48, 72 and 96 hours by incubating cells with 10 micrograms ml−1 resazurin. After 2 hours, fluorescence was measured at 590 nm using a micro-titer plate reader (2103 multilabel reader, Perkin Elmer).
Analysis of chromosomal aberrations
Exponentially growing MEFs were either collected and processed for immunoblotting as described below, or processed for metaphase spreads preparation. For this, cells were treated with 0.1μg ml−1 colcemid for 4 hr or overnight, trypsinized, swollen in hypotonic buffer (10mM Tris-HCl, pH7.5, 10 mM NaCl, 5 mM MgCl2) at 37°C for 5 minutes and fixed in 3:1 methanol:glacial acetid acid.
Immunoblotting
Cells were harvested by trypsinization, washed twice with cold HBSS, re-suspended in SDS-PAGE loading buffer and sonicated. Equal amounts of protein were analyzed by gel electrophoresis followed by western blotting. NuPAGE-Novex 10% Bis-Tris gels or Tris-Acetate 3-8% (Invitrogen) were run according to the manufacturer’s instructions.
Immunofluorescence
ES cells were grown on coverslips, γ-irradiated with 10Gy and fixed 6 hours later using 2% paraformaldehyde in PBS. Fixed cells were incubated with antibody in PBS containing 0.5% BSA and 0.15% glycine, mounted using Vectashield with 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and imaged with a Hamamatsu ORCA AG CCD camera on a Zeiss Axio Observer Z1 system. For quantification of RAD51 foci, images were converted into 8-bit gray-scale pictures using Image J software and the fluorescence intensity threshold was set based on a black&white intensity scale from 140-255. For each sample, the number of foci was counted in at least 20 individual nuclei.
MEFs were grown on coverslips and γ-irradiated with 10Gy. Cells were allowed to recover for the indicated times, washed in PBS and swollen in hypotonic solution (85.5 mM NaCl, 5 mM MgCl2, adjusted to pH 7.0) for 5 min. Cells were fixed with 4% paraformaldehyde (10′ RT), permeabilized by adding 0.03% SDS to the fixative and immunostained as described54. Dried coverslips were mounted on microscope slides using the ProLong Antifade kit (Invitrogen) supplemented with 1 mg ml−1 DAPI, and viewed with a Leica DMI6000B fluorescence microscope. Images were acquired with a Leica DFC350 FX R2 digital camera using LAS-AF software (Leica). Image brightness and contrast was adjusted using Photoshop CS3 (Adobe). To quantify RAD51 foci, we determined the frequency of nuclei with more than 10 RAD51 foci. At least 50 nuclei were analyzed for each sample.
Antibodies
Antibodies used for immunoblotting of ES cells: mouse monoclonal anti-mouse BRCA1 (GH118), affinity-purified rabbit polyclonal anti-mouse BRCA1 against peptide 452-469, 53BP1 (ab21083, Abcam) and p53 (IMX25, Monosan). Immunocytochemical staining of ES cells was performed using human RAD5154 and goat anti-rabbit Alexa 588 secondary antibody (Molecular Probes).
Antibodies used for immunobloting of MEFs: human RAD5154, SMC1 (BL308, Bethyl), 53BP1 (NB100-304, Novus), p53 (1C12, Cell Signaling), Chk2 (clone 7, Millipore) and α-tubulin54 (CRUK Monoclonal Antibody Service). Antibodies for imunofluorescence staining of MEFs: mouse monoclonal anti-phospho histone H2AX-Ser139 (Upstate Biotechnology) and RAD51 (H-92, Santa Cruz biotechnologies).
Gene targeting assays
R26CreERT2;Brca1SCo/Δ ES cells were electroporated with linearized Rb-neo 28 or Pim1-neo 29 targeting constructs. After drug selection with 200 μg ml−1 G418, colonies were picked, expanded and lysed in direct lysis reagent (Viagen) containing 100μg ml−1 proteinase K. Following heat-inactivation of proteinase K, genomic DNA was used for PCR analysis or digested with appropriate restriction enzymes and analyzed by Southern blotting as described28, 29.
Analysis of 53BP1 expression in human breast cancers
Details on analysis of 53BP1 expression in human breast cancers using tissue microarrays from the Yale and Helsinki cohorts or gene expression array datasets from Wang et al.33 and Richardson et al.35 are described in the Supplementary Information.
References
- 1.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 3.Boulton SJ. Cellular functions of the BRCA tumour-suppressor proteins. Biochem. Soc. Trans. 2006;34:633–645. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- 4.Collins N, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- 5.Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12-21 in familial breast and ovarian cancer involve the wild-type chromosome. Nature Genet. 1992;2:128–131. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- 6.Johannsson OT, et al. Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur. J. Cancer. 1997;33:362–371. doi: 10.1016/s0959-8049(97)89007-7. [DOI] [PubMed] [Google Scholar]
- 7.Foulkes WD, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 8.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl Acad. Sci. U. S. A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 10.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 11.Hakem R, de la Pompa JL, Elia A, Potter J, Mak TW. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 12.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 14.Morales JC, et al. 53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis. Proc Natl Acad Sci U S A. 2006;103:3310–3315. doi: 10.1073/pnas.0511259103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward IM, et al. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 17.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hameyer D, et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007;31:32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartz SR, et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol. 2006;26:9377–9386. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 23.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 24.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 25.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 26.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 28.Te Riele H, Robanus-Maandag E, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci U S A. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Te Riele H, Robanus-Maandag E, Clarke A, Hooper M, Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- 30.Holstege H, et al. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009;69:3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- 31.Manie E, et al. High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res. 2009;69:663–671. doi: 10.1158/0008-5472.CAN-08-1560. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 34.Alexe G, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007;67:10669–10676. doi: 10.1158/0008-5472.CAN-07-0539. [DOI] [PubMed] [Google Scholar]
- 35.Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Scully R, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 37.Bartek J, Hodny Z. SUMO boosts the DNA damage response barrier against cancer. Cancer Cell. 2010;17:9–11. doi: 10.1016/j.ccr.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 38.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 40.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010 doi: 10.1016/j.cell.2010.03.012. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silver DP, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 46.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rottenberg S, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci U S A. 2007;104:12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swisher EM, et al. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shafee N, et al. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–3250. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobs JJ, et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat. Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 51.Tarsounas M, et al. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 52.Dirac AM, Bernards R. Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J Biol Chem. 2003;278:11731–11734. doi: 10.1074/jbc.C300023200. [DOI] [PubMed] [Google Scholar]
- 53.Palmero I, Serrano M. Induction of senescence by oncogenic Ras. Methods Enzymol. 2001;333:247–256. doi: 10.1016/s0076-6879(01)33060-4. [DOI] [PubMed] [Google Scholar]
- 54.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.