Abstract
Antisense oligonucleotides (ASOs) can be used to alter the splicing of a gene and either restore production of a required protein or eliminate a toxic product. In this issue of Genes & Development, Hua and colleagues (pp. 1634–1644) show that ASOs directed against an intron splice silencer (ISS) in the survival motor neuron 2 (SMN2) gene alter the amount of full-length SMN transcript in the nervous system, restoring SMN to levels that could correct spinal muscular atrophy (SMA).
Keywords: Spinal muscular atrophy, SMN2, antisense oligonucleotide, splicing correction, spinal cord, mouse models
Spinal muscular atrophy (SMA)
SMA is an autosomal recessive disorder caused by loss or mutation of the survival motor neuron 1 gene (SMN1) and retention of the SMN2 gene (Lefebvre et al. 1995). The essential difference between SMN1 and SMN2 is a C-to-T transition in exon 7 (Lefebvre et al. 1995; Lorson et al. 1999; Monani et al. 1999; Cartegni and Krainer 2002; Kashima and Manley 2003). This change does not alter an amino acid, but rather disrupts a splice modulator. This splice modulator has been described as an exon splice enhancer (ESE) or an exon splice silencer (ESS), and may well act in both capacities (Cartegni and Krainer 2002; Kashima and Manley 2003). The net result of disruption of this splice modulator is that the majority of the transcript from SMN2 lacks exon 7, which encodes the C-terminal domain of SMN (Lefebvre et al. 1995; Lorson et al. 1999; Monani et al. 1999; Cartegni and Krainer 2002; Kashima and Manley 2003). The loss of the C-terminal sequence results in a SMN protein that does not oligomerize efficiently, and thus, like many oligomeric proteins, gets degraded rapidly (Lorson and Androphy 2000; Burnett et al. 2009). In ∼5% of SMA cases, one SMN1 allele has a small deletion, insertion, or point mutation. In some cases, such as in the Y272C mutant, these missense point mutations occur in the SMN oligomerization domain, and disrupt SMN's ability to oligomerize (Lorson et al. 1998). The amount of full-length SMN protein produced by these alleles is severely reduced (Coovert et al. 1997; Lefebvre et al. 1997; Burnett et al. 2009). Since SMN2 has a disrupted splice modulator, the gene still produces some full-length transcripts (∼10%), and thus some SMN proteins (Lefebvre et al. 1995, 1997; Coovert et al. 1997). Indeed, in every organism studied to date, complete loss of SMN is embryonic-lethal (Burghes and Beattie 2009). This situation can also be inferred in humans, as chromosomes that lack both SMN1 and SMN2 can occur in the human population, but have never been reported in a homozygous state (Burghes and Beattie 2009). The SMN2 gene has evolved recently, and occurs only in humans (chimpanzees have two SMN1 genes). Thus, SMA is caused by loss or mutation of SMN1, and the resulting insufficient levels of SMN protein produced by SMN2 (Lefebvre et al. 1995, 1997; Coovert et al. 1997). Consequently, the SMN2 gene is a major modifier of the severity of the SMA phenotype (Lefebvre et al. 1995; McAndrew et al. 1997; Burghes and Beattie 2009). The correlation of severity with SMN2 copy number is inverse: Patients with severe SMA have low SMN2 copy numbers, and mildly affected SMA patients have more SMN2 copies. Thus, in general, type I SMA patients have two copies of SMN2, type II SMA patients have three copies, and type III patients have four copies. However, there are exceptions in that it cannot be assumed that all SMN2 genes are intact. In addition, certain SMN2 alleles themselves contain a variation that increases incorporation of exon 7, making SMN2 an even more effective modifier (Prior et al. 2009; Vezain et al. 2010). Moreover, SMN protein that is lacking the exon 7-encoded amino acids cannot be considered nonfunctional, as it can associate with wild-type SMN and participate in the SMN complex (Le et al. 2005). In the case of missense alleles, those generating a mild phenotype can be defined as those that interact with wild-type SMN produced by SMN2 to give a functional SMN complex (Burghes and Beattie 2009; Workman et al. 2009). This situation is referred to as allelic complementation. Severe missense alleles are either inefficient at associating with wild-type SMN, or associate but do not result in a functional complex (fail to complement), and do not act in a dominant-negative manner (Burghes and Beattie 2009). SMN is expressed ubiquitously in cells, and is essential in the assembly of Sm proteins onto snRNA (Lefebvre et al. 1995; Meister et al. 2001; Pellizzoni et al. 2002). This function has been shown to be altered in SMA (Gabanella et al. 2007; Workman et al. 2009). However, it cannot be ruled out that other assembly reactions are also disrupted (Burghes and Beattie 2009). As such, SMN could be critical in the assembly of complexes important for transport of mRNA down axons (for review, see Burghes and Beattie 2009). Thus, the exact molecular mechanism by which reduction of SMN causes SMA is not currently known, and this has been discussed extensively elsewhere (Burghes and Beattie 2009). SMA has been modeled in mice (Hsieh-Li et al. 2000; Monani et al. 2000). In mice, as in other species, there is only one Smn gene, and knocking out this gene results in embryonic lethality (Schrank et al. 1997). Introduction of human SMN2 into animals lacking mouse Smn results in the rescue of embryonic lethality, with eight copies of SMN2 giving complete rescue and two copies of SMN2 giving severe SMA mice. Mouse deletions can result from either the loss of exon 7 (Hsieh-Li et al. 2000) or disruption of exon 2 (Schrank et al. 1997), which is essentially a null, whereas SMN lacking exon 7 does have some function. Thus, mice with two copies of SMN2 in an exon 7 deletion background have a milder phenotype. In addition, the copy number of SMN2 will affect the phenotype: Four copies of SMN2 yields a very mild phenotype (Hsieh-Li et al. 2000). Regardless of the mechanism by which SMA is caused, SMN2 is present in all SMA patients, and increasing SMN postnatally in animal models of SMA has been shown to be effective in correcting SMA (Foust et al. 2010; Passini et al. 2010).
Elements that regulate splicing of SMN and their modulation
Apart from the ESE/ESS that is altered between SMN1 and SMN2, there are numerous negative and positive elements within exon 7 and its flanking introns that affect the incorporation of exon 7 into the final transcript (Bebee et al. 2010; Lorson et al. 2010). These elements have been reviewed extensively, and consist of intron splice silencers (ISSs), intron splice enhancers (ISEs), ESEs, and ESSs (Bebee et al. 2010; Lorson et al. 2010). Blocking the binding of a protein to an ISS or enhancing the required splicing-positive proteins (SR) can increase the amount of full-length SMN produced by SMN2 (Cartegni and Krainer 2003; Skordis et al. 2003; Singh et al. 2006, 2009). Both approaches have been used previously. Krainer's group (Cartegni and Krainer 2003) reported that an antisense peptide nucleic acid (PNA; an artificially synthesized polymer composed of repeating N-[2-amionoethyl]-glycine units linked by peptide bonds) directed against the 5′ end of exon 7 and containing 10 copies of a RS peptide domain tail (the domain in SR proteins responsible for encouraging incorporation of exon) increased incorporation of SMN exon 7. Skordis et al. (2003) reported that an antisense oligonucleotide (ASO) that bound the same region of exon 7 and contained a tail capable of binding SR proteins enhanced incorporation of SMN exon 7 from the SMN2 gene in both minigenes and SMA patient fibroblasts. Subsequently, this antisense-tailed construct was placed in a U7snRNA expression vector and delivered to SMA fibroblasts, resulting in inclusion of exon 7 from SMN2 and an increase in the amount of SMN produced in these cells (Marquis et al. 2007). The U7snRNAs were then used to construct transgenic mice expressing the modifying antisense construct (Meyer et al. 2009). This transgene was crossed into the severe SMA mice. Despite relatively low expression of the U7snRNA construct, a marked increase in survival occurred, clearly indicating that sufficient functional SMN can be obtained by altering the splicing of SMN2 (Meyer et al. 2009).
The ASO: its chemistry and use in other disorders
The development of most effective ASO therapies for SMA requires careful selection of both the sequence to be used and the ASO backbone chemistry. The approaches to blocking or enhancement of SMN exon 7 inclusion are shown in Figure 1. Blocking of an ISS is one of the most effective methods for increasing incorporation of exon 7 of SMN2 (Singh et al. 2006, 2009; Hua et al. 2007, 2008). In particular, Singh et al. (2006) identified an ISS (ISS-N1) adjacent to exon 7 that binds hnRNPA1 and has a marked effect on incorporation of exon 7. Further studies using ASO tiling arrays across this region of the gene again identified this ISS as having a marked effect on incorporation of exon 7 (Hua et al. 2007, 2008). This element is the basis of the work reported in the current paper in this issue of Genes & Development (Hua et al. 2010) demonstrating that SMN2 can be modified very effectively in the nervous system to produce sufficient SMN. Systemic delivery of an ASO blocking ISS-N1 has resulted in restoration of SMN in tissues outside the nervous system (Hua et al. 2008). However, oligonucleotides do not cross the blood–brain barrier, and thus correction in the nervous system does not occur (Hua et al. 2008; Singh et al. 2009; Isis_Pharmaceuticals, http://www.isispharm.com). Although there are studies on reducing the size of the ASO targeting this ISS, the advantage of such shorter ASOs is unclear, as they reduce target specificity without resolving the issue of access to the nervous system (Singh et al. 2009).
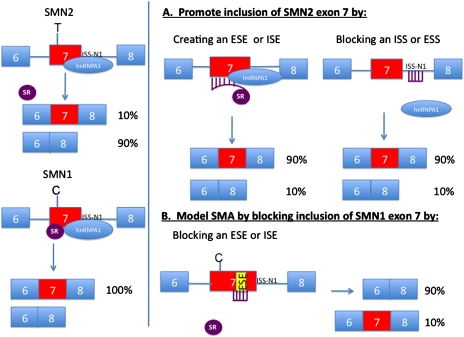
Figure 1.
Diagram of SMN1 and SMN2, and how splicing of the gene can be altered using ASOs. On the left side, the SMN2 (top) and SMN1 (bottom) genes are shown with the resulting transcripts. Note the C (SMN1)-to-T (SMN2) change is shown as disrupting a splice enhancer, but it may also act as a splice silencer (see the text). The SMN1 transcript binds both the splice activator ASF/SF2 (SR protein) and the hnRNPA1 protein, and the mRNA includes exon 7. SMN2 does not bind the splice activator, and hence the majority of the mRNA transcript lacks exon 7. The ISS-N1 is shown as binding hnRNPA1, a known negative regulator of splicing. (A) The SMN2 gene splicing is modified by either a synthetic ASO that allows binding of the SR protein to exon 7, or an ASO that blocks the binding of hnRNPA1 to the ISS-N1 site. (B) The ability to block the binding of a SR protein to a separate ESE in SMN1 exon7 is shown, essentially turning the SMN1 gene into a SMN2 gene.
Currently, there is limited information on the different chemistries of ASOs and their action on SMN2 in the nervous system. In Duchenne Muscular Dystrophy, ASOs have been used to induce exon skipping. This corrects the reading frame, and results in the production of some dystrophin with a phenotype similar to the milder Becker Muscular Dystrophy (Fletcher et al. 2006; van Deutekom et al. 2007). In the case of dystrophin, two main ASO types have been investigated: the 2-O-Methyl with a phosphorothioate backbone (Prosensa_Therapeutics, http://www.prosensa.eu), and the morpholinos (Fletcher et al. 2006; van Deutekom et al. 2007; AVI_BioPharma, http://www.avibio.com). Both appear effective in inducing exon skipping in dystrophin. Morpholinos have been delivered through the vascular system, and were shown to induce restoration of the dystrophin reading frame throughout the body (Fletcher et al. 2006). However, there is still debate over the exact optimal chemistry of the oligonucleotides to be used. This debate will extend to SMA, as the efficacy of 2-O-Methyl and morpholinos in comparison with 2-O-Methoxyethyl needs to be completely tested (Isis_Pharmaceuticals, http://www.isispharm.com). Furthermore, additional chemistry could be developed, and often the chemistries are proprietary to particular companies, making it difficult to evaluate the optimal product.
In the nervous system, ASOs have been delivered in mice and monkeys to reduce SOD1 (Miller et al. 2008). Mutations in SOD1 result in familial amyotrophic lateral sclerosis (ALS), and mouse models of ALS have been made on this basis. In general, SOD1 ALS is regarded as a gain-of-function mutation or neomorph, although there are some indications of wild-type SOD1 influencing the phenotype (Polymenidou and Cleveland 2008). However, loss of SOD1 in mice does not result in any major phenotype; thus, knockdown of SOD1 can be used to reduce the toxic load of mutant SOD1. Indeed, reduction in SOD1 levels in ALS mice does ameliorate the phenotype (Miller et al. 2008). This has led to the use of ASO gapmers to reduce SOD1 levels in ALS mice. In these gapmers, the middle portion of the ASO contains a mismatch to encourage RNaseH action. Delivery of gapmers by intracerberal ventricle injection (ICV) has been effective in reducing SOD1 levels and the ALS mouse phenotype. These studies have been extended to monkeys, in which uptake of the ASO into motor neurons as well as other neurons occurs in a relatively efficient manner. Indeed, these results have led to the initiation of a clinical trial of SOD1 ASOs in ALS patients that have SOD1 mutations (Muscular Dystrophy Association, http://www.MDA.org).
Delivery of ASOs in SMA mice
In the case of SMA, 2-O-Methyl ASOs directed against ISS-N1 have been administered to SMA mice (Williams et al. 2009). In particular, the ASO was administered to the so-called Δ7 mice, which lack mouse Smn and contain two copies of human SMN2, as well as a transgene that expresses high levels of SMNΔ7 (Le et al. 2005). These mice have an average lifespan of 13–14 d (Le et al. 2005). In the Williams et al. (2009) study, the investigators performed multiple ICV injections into the Δ7 mice at different time points. Perhaps not surprisingly, this resulted in death in both SMA and control groups, making it difficult to draw conclusions about the potential of the ASOs (Williams et al. 2009). In addition, there was no assessment of the ASO concentration or stability in vivo. Interestingly, delivery of the naked 2-O-Methyl ASO directed against ISS-N1 was more effective than an ASO complexed with lipids (Williams et al. 2009). Williams et al. (2009) did report a modest increase in SMN levels. In this issue of Genes & Development, Hua et al. (2010) present convincing findings that demonstrate essential preclinical data for the use of 2-O-Methoxyethyl phosophorothioate oligonucleotides (MOEs) as opposed to 2-O-Methyl phosphorothioate oligonucleotides for modification of SMN2 splicing to produce more SMN in SMA. Previously, Hua et al. (2008) optimized the size of the MOE ASO specific to ISS-N1 to an 18mer, and reported that systemic introduction of this ASO modified the splicing of SMN2 in a number of tissues. Importantly, splicing of SMN2 was not reported as altered in neural tissues such as the brain and spinal cord, as the oligonucleotide does not cross the blood–brain barrier (Hua et al. 2008). In the current study, Hua et al. (2010) used ICV dosing in a type III SMA mouse model, as well as single-dose ICV injections into neonatal and embryonic mice. The SMA mouse used in these studies has four copies of SMN2, and a deletion that removes exon 7 of the mouse Smn gene (Hsieh-Li et al. 2000). Thus, the majority of SMN protein produced is from SMN2 and is of human origin. When four copies of SMN2 are present, SMA mice show necrosis of the tail and ears, but no other marked phenotype (Hsieh-Li et al. 2000). On the other hand, mice with two copies of SMN2 in the presence of the mouse Smn allele deleted for mouse exon 7 show a marked phenotype (Hsieh-Li et al. 2000). These mice die, on average, at 11 d, and are very similar to SMNΔ7 mice showing a clear motor neuron phenotype (Le et al. 2005). In type 1 SMA patients, there have been reports of cases with distal necrosis associated with anomalous vascular perfusion, and thus it can be viewed that the mouse phenotype has some similarity to what is observed in humans (Araujo Ade et al. 2009). However, this phenotype has not been reported in milder SMA cases, and the exact origins of this phenotype remain unclear, although in mice it is clearly dependent on SMN levels (i.e., eight-copy SMN2 mice do not show this phenotype) (Monani et al. 2000). Regardless of the exact phenotype in the type III SMA mouse, the model can and has been used to evaluate the effectiveness of the MOE ISS-N1 ASO in modifying the SMN2 gene to produce more SMN protein (Hua et al. 2010). Hua et al. (2010) show that the ASO against ISS-N1—when introduced via ICV and an osmotic pump into adult mice containing SMN2—altered the splicing of SMN2 such that more full-length transcript was produced. A dose of 50 μg per day resulted in 90% inclusion of SMN exon 7 in spinal cord samples (Hua et al. 2010). Furthermore, Hua et al. (2010) demonstrated that SMN protein levels were increased significantly with 50 μg of ASO per day, resulting in a 4.3-fold increase in SMN. Similar effects were also seen in the brain. Immunostaining with an antibody that specifically recognizes human SMN, and with an antibody directed against the ASO, demonstrated that the ASO was found in the motor neurons of the spinal cord, showing that these motor neurons had an increase in human SMN in those neurons. Thus, 50 μg per day resulted in an effective dose throughout the spinal cord. The ASO is extremely stable, and, after a 7-d treatment, the effect could still be seen half a year later. Thus, these ASOs, which are not gapmers, are even more stable, and give a long-term effect. Importantly, there was no indication of toxicity at the doses used. Hua et al. (2010) also compared their 18mer MOE to the previously reported 20mer with 2-O-Methyl chemistry. Somewhat surprisingly, the 20mer 2-O-Methyl ASO was not effective in altering splicing of SMN2 in the nervous system, and also induced inflammation. Whether this would be the case with an 18mer 2-O-Methyl or whether morpholino chemistry could hold additional benefits is not known. Hua et al. (2010) then looked at correction of the tail and ear phenotype of type III SMA mice by a single-dose injection of the ASO via ICV into either day 15 embryonic mice or neonatal mice. A single 20-μg dose of the MOE ASO in embryonic mice resulted in considerable rescue of both the tail and ear phenotype, whereas neonatal injection had a less pronounced effect. As the tail and ear phenotype are SMN-dependent phenotypes, this could indicate an effective increase in SMN levels. Indeed, after a postnatal day 1 (P1) injection, spinal cord tissue showed an 89% exon 7 inclusion. Since the splice correction was limited to the nervous system, the tail and ear necrosis phenotypes do appear related to the nervous system. The next step is to repeat this study in more severely affected SMA mice that have motor neuron defects. Yet, from previous studies, certain requirements can be inferred. An approximately threefold increase in SMN levels from SMN2 in the nervous system is required in order to get a major impact on the SMA phenotype (Burghes and Beattie 2009). In adult mice, Hua et al. (2010) obtained a 4.3-fold increase in SMN levels and extensive distribution throughout the nervous system using the MOE ASO. Thus, it is expected that phenotypic correction can be obtained postnatally. Recently, two studies have investigated the use of AAV for delivery of SMN to SMA mice (Foust et al. 2010; Passini et al. 2010). In the first study, AAV8-hSMN or a self-complimentary AAV8-hSMN vector (scAAV8-hSMN) was used that allows earlier expression and appears to transduce more motor neurons more efficiently (Passini et al. 2010). The viral vector was introduced by ICV combined with intraspinal injections (Passini et al. 2010). The injections were performed at P1/P2, and, in particular for the scAAV8-hSMN, resulted in a major improvement in survival from 14 d to 157 d and correction of motor phenotypes (Passini et al. 2010). The SMN was confined to the nervous system, and the rescued mice had shorter tails and some mild necrosis (Passini et al. 2010). In the second case, scAAV9-hSMN was delivered via the vascular system to a large proportion of motor neurons in P1 or P2 animals (Foust et al. 2010). It has been shown previously that scAAV9 can be introduced into the vascular system, transverse the blood–brain barrier, and transduce motor neurons (Duque et al. 2009; Foust et al. 2009). In the case of scAAV9-hSMN, the SMA mice survive >200 d, and show rescued neuromuscular junction physiology and normal behavior (Foust et al. 2010). However, again, the tails are shorter and ear necrosis is present (Foust et al. 2010). Thus, one can obtain substantial correction of SMA motor phenotype by postnatal induction of SMN, and the necrosis and/or short tail phenotype is not necessarily linked to rescue of the motor phenotypes. Furthermore, ASOs can achieve the level of SMN required for rescue, and we would predict that, when introduced into the more severe SMNΔ7 mice, they will impart substantial rescue of motor phenotypes.
Two methods to restore high SMN levels in SMA mice have been described: scAAV vectors expressing SMN delivered by ICV and intrathecal injection or through the vascular system, and delivery of ASOs delivered by ICV through pumps (Foust et al. 2010; Hua et al. 2010; Passini et al. 2010). One can debate the advantages and disadvantages of both approaches; presumably, AAV will be administered once at a particular time point, whereas ASOs will be delivered continuously via pumps. In the case of ASOs, it could be argued that one could withdraw the treatment; however, the relative stability of the current oligonucleotides indicates that both the ASO and the effect on SMN2 will remain long after the removal of the ASO. It is currently unclear if a patient requires high levels of SMN throughout life, or only during a certain period in development. Once adulthood is reached, it is possible that SMN2 could produce sufficient levels of SMN for motor neurons. With regard to both the AAV and ASO approaches, further safety studies in primates and larger organisms are required before clinical trials can be undertaken. In addition, a number of studies examining drug compounds that increase SMN produced by SMN2 are under way. Indeed, the first compounds developed through the use of high-throughput screens have a weak effect in SMA mice, most likely due to the relatively low levels of SMN induction (Butchbach et al. 2010). Certainly a drug candidate that elevated SMN to levels similar to what has been obtained with ASOs and AAV would be a welcome addition.
In conclusion, ASOs that increase the expression of SMN from SMN2 in the nervous system have been developed, and are now poised to move into larger animal studies and, eventually, into clinical trials for SMA. What else could this technology be applied to? Certainly there are other disorders caused by disruption of splicing. For instance, Familial Dysautonomia is a recessive condition in which the major mutation alters incorporation of exon 20 of the IKBKAP (inhibitor of κ light-polypeptide gene enhancer in B cells,kinase complex-associated protein) gene, and the drug compound kinten can enhance inclusion of exon 20 in cells from patients (Hims et al. 2007). Thus, ASOs directed against an appropriate element could also encourage incorporation of exon 20, and act as a treatment for this disorder. Disruption of a gene can also be considered for treatment in reducing the toxic effect of a gene, or in studying the basic function of that gene. Blocking an ESE or ISE (Fig. 1) could cause an exon to be skipped, disrupting the reading frame and translation of the transcript, and thereby diminishing the toxic protein. The ability to alter the splicing pattern of genes can be used in many ways. The study by Hua et al. (2010) in this issue of Genes & Development presents a system whereby ASOs can be developed and delivered in mice to enhance or disrupt a particular transcript in the nervous system of an animal. Thus, apart from therapeutics in a specific condition, there are many additional applications of efficient ASO alteration of splicing. The understanding of elements such as ISE, ESE, ESS, and ISS, along with ASOs against the element, allows a wide region of manipulation of genes that can be used for both the development of treatments and basic understanding of gene function.
Acknowledgments
We thank the many people in the SMA field over the years for many discussions, and we apologize if we did not include certain references; this is due to the necessity of being brief. We hope that the future holds true treatments for SMA patients. Funding in the Burghes laboratory has been generously provided by NIH grants (NS038650, HD060586, and RC2 NS069476), as well as MDA; Families of SMA; the Madison, Matthew, Preston, and Cade and Katelyn funds; and the SMA Angels (Savannah).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1961710.
References
- Araujo Ade Q, Araujo M, Swoboda KJ 2009. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J Pediatr 155: 292–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee TW, Gladman JT, Chandler DS 2010. Splicing regulation of the survival motor neuron genes and implications for treatment of spinal muscular atrophy. Front Biosci 15: 1191–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE 2009. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH 2009. Regulation of SMN protein stability. Mol Cell Biol 29: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach ME, Singh J, Thorsteinsdottir M, Saieva L, Slominski E, Thurmond J, Andresson T, Zhang J, Edwards JD, Simard LR, et al. 2010. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet 19: 454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR 2003. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 10: 120–125 [DOI] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH 1997. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 6: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Duque S, Joussemet B, Riviere C, Marais T, Dubreil L, Douar AM, Fyfe J, Moullier P, Colle MA, Barkats M 2009. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther 17: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher S, Honeyman K, Fall AM, Harding PL, Johnsen RD, Wilton SD 2006. Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J Gene Med 8: 207–216 [DOI] [PubMed] [Google Scholar]
- Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK 2009. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 27: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AH, et al. 2010. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol 28: 271–274 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L 2007. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2: e921 doi: 10.1371/journal.pone.0000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hims MM, Ibrahim EC, Leyne M, Mull J, Liu L, Lazaro C, Shetty RS, Gill S, Gusella JF, Reed R, et al. 2007. Therapeutic potential and mechanism of kinetin as a treatment for the human splicing disease familial dysautonomia. J Mol Med 85: 149–161 [DOI] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H 2000. A mouse model for spinal muscular atrophy. Nat Genet 24: 66–70 [DOI] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR 2007. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5: e73 doi: 10.1371/journal.pbio.0050073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR 2010. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev (this issue). doi: 10.1101/gad.1941310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463 [DOI] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH 2005. SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 14: 845–857 [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80: 155–165 [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 16: 265–269 [DOI] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ 2000. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet 9: 259–265 [DOI] [PubMed] [Google Scholar]
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ 1998. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 19: 63–66 [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B 1999. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci 96: 6307–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Rindt H, Shababi M 2010. Spinal muscular atrophy: Mechanisms and therapeutic strategies. Hum Mol Genet 19: R111–R118 doi: 10.1093/hmg/ddq147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis J, Meyer K, Angehrn L, Kampfer SS, Rothen-Rutishauser B, Schumperli D 2007. Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol Ther 15: 1479–1486 [DOI] [PubMed] [Google Scholar]
- McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH 1997. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet 60: 1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U 2001. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol 3: 945–949 [DOI] [PubMed] [Google Scholar]
- Meyer K, Marquis J, Trub J, Nlend Nlend R, Verp S, Ruepp MD, Imboden H, Barde I, Trono D, Schumperli D 2009. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum Mol Genet 18: 546–555 [DOI] [PubMed] [Google Scholar]
- Miller TM, Smith RA, Kordasiewicz H, Kaspar BK 2008. Gene-targeted therapies for the central nervous system. Arch Neurol 65: 447–451 [DOI] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD 1999. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet 8: 1177–1183 [DOI] [PubMed] [Google Scholar]
- Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossol W, Prior TW, et al. 2000. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn−/− mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet 9: 333–339 [DOI] [PubMed] [Google Scholar]
- Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O'Riordan CR, Klinger KW, Shihabuddin LS, Cheng SH 2010. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest 120: 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298: 1775–1779 [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW 2008. Motor neuron disease: The curious ways of ALS. Nature 454: 284–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ, Burghes AH, Kissel JT 2009. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet 85: 408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci 94: 9920–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ, Singh RN 2006. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol 26: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN 2009. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol 6: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F 2003. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci 100: 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, et al. 2007. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med 357: 2677–2686 [DOI] [PubMed] [Google Scholar]
- Vezain M, Saugier-Veber P, Goina E, Touraine R, Manel V, Toutain A, Fehrenbach S, Frebourg T, Pagani F, Tosi M, et al. 2010. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum Mutat 31: E1110–E1125 doi: 10.1002/humu.21173 [DOI] [PubMed] [Google Scholar]
- Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ 2009. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci 29: 7633–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E, Saieva L, Carrel TL, Crawford TO, Liu D, Lutz C, Beattie CE, Pellizzoni L, Burghes AH 2009. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet 18: 2215–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]