Fig. 5.
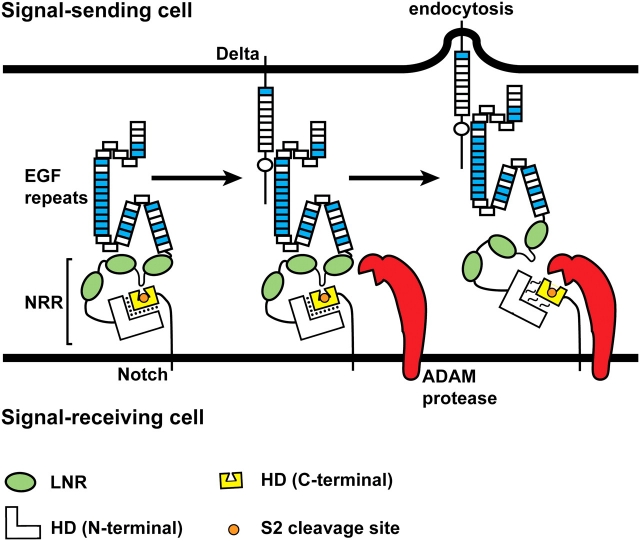
Ligand endocytosis makes Notch sensitive to the S2 proteolytic cleavage. The negative regulatory region (NRR) consists of the LNR motifs (green ovals) and the N-terminal and C-terminal parts of the heterodimerization domain (HD). At the cell membrane, the S2 cleavage site (orange circle) is buried in a cleft in the C-terminal part of the HD domain (yellow). A loop between the first two LNR motifs functions as a plug to ensure that the ADAM protease cannot gain access to the S2 cleavage site (Gordon et al. 2007). Binding of ligand from the neighboring signal-sending cell to the Notch receptor and endocytosis of the Notch-bound ligand into the signal-sending cell lead to the disengagement of the LNR motifs from the HD domain and relaxation of the interaction between the C-terminal and N-terminal parts of the HD domain. As a result, the S2 cleavage site is exposed, and Notch becomes a substrate for cleavage by the ADAM protease. The force generated by ligand endocytosis is thought to cause the above-mentioned conformational changes in the extracellular domain of Notch, but it is also possible that allosteric changes—independent of force—are involved (Gordon et al. 2008). The folds drawn in the Notch ECD are based on the reports that the linkage between non-Ca+//0-binding EGF-like repeats and their neighboring EGF-like repeats can be flexible, whereas the linkage between two Ca+//0-binding EGF-like repeats is rigid (Downing et al. 1996; Hambleton et al. 2004; Irvine 2008). See Figure 3 for the distribution of Ca+//0-binding EGF-like repeats in Notch receptors. Although not shown in this schematic, the three-dimensional structure of the Notch ECD might also change upon ligand binding and endocytosis