Abstract
One obstacle to wider use of rapid liquid culture-based tuberculosis diagnostics such as the microscopic observation drug susceptibility (MODS) assay is concern about cross-contamination. We investigated the rate of laboratory cross-contamination in MODS, automated MBBacT, and Lowenstein–Jensen (LJ) cultures performed in parallel, through triangulation of microbiologic (reculturing stored samples), molecular (spoligotype/RFLP), and clinical epidemiologic data. At least 1 culture was positive for Mycobacterium tuberculosis for 362 (11%) of 3416 samples; 53 were regarded as potential cross-contamination suspects. Cross-contamination accounted for 17 false-positive cultures from 14 samples representing 0.41% (14/3416) and 0.17% (17/10 248) of samples and cultures, respectively. Positive predictive values for MODS, MBBacT (bioMérieux, Durham, NC), and LJ were 99.1%, 98.7%, and 99.7%, and specificity was 99.9% for all 3. Low rates of cross-contamination are achievable in mycobacterial laboratories in resource-poor settings even when a large proportion of samples are infectious and highly sensitive liquid culture-based diagnostics such as MODS are used.
Keywords: Tubercolosis, Multidrug resistance, Cross-contamination, Microscopic observation drug susceptability assay, MODS, Microbacterial culture
1. Background
In mycobacteriology reference-level laboratories in the industrialized world, cross-contamination is estimated to account for between 0.5% and 6% of positive results (Bauer et al., 1997; Nivin et al., 1998; Burman and Reves 2000; de Boer et al., 2002; Jasmer et al., 2002; Ruddy et al., 2002) with significant associated cost implications (Northrup et al., 2002), and higher levels might be anticipated in high tuberculosis (TB)-burden resource-limited settings where laboratory facilities are less sophisticated and a greater proportion of samples contain Mycobacterium tuberculosis. The microscopic observation drug susceptibility (MODS) assay in which 2 sputum samples are cultured on the same 24-well tissue-culture plate in Middlebrook 7H9 medium (Caviedes et al., 2000; Moore et al., 2004) has been proposed as a potential tool to bring low-tech sensitive TB diagnosis to the developing world where the need is most urgent; however, the risk of cross-contamination in this simple low-tech method using liquid culture medium and multiple samples in a single 24-well plate has not been previously determined.
Examining samples obtained in a large community-based study in urban Lima, Peru, we determined the rate of M. tuberculosis cross-contamination in sputum cultures performed in parallel in MODS, MBBacT-automated mycobacterial culture system (bioMérieux, Durham, NC), and Lowenstein–Jensen (LJ) solid media by first defining and then investigating contamination suspect positive cultures.
2. Patients, materials, and methods
2.1. Sample collection
After written informed consent, 1923 patients undergoing investigation for TB at health centers in Lima, Peru, were recruited over an 18-month period into an operational evaluation of the MODS assay. Two sputum samples were requested from each participant, and 3416 samples were obtained for auramine stain microscopy and parallel culture by all of LJ, automated MBBacT, and MODS. The study protocol and informed consent forms were approved by all of the following: Ethics Committees of Universidad Peruana Cayetano Heredia, Lima, Peru; Asociación Benéfica PRISMA, Lima, Peru; Johns Hopkins Bloomberg School of Public Health, Baltimore, MD; Imperial College London, UK; and Dirección de Salud III Lima Norte and Dirección de Salud II Lima Este (Regional Ministry of Health), Peru.
2.2. Laboratory
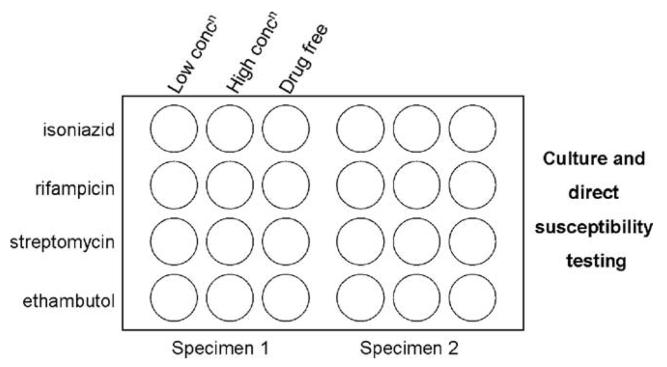
After decontamination by the N-acetyl-L-cysteine (NALC)-NaOH method (WHO, 1998) and auramine smear microscopy, all samples were divided into 3 aliquots for culture i) on an LJ slant, ii) in the MBBacT colorimetric, automated mycobacterial culture system, and iii) in 12 wells of a 24-well tissue-culture plate in the MODS assay (Fig. 1).
Fig. 1.
Schematic of sample layout on MODS plate (2 samples per plate—no plate contained 2 samples from the same patient).
Lowenstein–Jensen cultures were examined twice weekly from day 7 to 60; after which, the absence of typical colonies was regarded as a negative result. Ziehl-Neelson (ZN) smears were made from characteristic colonies appearing before day 60 to confirm the presence of acid fast bacilli. MBBacT cultures were automatically monitored continuously for 42 days and determined as positive or negative in accordance with manufacturer's recommendations. ZN staining of an aliquot of culture media from any MBBacT bottle reported as positive was performed to confirm the presence of acid fast bacilli.
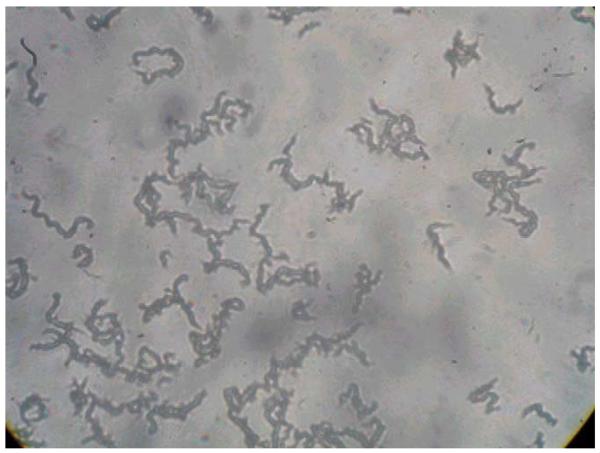
Microscopic observation drug susceptibility cultures were examined every weekday from day 4 to 15, on alternate days from day 16 to 25, and twice weekly from day 26 to 40 under an inverted light microscope. Positive cultures were identified by the characteristic cording morphology of M. tuberculosis growth in liquid media (in drug-free control wells) (Fig. 2) as described previously (Caviedes et al., 2000; Park et al., 2002; Moore et al., 2004). Nontuberculous mycobacteria (NTM) were recognized by their lack of cording or (in the case of Mycobacterium chelonae that uniquely among NTM does form cords) rapid overgrowth of wells by day 5.
Fig. 2.
Characteristic cordlike tangles of M. tuberculosis in MODS (original magnification ×40).
In the event of bacterial or fungal overgrowth in any of the 3 cultures, the original stored decontaminated sample was decontaminated a second time by the same NALC–NaOH method, and the affected culture method was repeated. In the event of repeated bacterial/fungal over-growth, the culture was abandoned.
Additional molecular confirmation of the presence of M. tuberculosis was performed for all isolates. Fingerprinting of every isolate was performed by spoligotyping (Goyal et al., 1997), with subsequent selective restriction fragment length polymorphism (RFLP) typing (van Embden et al., 1993) where further discriminatory data were required.
2.3. Defining contamination suspects
The process for determining whether a positive M. tuberculosis culture was the result of cross-contamination involved identifying and then thoroughly investigating contamination suspects. Contamination suspect cultures were identified by a process of exclusion: positive cultures regarded as unlikely to be due to cross-contamination were those in which a) all 3 culture methods for a sample were positive, or if there was fungal/bacterial overgrowth of 1 method, the remaining 2 were positive; b) all 3 methods were culture positive for the patient's other sputum sample (the protocol required submission of 2 sputum samples per patient); or c) the auramine smear was positive.
2.4. Investigating contamination suspects
The approach to distinguishing between true and false positives from among the remaining smear-negative samples, which were culture-positive in only 1 or 2 of the 3 methods (contamination suspects), involved the triangulation of investigations using conventional microbiology, molecular epidemiology, and conventional clinical epidemiology.
The procedures undertaken are described in detail in the Results section below, but briefly:
1) All the stored decontaminated sputum samples for this group of contamination suspects were recultured by all 3 methods to determine whether M. tuberculosis could be isolated again. In this rule-out step, any positive culture with a spoligotype matching the original was regarded as sufficient evidence that the original culture had been a true positive. However, because most samples had been stored for more than 12 months at −70 °C, negative cultures were not regarded as necessarily indicative that the original culture had been a false positive;
2) The molecular fingerprints of all isolates from cultures setup contemporaneously with the contamination suspect culture were compared. Initial molecular evaluation of strain diversity was performed for all isolates by spoligotyping—thus, for a patient with 2 samples, which were culture positive in 2 of the 3 methods, there were 4 available spoligotypes. For strains from contamination suspects with non-unique spoligotypes and for which the date of sample processing of an identical strain overlapped within 2 days, subsequent IS6110 RFLP typing was performed to enhance discrimination;
3) All patients contributing contamination suspect samples were followed up to determine clinical outcome. Smear-negative, culture-positive TB is managed on a case by case basis in the National Tuberculosis Program (NTP), and all such study patients (which included all those with contamination suspect samples) were managed by one of us (JCS, Ministry of Health TB physician) in a dedicated clinic. Thus, clinical outcome data and information on any subsequent TB diagnostic testing or treatment elsewhere was collected, and follow-up samples were sought for repeat culture, again in triplicate.
3. Results
3.1. Identification of contamination suspects
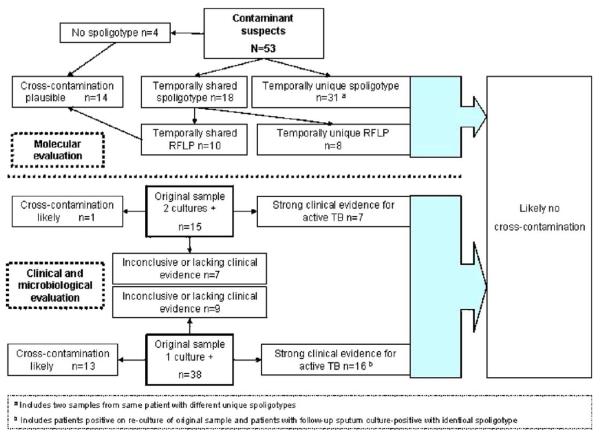
At least 1 culture was positive for M. tuberculosis in 362 (11%) of the 3416 samples (Fig. 3). For 79% (n = 285) of these samples, all 3 culture methods were positive, whereas for a further 7% (n = 24), the sample was i) culture positive by 2 of the 3 methods with bacterial or fungal overgrowth of the 3rd method (n = 4), ii) one of a pair of samples from the same patient, the other of which was culture positive in all 3 methods (n = 16), or iii) none of the above but auramine smear positive (n = 4). Thus, 53 smear-negative samples (from 47 patients) yielded only 1 (n = 38) or 2 (n = 15) positive cultures (68 cultures in total) and lacked a 2nd corresponding patient sample, which was either smear positive or culture positive in all 3 methodologies.
Fig. 3.
Rule-out sequence to define contamination suspect positive cultures (n = 53) from 362 culture-positive samples. *Includes 55 samples in which 1(n = 52), 2 (n = 2), or all 3 (n = 1) methods were irretrievably overgrown with fungi or bacteria; # 3 of the 4 were auramine positive; $either positive or negative culture.
3.2. Investigation of contamination suspects
3.2.1. Recovery of M. tuberculosis from stored decontaminated sputum samples
Aliquots of frozen decontaminated sputum were available for reculture for 51 of the 53 suspect samples. Three samples yielded a positive culture in MODS alone, and the remainder were culture negative in all 3 methods (Table 1). This 2nd retrieval of M. tuberculosis from the same sample was regarded as definitive evidence against cross-contamination.
Table 1.
Individual data for 53 contamination suspect samples
| Sample no. |
Original culture profile | Repeat culture profilea | Unique spoligotype?b |
If nonunique spoligotype, unique RFLP?c |
Clinical follow-up achieved |
Follow-up culture resultd | Microbiologic assessment (reculture) |
Molecular assessment |
Clinical assessment (including follow-up investigations) |
Overall assessment | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|||||||||||
| MODS | MBBacT | LJ | MODS | MBBacT | LJ | MODS | MBBacT | LJ | 1 = Consistent with cross-contamination, 0 = regarded as excluding cross-contamination |
|||||||
| 1 | + | + | − | − | − | − | N | N e | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 2 | + | + | − | + | − | − | Y | − | N | NA | NA | NA | 0 | 0 | 1 | Not cross-contamination |
| 3 | + | + | − | − | − | − | Y | − | Y | − | − | − | 1 | 0 | 1 | Not cross-contamination |
| 4 | + | + | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 5 | + | + | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 6 | + | − | − | − | − | − | N | Y | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 7 | + | − | − | + | − | − | N | N | Y | NA | NA | NA | 0 | 1 | 1 | Not cross-contamination |
| 8 | + | − | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 9 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 10 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 11 | + | − | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 12 | + | − | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 13 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 14 | + | − | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 15 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 16 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 17 | − | + | − | − | − | − | Y | − | Y | − | − | − | 1 | 0 | 1 | Not cross-contamination |
| 18 | − | − | + | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 1 | Not cross-contamination |
| 19 | + | + | − | − | − | − | N | Y | Y | OT− | OT− | OT− | 1 | 0 | 0 | Not cross-contamination |
| 20 | + | + | − | − | − | − | N | Y | Y | OT− | OT− | OT− | 1 | 0 | 0 | Not cross-contamination |
| 21 | + | + | − | − | − | − | Y | − | Y | − | − | − | 1 | 0 | 0 | Not cross-contamination |
| 22 | + | + | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 0 | Not cross-contamination |
| 23 | + | + | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 0 | Not cross-contamination |
| 24 | + | − | + | + | − | − | N | Y | Y | NA | NA | NA | 0 | 0 | 0 | Not cross-contamination |
| 25 | + | − | − | − | − | − | N | Y | Y | OT− | OT− | OT− | 1 | 0 | 0 | Not cross-contamination |
| 26 | + | − | − | − | − | − | N | Y | Y | OT+ | NA | OT− | 1 | 0 | 0 | Not cross-contamination |
| 27 | + | − | − | − | − | − | N | N | Y | − | − | − | 1 | 1 | 0 | Not cross-contamination |
| 28 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 0 | Not cross-contamination |
| 29 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 0 | Not cross-contamination |
| 30 | + | − | − | − | − | − | Y | − | Y | NA | NA | NA | 1 | 0 | 0 | Not cross-contamination |
| 31 | + | − | + | − | − | − | Y | − | Y | + | + | + | 1 | 0 | 0 | Not cross-contamination |
| 32 | + | − | − | − | − | − | Y | − | Y | − | − | − | 1 | 0 | 0 | Not cross-contamination |
| 33 | + | − | − | − | − | − | Y | − | Y | − | − | − | 1 | 0 | 0 | Not cross-contamination |
| 34 | + | − | − | − | − | − | Y | − | Y | + | − | + | 1 | 0 | 0 | Not cross-contamination |
| 35 | + | − | − | − | − | − | Y | − | Y | + | + | + | 1 | 0 | 0 | Not cross-contamination |
| 36 | + | − | − | − | − | − | NA | − | Y | NA | NA | NA | 1 | NA | 0 | Not cross-contamination |
| 37 | − | + | − | − | − | − | N | Y | Y | − | − | − | 1 | 0 | 0 | Not cross-contamination |
| 38 | − | − | + | − | − | − | N | Y | Y | − | − | − | 1 | 0 | 0 | Not cross-contamination |
| 39 | − | − | + | − | − | − | Y | − | Y | − | − | − | 1 | 0 | 0 | Not cross-contamination |
| 40 | + | + | − | − | − | − | N | N | Y | − | − | − | 1 | 1 | 1 | Probable cross-contamination |
| 41 | + | + | − | − | − | − | Y | − | Y | OT− | OT− | OT− | 1 | 1f | 1 | Probable cross-contamination |
| 42 | + | + | − | − | − | − | Y | − | N | NA | NA | NA | 1 | 1f | 1 | Probable cross-contamination |
| 43 | + | − | − | − | − | − | Y | Y | Y | NA | NA | NA | 1 | 1g | 1 | Probable cross-contamination |
| 44 | + | − | − | − | − | − | Y | Y | Y | NA | NA | NA | 1 | 1g | 1 | Probable cross-contamination |
| 45 | + | − | − | − | − | − | N | N | Y | NA | NA | NA | 1 | 1 | 1 | Probable cross-contamination |
| 46 | + | − | − | − | − | − | N | N | Y | NA | NA | NA | 1 | 1 | 1 | Probable cross-contamination |
| 47 | + | − | − | − | − | − | N | N | Y | − | − | − | 1 | 1 | 1 | Probable cross-contamination |
| 48 | + | − | − | − | − | − | N | N | Y | NA | NA | NA | 1 | 1 | 1 | Probable cross-contamination |
| 49 | + | − | − | − | − | − | N | N | Y | − | − | − | 1 | 1 | 1 | Probable cross-contamination |
| 50 | + | − | − | − | − | − | N | N | Y | − | − | − | 1 | 1 | 1 | Probable cross-contamination |
| 51 | + | − | − | − | − | − | NA | − | Y | NA | NA | NA | 1 | NA | 1 | Probable cross-contamination |
| 52 | − | + | − | − | − | − | NA | − | Y | OT− | OT− | OT− | 1 | NA | 1 | Probable cross-contamination |
| 53 | − | − | + | − | − | − | NA | − | Y | − | − | − | 1 | NA | 1 | Probable cross-contamination |
Though evidence from 1 approach may have implied that cross-contamination was unlikely, in some instances, evidence to the contrary was regarded as superior (e.g., unique and different molecular fingerprints were observed for 2 cultures from the same participant in whom there was no clinical suspicion of disease—she had attended the NTP asymptomatically for an insurance screen). NA = not available.
+ = reculture of original sample positive; − = reculture of original sample negative.
Unique spoligotype for date of sample processing and 2 days at either side (5-day window); 4 patients had 2 samples with identical spoligotypes (11,12; 10,18; 32,33; 43,44) unique to each patient.
Strains with contemporary matching spoligotypes were also typed by RFLP to further aid discrimination; thus, “unique RFLP” refers to differentiation from strains with shared spoligotype.
All results are in duplicate (i.e., 2 samples processed); OT = follow-up sample taken on TB therapy.
Cultured strains from samples 45 and 1, but no other contemporarily processed samples shared identical spoligotypes and RFLP patterns; 45 probably cross-contaminated from 1.
Spoligotypes for 2 isolates from the same sample differ.
Although spoligotypes for these 2 samples from the same patient are temporally unique, they differ from each other, as do their RFLP patterns.
3.2.2. Molecular evaluation
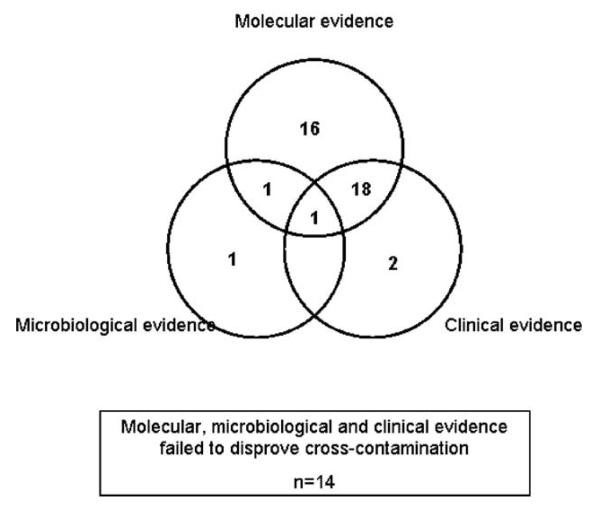
Of the 68 original isolates (from all 53 contamination suspect samples), it was possible to obtain spoligotypes for 64 (from 49 cultured patient samples). Reasons for lack of a spoligotype included loss or absence of stored isolate (n = 3) or technical failure (n = 1). The spoligotypes of the 3 isolates recovered from stored frozen sputum (Table 1) matched those of the strain from the original culture in all 3 cases. Compared with all other positive isolates from the same 5-day period (2 days either side of the date of processing of the contamination suspect), 31 samples were found to have unique spoligotypes, indicative of a very low probability of cross-contamination. Of the remaining 18, which shared a spoligotype with a contemporary isolate, subsequent RFLP fingerprinting confirmed identical strain identity in 10, which were thus deemed, on molecular grounds, highly suspicious of episodes of cross-contamination. The other 8 had RFLP fingerprints that differed from their contemporary spoligotype match. Of the 15 samples culture positive only in MODS and MBBacT (n = 13) or MODS and LJ (n = 2), both isolates showed identical spoligotypes (evidence against cross-contamination) in all except 2 samples (Fig. 4).
Fig. 4.
Clinical, microbiologic, and molecular approach to determining cross-contamination (by sample).
Forty-one patients contributed a single contamination suspect sample, but 6 patients each contributed 2 samples for which all but 1 pair shared identical spoligotypes. Overall, on molecular grounds, cross-contamination was deemed unlikely for 39 samples and still plausible for the other 14 (Fig. 4).
3.2.3. Clinical evaluation
Follow-up information was available for 39 of the 47 patients (accounting for 43 of the 53 samples), 10 of whom had microbiologic confirmation of TB (either positive sputum smear elsewhere or repeat culture in our laboratory) and 19 of whom had been commenced on TB treatment—all 19 reported symptom improvement and had objective increases in weight on therapy (range, 2–8 kg), thus the 9 without microbiologic confirmation were regarded as having probable TB. A further four samples deemed true positives came from 3 patients who refused treatment or further investigation despite continuing constitutional and respiratory symptoms, ongoing weight loss (4–13 kg), and a history of prior TB treatment and/or contact (Table 1).
Although the phenomenon of self-cure is well recognized, we regarded reported recovery without TB treatment as suspicious that the original result had been falsely positive. The study was conducted as an operational evaluation within the NTP in health centers; thus, all NTP-received samples from consenting adults had also been processed in the study—however, a proportion did not fulfill the NTP criteria for a TB suspect and did not receive treatment (e.g., paucisymptomatic index-case contacts, insurance screening examinations), and in such instances, a lack of symptoms or reported recovery from symptoms at the follow-up visit was also deemed highly suggestive, that the original culture had been falsely positive.
Overall, on clinical grounds, cross-contamination was deemed likely for 14 samples, highly unlikely for 23 samples and indeterminate (consistent but not probable) for 16 samples (Fig. 4).
3.2.4. Resolving true/false-positive allocation
Combining the evidence from the 3 distinct approaches, the final assessment inferred that cross-contamination probably accounted for 17 false-positive cultures from 14 samples (Table 1), representing overall false-positive percentages of 0.41% (14/3416) and 0.17% (17/10 248) for samples and cultures, respectively. The number (percentage) of cultures that were deemed false positive because of cross-contamination was 12 (3.3%), 4 (1.1%), and 1 (0.3%) for MODS, MBBacT, and LJ representing specificities of 99.6%, 99.9%, and 99.9%, respectively (data not shown). Positive predictive values for MODS, MBBacT, and LJ were 96.2%, 98.7%, and 99.7%, respectively. The contributions to “cross-contamination rule-out” of each element of the triangulating approach are shown in Fig. 5.
Fig. 5.
Contribution by method to “rule-out” of cross-contamination for 39 samples deemed true-positive cultures.
3.2.5. The effect of more rigorous definition of culture positivity in MODS
A MODS plate has thus far been defined as positive in this study if there was a characteristic and confirmed growth noted in any of the 4 control wells (Fig. 1), regardless of the appearance in the other wells. Of the 12 “probable MODS cross-contamination”, growth was almost universally noted in only 1 (n =10) or 2 (n = 1) wells. By contrast, of the 285 samples culture positive in all 3 methods (Fig. 3), mycobacterial growth was detected in 1, 2, 3, and 4 wells for 1, 2, 1, and 281 samples, respectively. A receiver-operator characteristic curve using these data confirmed the modest but detectable effect of the 4 potential case definitions—growth required in 1, 2, 3, or 4 wells—for a positive culture (not shown). By revising the case definition for a positive MODS culture to detection of mycobacterial growth in at least 2 of the 4 control wells, the number of false-positive cultures is reduced to 3. This equates to overall study and MODS-specific false-positive culture rates of 1.4% and 0.75% with a MODS positive predictive value of 99.1% and specificity of 99.9%.
4. Discussion
The key finding of this head-to-head study of MODS with MBBacT and LJ culture was the similarly low proportion of positive cultures determined to be false positive because of cross-contamination and high specificity (99.9%) of all 3 methods. In contrast to accepted wisdom and some data (Small et al., 1993; Gascoyne-Binzi et al., 2001), we found no significant difference between the cross-contamination rates in liquid (MBBacT and MODS) and solid (LJ) media, despite the greater detection sensitivity of the former.
There were 2 major strengths of this study: first, we were able to evaluate 2 samples from most patients by all 3 culture methods, and second we were then able to investigate the likelihood that a positive culture represented cross-contamination by a triangulating approach of microbiologic, molecular, and clinical (conventional “shoe-leather”) epidemiology.
We applied strict criteria in requiring smear-negative samples (or the patient's other sample) to be culture positive in all 3 methods to be above suspicion of cross-contamination. Multiple contamination episodes affecting a patient's cultures have rarely been reported (Bauer et al., 1997), and we cannot exclude the possibility that occasional samples regarded as meeting this criterion resulted from cross-contamination. Patient follow-up was incomplete, which prevented “rule-out” of cross-contamination on clinical grounds in 11 untraceable individuals. All had provided false addresses, a phenomenon we have described previously in TB suspects (Ouyang et al., 2005) and a potentially important obstacle to efficient TB case management.
Reported mycobacterial cross-contamination rates vary widely, partly reflecting reporting bias and nonstandardized ascertainment methodologies—pseudooutbreak reports have reached rates of 65% (de Ramos et al., 1999), whereas comprehensive molecular epidemiologic studies elsewhere have demonstrated underlying rates of 0.1% to 4% (Bauer et al., 1997; Burman et al., 1997; de Boer et al., 2002; Jasmer et al., 2002; Ruddy et al., 2002). Our findings, the 1st reported from a resource-limited, high TB-burden setting, fall within this range for all 3 methodologies. The potential for cross-contamination is greater in laboratories handling larger numbers of culture-positive samples and manipulating isolates (Carroll et al., 2003). That we were able to maintain such low-level cross-contamination probably reflects both the unusually bio-secure nature of our developing world laboratory facility (P3 level containment) and the experience of our mycobacterial laboratory staff.
Because the cross-contaminating inoculum is usually small (droplet), most false-positive cultures have low colony counts (MacGregor et al., 1975) and/or require prolonged incubation for detection. The differential cross-contamination rates determined by the number of MODS wells with confirmed growth is a useful check—the presence of characteristic tangles in at least 2 wells is highly unlikely to be a false-positive or cross-contamination event.
Cross-contamination can occur at every stage from specimen collection to strain handling for indirect drug susceptibility testing (DST) or species determination (in which the concentration of M. tuberculosis has been greatly amplified from the source sample) (Van Duin et al., 1998) and is further favored by inadequate environmental controls (Segal-Maurer et al., 1998). The contribution of external soiling of the much handled sputum pot may also be underestimated (Allen and Darrell 1983). Cross-contamination may occur directly (sample-sample, sample-culture media, isolate-sample, isolate-culture media) or indirectly (e.g., contaminated stock solution) (Van Duin et al., 1998), though modification of laboratory practice (e.g., avoiding common flasks for reagent dispensing, waiting 5 min after centrifuging to allow settlement of potential tube aerosols) can reduce the incidence of false-positive cultures (Breese et al., 2001; Carroll et al., 2002). The MODS methodology inherently protects against cross-contamination from the point of sample inoculation onward because the culture is then sealed within a transparent plastic bag. Direct DST obviates the need for further culture manipulation, reducing the potential for cross-contamination and biohazard for laboratory staff. Cross-contamination between adjacent samples cultured on the same plate has not been observed, concordant with the observation that in > 1100 MODS cultures over a 3-year period, contaminating growth was never seen in a row of adjacent sample-free control wells containing media alone (Luz Caviedes, unpublished data).
New molecular techniques for strain differentiation (Burman et al., 1997; Poynten et al., 2002; Small et al., 1993; Bauer et al., 1997; de Ramos et al., 1999; Nivin et al., 2000; Filho et al., 2002; Gascoyne-Binzi et al., 2001) with superior sensitivity and discriminatory power have rendered conventional methods such as phage testing (Sula and Sulova 1979; Maurer et al., 1984; Jones 1988) obsolete. Spoligotyping is a useful 1st-line tool (de Ramos et al., 1999; Nivin et al., 2000) in a 2-stage algorithm (as used in this study) in which subsequent RFLP examination may enhance discrimination of suspicious strains sharing the same spoligotype.
Methodologies with low innate cross-contamination risk are crucial in resource-limited high-burden settings, where fluctuations in case detection or emergence of unusual phenotypes (Smith and Vance 1991; Nitta et al., 1996; Wurtz et al., 1996; Bearman et al., 2002) or genotypes (Nivin et al., 1998; Anonymous 2000) are much less likely to raise the “cross-contamination alarm”. Designing result readouts so that positive results for sequentially processed samples are easily seen (Bauer et al., 1997) is one potential strategy to aid detection.
In summary, through a vigorous 3-pronged approach to investigation of potential false-positive cultures, we have clearly defined that the cross-contamination rates in MODS, LJ, and MBBacT were all equally low. The finding that, even in this resource-poor setting, the use of the highly sensitive but technically simple liquid culture assay MODS is associated with a specificity of 99.9% addresses a key concern and removes an important obstacle to the wider implementation of this inexpensive methodology.
Acknowledgements
The following are thanked for their contributions of ensuring the success of this study: Adolfo Orellana, Milciades Reátegui Sanchez, Yuri Garcia, Maria Burga, Felipe Castillo, Fanny Garcia, Eleana Sanchez, Yrma Chuquiruna, Sonia Lopez, Rosmery Gutierrez, Raul Miranda Arrostigue, Betty Cajaleón Ascencios, José Calderón Yberico, Luis Rivera Pérez, Walter Ramos Maguiña, Luz Vásquez Chávez, Félix Pari Loayza, Ruth Flores Escobar, Jesús Castillo Diaz, Alicia Vigo Alegria.
References
- Allen BW, Darrell JH. Contamination of specimen container surfaces during sputum collection. J Clin Pathol. 1983;36:479–481. doi: 10.1136/jcp.36.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous Misdiagnoses of tuberculosis resulting from laboratory cross-contamination of Mycobacterium tuberculosis cultures—New Jersey, 1998. MMWR Morb Mortal Wkly Rep. 2000;49:413–416. [PubMed] [Google Scholar]
- Bauer J, Thomsen VO, et al. False-positive results from cultures of Mycobacterium tuberculosis due to laboratory cross-contamination confirmed by restriction fragment length polymorphism. J Clin Microbiol. 1997;35:988–991. doi: 10.1128/jcm.35.4.988-991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearman G, Vaamonde C, et al. Pseudo-outbreak of multidrug-resistant Mycobacterium tuberculosis associated with presumed laboratory processing contamination. Infect Control Hosp Epidemiol. 2002;23:620–622. doi: 10.1086/501982. [DOI] [PubMed] [Google Scholar]
- Breese PE, Burman WJ, et al. The effect of changes in laboratory practices on the rate of false-positive cultures for Mycobacterium tuberculosis. Arch Pathol Lab Med. 2001;125:1213–1216. doi: 10.5858/2001-125-1213-TEOCIL. [DOI] [PubMed] [Google Scholar]
- Burman WJ, Reves RR. Review of false-positive cultures for Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin Infect Dis. 2000;31:1390–1395. doi: 10.1086/317504. [DOI] [PubMed] [Google Scholar]
- Burman WJ, Stone BL, et al. The incidence of false-positive cultures for Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:321–326. doi: 10.1164/ajrccm.155.1.9001331. [DOI] [PubMed] [Google Scholar]
- Carroll NM, Richardson M, et al. Reduction of the rate of false-positive cultures of Mycobacterium tuberculosis in a laboratory with a high culture positivity rate. Clin Chem Lab Med. 2002;40:888–892. doi: 10.1515/CCLM.2002.157. [DOI] [PubMed] [Google Scholar]
- Carroll NM, Richardson M, et al. Criteria for identification of cross-contamination of cultures of Mycobacterium tuberculosis in routine microbiology laboratories. J Clin Microbiol. 2003;41:2269. doi: 10.1128/JCM.41.5.2269-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviedes L, Lee TS, et al. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The Tuberculosis Working Group in Peru. J Clin Microbiol. 2000;38:1203–1208. doi: 10.1128/jcm.38.3.1203-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer AS, Blommerde B, et al. False-positive Mycobacterium tuberculosis cultures in 44 laboratories in The Netherlands (1993 to 2000): incidence, risk factors, and consequences. J Clin Microbiol. 2002;40:4004–4009. doi: 10.1128/JCM.40.11.4004-4009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ramos CM, Soini H, et al. Extensive cross-contamination of specimens with Mycobacterium tuberculosis in a reference laboratory. J Clin Microbiol. 1999;37:916–919. doi: 10.1128/jcm.37.4.916-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho LA, Kritski AL, et al. Mycobacterium tuberculosis typing: usefulness of DRE-PCR to confirm cross-contamination in the mycobacteriology laboratory of a general reference hospital for AIDS. Int J Tuberc Lung Dis. 2002;6:150–154. [PubMed] [Google Scholar]
- Gascoyne-Binzi DM, Barlow RE, et al. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J Clin Microbiol. 2001;39:69–74. doi: 10.1128/JCM.39.1.69-74.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal M, Saunders NA, et al. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmer RM, Roemer M, et al. A prospective, multicenter study of laboratory cross-contamination of Mycobacterium tuberculosis cultures. Emerg Infect Dis. 2002;8:1260–1263. doi: 10.3201/eid0811.020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD., Jr Bacteriophage typing of Mycobacterium tuberculosis cultures from incidents of suspected laboratory cross-contamination. Tubercle. 1988;69:43–46. doi: 10.1016/0041-3879(88)90039-6. [DOI] [PubMed] [Google Scholar]
- MacGregor RR, Clark LW, et al. The significance of isolating low numbers of Mycobacterium tuberculosis in culture of sputum specimens. Chest. 1975;68:518–523. doi: 10.1378/chest.68.4.518. [DOI] [PubMed] [Google Scholar]
- Maurer JR, Desmond EP, et al. False-positive cultures of Mycobacterium tuberculosis. Chest. 1984;86:439–443. doi: 10.1378/chest.86.3.439. [DOI] [PubMed] [Google Scholar]
- Moore DA, Mendoza D, et al. Microscopic observation drug susceptibility assay, a rapid, reliable diagnostic test for multidrug-resistant tuberculosis suitable for use in resource-poor settings. J Clin Microbiol. 2004;42:4432–4437. doi: 10.1128/JCM.42.10.4432-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta AT, Davidson PT, et al. Misdiagnosis of multidrug-resistant tuberculosis possibly due to laboratory-related errors. JAMA. 1996;276:1980–1983. [PubMed] [Google Scholar]
- Nivin B, Fujiwara PI, et al. Cross-contamination with Mycobacterium tuberculosis: an epidemiological and laboratory investigation. Infect Control Hosp Epidemiol. 1998;19:500–503. doi: 10.1086/647856. [DOI] [PubMed] [Google Scholar]
- Nivin B, Driscoll J, et al. Use of spoligotype analysis to detect laboratory cross-contamination. Infect Control Hosp Epidemiol. 2000;21:525–527. doi: 10.1086/501799. [DOI] [PubMed] [Google Scholar]
- Northrup JM, Miller AC, et al. Estimated costs of false laboratory diagnoses of tuberculosis in three patients. Emerg Infect Dis. 2002;8:1264–1270. doi: 10.3201/eid0811020387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H, Chepote F, et al. Failure to complete the TB diagnostic algorithm in urban Perú: a study of contributing factors. Trop Doc. 2005;35:120–121. doi: 10.1258/0049475054037002. [DOI] [PubMed] [Google Scholar]
- Park WG, Bishai WR, et al. Performance of the microscopic observation drug susceptibility assay in drug susceptibility testing for Mycobacterium tuberculosis. J Clin Microbiol. 2002;40:4750–4752. doi: 10.1128/JCM.40.12.4750-4752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynten M, Andresen DN, et al. Laboratory cross-contamination of Mycobacterium tuberculosis: an investigation and analysis of causes and consequences. Intern Med J. 2002;32:512–519. doi: 10.1046/j.1445-5994.2002.00271.x. [DOI] [PubMed] [Google Scholar]
- Ruddy M, McHugh TD, et al. Estimation of the rate of unrecognized cross-contamination with Mycobacterium tuberculosis in London microbiology laboratories. J Clin Microbiol. 2002;40:4100–4104. doi: 10.1128/JCM.40.11.4100-4104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal-Maurer S, Kreiswirth BN, et al. Mycobacterium tuberculosis specimen contamination revisited: the role of laboratory environmental control in a pseudo-outbreak. Infect Control Hosp Epidemiol. 1998;19:101–105. doi: 10.1086/647774. [DOI] [PubMed] [Google Scholar]
- Small PM, McClenny NB, et al. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol. 1993;31:1677–1682. doi: 10.1128/jcm.31.7.1677-1682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WB, Vance DW., Jr Specimen cross-contamination by a strain of Mycobacterium tuberculosis lacking nitrate reductase activity. Diagn Microbiol Infect Dis. 1991;14:523–526. doi: 10.1016/0732-8893(91)90012-5. [DOI] [PubMed] [Google Scholar]
- Sula L, Sulova J. Accidental contamination of diagnostic cultures of mycobacteria and phage identification of the contaminating strain. Tubercle. 1979;60:159–162. doi: 10.1016/0041-3879(79)90016-3. [DOI] [PubMed] [Google Scholar]
- Van Duin JM, Pijnenburg JE, et al. Investigation of cross contamination in a Mycobacterium tuberculosis laboratory using IS6110 DNA fingerprinting. Int J Tuberc Lung Dis. 1998;2:425–429. [PubMed] [Google Scholar]
- van Embden JD, Cave MD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Laboratory Services in Tuberculosis Control. Part III—Culture, 41. World Health Organization; Geneva: 1998. [Google Scholar]
- Wurtz R, Demarais P, et al. Specimen contamination in mycobacteriology laboratory detected by pseudo-outbreak of multidrug-resistant tuberculosis: analysis by routine epidemiology and confirmation by molecular technique. J Clin Microbiol. 1996;34:1017–1019. doi: 10.1128/jcm.34.4.1017-1019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]