Abstract
Mdm2 has been well characterized as a negative regulator of the tumor suppressor p53. Recent studies have shown that Mdm2 is activated in response to a variety of oncogenic pathways independent of p53. Although its role as an oncogene via suppression of p53 function remains clear, growing evidence argues for p53-independent effects, as well as the remarkable possibility that Mdm2 has tumor suppressor functions in the appropriate context. Hence, Mdm2 is proving to be a key player in human cancer in its own right, and thus an important target for therapeutic intervention.
Keywords: Mdm2, tumorigenesis, p53, oncogene, tumor suppressor, ubiquitination
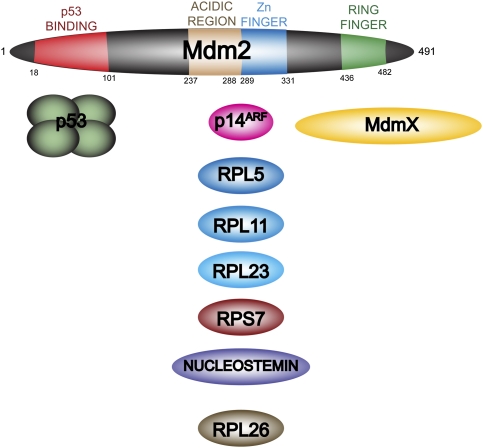
Mdm2 was originally identified as being gene-amplified on double-minute chromosomes in transformed mouse fibroblasts (Fakharzadeh et al. 1991). It subsequently was shown to interact with the tumor suppressor protein p53, and to negatively regulate its function. This occurs through two main mechanisms. First, the direct binding of Mdm2 to the N-terminal end of p53 inhibits the transcriptional activation function of p53 (Momand et al. 1992; Oliner et al. 1993). Second, Mdm2 possesses E3 ubiquitin ligase activity that targets p53 for modification and subsequent degradation through the 26S proteasome (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997). Human Mdm2 is a protein of 491 amino acids. Four regions of Mdm2 have been defined (Fig. 1; see Marine et al. 2007; Wade et al. 2010). At the N terminus is the main binding site for p53. In the central part of the protein are an acidic region and a zinc finger, whereas the C terminus contains a RING finger. A number of proteins have been shown to interact with Mdm2 in the middle of the protein, and these will be described in detail below. The RING finger is responsible for the ubiquitin ligase activity of Mdm2, and also serves as a binding site for a closely related partner, MdmX (Tanimura et al. 1999).
Figure 1.
Mdm2 contains multiple protein-binding regions. Human Mdm2 consists of 491 amino acids. There are four previously characterized regions of the protein. At the N terminus, residues 18–101 are the main p53-binding region, and are the site that is targeted by several known Mdm2 inhibitors, including nutlin-3. In the central region is an acidic domain adjacent to a zinc finger (residues 237–331). This part of the protein interacts with a variety of regulatory factors, including the tumor suppressor p14ARF as well as multiple ribosomal or nucleolar proteins. The C terminus contains a RING finger that has been shown to be responsible for the E3 ubiquitin ligase activity, as well as the binding of the closely related MdmX.
The intimate relationship between Mdm2 and p53 was clearly revealed in studies in which Mdm2 was knocked out in the mouse germline. Homozygous deletion of Mdm2 results in lethality at the blastocyst stage, due to inappropriate apoptosis. Remarkably, deletion of p53 completely rescues this phenotype (Jones et al. 1995; Montes de Oca Luna et al. 1995). It should be noted that loss of MdmX likewise results in a p53-dependent embryonic-lethal phenotype, albeit at a later stage of development, and, in this case, because of loss of cellular proliferation (Parant et al. 2001; Finch et al. 2002). Mdm2 itself is in turn regulated by p53, as there are p53 response elements located in the promoter of the Mdm2 gene (Barak et al. 1993; Juven et al. 1993; Perry et al. 1993). Thus, a negative feedback loop exists between p53 and Mdm2 that has been confirmed by elegant studies at the single-cell level (see Lahav 2008). In contrast, the evidence to date suggests that MdmX does not appear to be transcriptionally regulated by p53 (see Marine et al. 2007).
The focus of the current discussion is the role of Mdm2 in regulating cell proliferation as it relates to tumorigenesis. To address this, some brief background into Mdm2 and its biochemical functions is needed. The role of Mdm2 in regulating p53 by ubiquitination, as well as its interaction with MdmX, has been summarized recently in several excellent reviews (see Brooks and Gu 2006; Marine et al. 2006; Toledo and Wahl 2007; Kruse and Gu 2009; Marine and Lozano 2010; Wade et al. 2010). By its very nature, this discussion can only highlight a subset of the important observations that have been published recently. I apologize for the need to be selective, and wish to emphasize that this review should by no means be considered exhaustive of the current literature.
Mdm2 regulates p53
A variety of stress signals results in a disruption of the Mdm2–p53 interaction, leading to activation of p53-dependent cellular responses. The best-characterized of these is the effect of damage to cellular DNA (Fig. 2). Post-translational modification of both p53 and Mdm2 disrupt their interaction and lead to p53-dependent effects on cellular gene expression (see Kruse and Gu 2009; Vousden and Prives 2009; Marine and Lozano 2010). Phosphorylation of Mdm2 has been shown to inhibit its activity as an E3 ligase toward p53 (see Marine and Lozano 2010; Wade et al. 2010). It has been proposed that this is due to a disruption of higher-order oligomers of Mdm2 that are necessary for this activity (Cheng et al. 2009). In a similar manner, p53 itself is both phosphorylated and acetylated on specific residues in response to genotoxic stress (see Kruse and Gu 2009). It was originally assumed that these modifications also played an obligatory role in p53 function. More recently, it has been shown that mere disruption of the p53–Mdm2 interaction using chemical inhibitors is sufficient for p53 to act as a transcription factor (Vassilev et al. 2004). This apparently occurs in the absence of phosphorylation on many of the known sites on p53 (Thompson et al. 2004). Likewise, ablation of Mdm2 levels using an RNAi approach resulted in p53-dependent transcriptional activity in the absence of either phosphorylation or acetylation on specific relevant sites (Giono and Manfredi 2007). An elegant study from the Gu laboratory (Tang et al. 2008) has since clearly shown that acetylation is absolutely required for p53 activation, but only in the presence of Mdm2. Thus, the main function of modification of p53 is a disruption of the interaction with Mdm2, thereby preventing the ability of Mdm2 to repress p53 activity.
Figure 2.
Mdm2 regulates the p53-dependent response to DNA damage. Prior to DNA damage, Mdm2 interacts with both p53 and the ribosomal protein RPL26, leading to their ubiquitination and targeting for proteasomal degradation. Mdm2 bound to p53 has been localized to regulatory regions of a number of p53 target genes, leading to repression of their expression. Upon DNA damage, both Mdm2 and p53 become post-translationally modified such that they no longer interact. p53 is now capable of influencing gene expression. Recently, an additional mechanism for regulation of p53 expression by Mdm2 has been elucidated. It has been suggested that Mdm2 interacts with the mRNA encoding p53 itself and suppresses its translation. In addition, the ribosomal protein RPL26 has been shown to be an obligate effector of p53 translation as well. Thus, upon DNA damage, the interaction of RPL26 with Mdm2 is also disrupted, leading to increased levels and its association with p53 mRNA, facilitating its translation and increasing p53 levels.
Mdm2 has also been shown to affect translation of p53 by two mechanisms. First, it has been shown to interact directly with the messenger RNA (mRNA) encoding p53 itself and to suppress its translation (Candeias et al. 2008). Second, the ribosomal protein RPL26 has been shown to play an obligate role in p53 translation in response to genotoxic stress (Takagi et al. 2005). Mdm2 is capable of targeting RPL26 for ubiquitin targeted degradation (Ofir-Rosenfeld et al. 2008). Upon DNA damage, this effect of Mdm2 on RPL26 is inhibited, leading to enhanced association of RPL26 with p53 mRNA and increased expression of p53 protein (Takagi et al. 2005).
In contrast to DNA damage signaling, two other means for up-regulation of p53 activity rely on direct protein–protein interactions with Mdm2 (Fig. 3). In response to oncogenic signaling, the p14ARF protein is increased transcriptionally. The p14ARF protein binds to the central domain of Mdm2, including the acidic region, leading to inhibition of the ability of Mdm2 to act on p53 (see Sherr 2006). In a similar manner, ribosomal or nucleosomal stress leads to the binding of a select group of proteins to the similar region on Mdm2, and likewise prevents its functions toward p53. These include the ribosomal proteins RPL5, RPL11, RPL23, and RPS7 (Lohrum et al. 2003; Zhang et al. 2003; Dai et al. 2004; Zhu et al. 2009), as well as nucleophosmin (NPM) (Colombo et al. 2002) and nucleostemin (NS) (Dai et al. 2008). In each of these cases, their binding to Mdm2 appears to be sufficient for p53 to function as a transcription factor, and affect its downstream target genes. The interested reader is directed to a recent excellent review on this topic (Zhang and Lu 2009).
Figure 3.
The interaction of Mdm2 and p53 is disrupted in response to oncogenic activation as well as ribosomal stress. The interaction of Mdm2 with p53 is also disrupted via the binding of specific regulatory proteins. In response to oncogenic activation, p14ARF interacts with the central region of Mdm2, thereby inhibiting its activity toward p53. Nucleolar or ribosomal stress has been suggested to lead to the release of several proteins that also interact with Mdm2, preventing it from negatively regulating p53.
Mdm2 levels are transcriptionally up-regulated in response to a variety of oncogenic and tumor suppressor pathways
Transcription of the Mdm2 gene is controlled by two distinct promoters, referred to as P1 and P2 (Barak et al. 1994; Zauberman et al. 1995). The P1 promoter controls basal expression of Mdm2, and is situated upstream of the first exon of the Mdm2 gene. The P2 promoter is highly regulated, is responsible for inducible expression of Mdm2, and is found in the first intron (Fig. 4A). As the start site for translation is contained in exon 2, the transcripts expressed from both the P1 and P2 promoters encode identical full-length Mdm2 proteins. However, there are differences in the 5′ untranslated regions of these transcripts that allow them to be distinguished experimentally (Fig. 4B). The p53 response elements are located upstream of the P2 promoter, and hence the p53-inducible expression of Mdm2 can be monitored by expression levels of this specific transcript (Fig. 4A). There is also evidence that these two transcripts may differ in their propensity for alternative splicing as well as in their translatability (Brown et al. 1999; Cheng and Cohen 2007).
Figure 4.
Mdm2 is transcriptionally regulated by multiple pathways. (A) There are two promoters that function in the regulation of Mdm2 expression. The P1 promoter controls basal expression of Mdm2 and is upstream of exon 1a. The P2 promoter is further downstream and uses a start site at an alternate first exon, 1b. The P2 promoter is inducible, and is regulated by response elements for a variety of transcription factors, including RXR, AP-1, the Ets family, Smad2, and Smad3, as well as p53 itself. A cluster of nGGGGC boxes act downstream from the Erk pathway. Three GC boxes control expression of Mdm2, with one of these containing the site of a single-nucleotide polymorphism (SNP309) that influences the binding of the Sp-1 transcription factor. (B) The two transcripts from the P1 and P2 promoters have distinct 5′ untranslated regions (encoded by either exon 1a or exon 1b) and are different lengths. It should be noted that sequences corresponding to exon 1b are not present in the mature P1 transcript. Nevertheless, the start site of translation is contained in a shared exon 2. Thus, the Mdm2 protein that is expressed is identical between the two transcripts. There has been a suggestion, however, that further splicing of the transcript may be influenced by promoter selection.
A number of other signaling pathways lead to the interaction of specific transcription factors with sequences contained within the first intron of the Mdm2 gene, leading to the activation of transcription via the P2 promoter in addition to p53 (Fig. 4A). Signaling via Ras and the Erk pathway results in the binding of both AP-1 and Ets family members to a response element that is immediately adjacent to that of p53 (Figs. 4A, 5; Ries et al. 2000; Phelps et al. 2003). A cluster of GC boxes lie further upstream, and have been implicated in the differential expression of Mdm2 due to an interesting polymorphism, SNP309 (Bond et al. 2004). When this polymorphism is a guanine, it results in the ability of Sp1 to regulate Mdm2 expression. In contrast, a thymine in this position abrogates this, with a concomitant reduction in Mdm2 levels. It has been shown that SNP309 correlates with the incidence of tumorigenesis in certain patient populations, thereby emphasizing the role of Mdm2 as an oncogenic modifier in human cancer (Bond et al. 2005). TGFβ has been shown to regulate Mdm2 expression via the interaction of Smad2 and Smad3 with an element in this intron as well (Figs. 4A, 5; Araki et al. 2010). Finally, levels of Mdm2 appear to be elevated in retinal cone cells due to expression of a tissue-specific RXR that can activate expression of the Mdm2 gene through the P2 promoter (Xu et al. 2009).
Figure 5.
Oncogenic pathways enhance Mdm2 activity. Growth factor signaling via tyrosine kinase receptors and Ras lead to the activation of AP-1 and Ets family members, which have been shown to transcriptionally up-regulate Mdm2. TGFβ signaling has also been shown to increase Mdm2 via the Smad transcription factors. Phosphatidyl-3 kinase (PI3K) signaling through the Akt kinase leads to phosphorylation of Mdm2 on sites distinct from those modified in response to DNA damage. This results in nuclear import of Mdm2 and enhancement of its ability to inhibit p53 (Zhou et al. 2001).
Mdm2 as an oncogene: inactivation of the wild-type tumor suppressor function of p53
The initial identification of Mdm2 as the protein product of a gene that was amplified on double-minute chromosomes hinted at a possible role in oncogenesis. This was validated after the demonstration that Mdm2 interacts directly with p53 and inhibits its ability to function as a transcriptional activator. In early studies, Mdm2 was shown to be overexpressed in soft tissue sarcomas, and its increased level was found to be mutually exclusive with mutation of p53 (Oliner et al. 1992). Mdm2 has since been shown to be overexpressed by either gene amplification or other as-yet-uncharacterized mechanisms in a variety of human tumors (see Rayburn et al. 2005). Targeted expression of Mdm2 in specific tissues in the mouse leads to increased tumorigenesis, consistent with a role as an oncogene (Jones et al. 1998). Likewise, in mouse models in which p53 inactivation has been shown to play a role, there is evidence that this can also occur through enhanced expression of Mdm2 (Eischen et al. 1999). Given its well-studied role in inhibiting p53 function through multiple mechanisms, it is reasonable that the basis for its oncogenic activity is via p53, a notion that is consistent with correlative studies in human tumors. It nevertheless remains unclear how this may occur. It is generally thought that basal levels of p53 are indeed controlled by Mdm2. However, upon p53 activation by DNA damage and subsequent post-translational modification of both partners, the interaction between p53 and Mdm2 is disrupted (see Kruse and Gu 2009; Vousden and Prives 2009; Marine and Lozano 2010; Wade et al. 2010). Thus, the notion that mere overexpression of Mdm2 under such conditions would disable p53 is difficult to reconcile with such a model. It has been speculated that an initiating event in oncogenesis triggers a response similar, if not identical, to genotoxic stress (see Halazonetis et al. 2008). One possibility is that high levels of Mdm2 may saturate the protein-modifying cascades. Alternatively, the DNA damage response during cancer development may have aspects, yet to be elucidated, that are distinct from the well-characterized genotoxic response pathways.
The ability of Mdm2 to target p53 for proteolytic degradation has also been shown to be inhibited by p14ARF as well as a subset of ribosomal proteins (Fig. 3; see Sherr 2006; Zhang and Lu 2009). If these latter regulators are limiting in the cell, it can readily be hypothesized that high levels of Mdm2 can overcome their effects. Thus, if the initiating event involves up-regulation of p14ARF, as has been shown in some settings, overexpression of Mdm2 could easily account for the blocking of this signal and the prevention of p53 activation. A role for ribosomal proteins as tumor suppressors has been found in zebrafish (Amsterdam et al. 2004). Furthermore, in the tumors that arise due to mutation of certain ribosomal proteins in zebrafish, there is decreased p53 expression (MacInnes et al. 2008). This is all consistent with the idea that both p14ARF and ribosomal proteins may be the relevant targets for the overexpressed Mdm2 found in certain tumor types.
Alternately, there may be functions of p53 in the basal setting that are relevant for its ability to suppress tumorigenesis. In this case, pathologically high levels of Mdm2 would be involved in preventing these activities. It has been shown that overexpression of p53 in the absence of a specific activating signal results in altered target gene expression, yet this has generally been found in cell culture. It has been argued that the growth of cells in culture results in oxidative stress or some other mechanism that sends a signal to p53 to be active. Thus, p53 regulation of basal target gene expression in the absence of an upstream signal may not occur in vivo. Comparison of p53 target gene expression in wild-type and p53-null mice confirms this (Macleod et al. 1995; Parker et al. 1995; Leveillard et al. 1998; Burns et al. 2001), although there are exceptions (Miyashita et al. 1994; Macleod et al. 1995). Further, Perry's laboratory (Mendrysa and Perry 2000) showed that p53 controls Mdm2 gene expression only after cells are established in vitro, and not in vivo or at the initial introduction of cells in culture. There are isolated examples of p53 targets that do not conform to this. For example, the expression of LIF (leukemia inhibitory factor), a cytokine involved in early embryo implantation, has been shown to be controlled by p53. In vivo, it was shown that p53-null mice lack sufficient levels of LIF, leading to defects in maternal reproduction (Hu et al. 2007). The challenge is to determine whether there are other such targets of p53 that play key roles in the basal setting in vivo, and thereby contribute to the tumor suppressor activity of p53 in the organism.
Mdm2 as an oncogene: p53-independent activities that control proliferation, apoptosis, and the epithelial-to-mesenchymal transition
As already noted, targeted ectopic expression of Mdm2 to specific tissues in the mouse leads to cellular proliferation and tumor development. Intriguingly, in some cases, these effects were seen even in the absence of p53. Indeed, several oncogenic pathways lead to up-regulation of Mdm2 levels (Fig. 5) that can result in both p53-dependent and -independent effects. It should be noted that these pathways may also impact p53 directly as well. In an early study of a transgenic mouse in which ectopic Mdm2 was expressed ubiquitously, survival was not affected by p53 status. However, the spectrum of tumors that was observed was different in wild-type and p53-null backgrounds (Jones et al. 1998). Targeted expression of Mdm2 in the mammary gland gave the fascinating phenotype of increased ploidy, suggesting that Mdm2 regulates cell cycle progression. This latter finding was seen in p53 knockout mice as well (Lundgren et al. 1997). Splice variants that fail to bind p53 can also be shown to promote tumorigenesis (Fridman et al. 2003; Steinman et al. 2004). Although early studies suggested that p53 mutation and Mdm2 overexpression were not found in the same cancer sample, tumor types have since been identified in which overexpression of Mdm2 is not mutually exclusive with mutation of p53 (Cordon-Cardo et al. 1994). Intriguingly, tumor latency was shorter in Mdm2/p53 double-knockout mice as compared with p53-null mice that are heterozygous for the Mdm2 deletion. In addition, homozygous deletion of Mdm2 led to a change in tumor spectrum with an increased incidence of sarcomas (McDonnell et al. 1999). Taken together, such findings support a role for Mdm2 in tumorigenesis that is independent of its effects on p53.
Consistent with these results, Mdm2 has been shown to regulate the expression of certain proteins that contribute to key aspects of cell proliferation, apoptosis, and tumor invasion and metastasis independent of its interaction with p53. For example, Mdm2 has been shown to target the product of the retinoblastoma susceptibility gene (pRB), as well as the transcription factor E2F-1 and its binding partner, DP-1, among others (see Ganguli and Wasylyk 2003; Zhang and Zhang 2005). More recently, several novel targets for Mdm2 have been identified. In response to signaling via the Erk pathway, the transcription factor Foxo3a is phosphorylated. In this modified form, Foxo3A is a target for Mdm2-dependent ubiquitination and degradation. As Foxo3A regulates expression of genes that encode cell cycle regulators such as p27, this leads to an Mdm2-mediated control of cell cycle progression in response to oncogenic growth factor signaling or Ras activation (Yang et al. 2008; Fu et al. 2009). Mdm2 has also been shown to regulate the expression of the anti-apoptotic protein XIAP by a novel mechanism. Binding of Mdm2 to the mRNA that encodes XIAP enhances its translation, leading to the increased expression of XIAP. This leads to an Mdm2-dependent prevention of cell death via caspase-mediated apoptosis (Gu et al. 2009).
Most notably, Mdm2 has been shown recently to target E-cadherin for degradation via the 26S proteasome (Yang et al. 2006). E-cadherin has a well-characterized role in the epithelial-to-mesenchymal transition that is a critical step on the way to the metastasis of solid tumors of epithelial origin (see Thiery et al. 2009). Previous studies have shown that transcriptional repression of E-cadherin expression via regulators such as Slug or Twist contributed to the loss of cell polarity and cell–cell adhesion, as well as enhanced motility of epithelial tumor cells (see Polyak and Weinberg 2009; Thiery et al. 2009). Although transcriptional repression is a means for down-regulation of specific targets, levels of expression are also determined by mRNA stability as well as the half-life of the protein of interest. This recent study indicates that Mdm2 may play a critical role in loss of E-cadherin protein expression during the epithelial-to-mesenchymal transition that reinforces the effects of transcriptional repression (Yang et al. 2006). Thus, full down-regulation of E-cadherin could be achieved via a cooperation between Mdm2 and transcriptional repressors such as Slug and Twist. However, whether Mdm2 plays any obligate role in the epithelial-to-mesenchymal transition still needs to be directly tested.
Transcriptional regulation of p14ARF lies downstream from the RB pathway (Fig. 6). Inactivation of this pathway increases p14ARF expression and can suppress Mdm2 activity toward p53 (see Gil and Peters 2006; Sherr 2006; Kruse and Gu 2009). It remains to be determined whether p14ARF influences p53-independent effects of Mdm2.
Figure 6.
Tumor suppressor pathways control Mdm2 function. The p14ARF protein is a negative regulator of Mdm2 function and is transcriptionally controlled by the E2F family. The RB pathway regulates E2F function. Multiple components of this pathway have been shown to play a role in human cancer as either oncogenes (Cyclin D, Cdk4, and Cdk6) or tumor suppressors (p16INK4A and pRB). TGFβ signaling can also exert a suppressor effect via transcriptional up-regulation of the cyclin-dependent kinase inhibitor p15INK4B. It should also be noted that TGFβ signaling has been implicated in regulating p14ARF expression more directly via the direct interaction of Smad proteins with the ARF genomic locus (Freeman-Anderson et al. 2009).
The finding that homozygous deletion of Mdm2 results in an early embryonic lethality that is completely rescued by loss of p53 (Jones et al. 1995; Montes de Oca Luna et al. 1995) has been used to argue against the importance of p53-independent effects of Mdm2. Nevertheless, it is unclear whether the physiological role of basal levels of Mdm2 in development is relevant to its pathological effects when overexpressed during tumorigenesis.
Mdm2 as a tumor suppressor: mutant p53 protein stability and a role in nuclear export of wild-type p53
Given its role as a negative regulator of p53, as well as its overexpression in human tumors, the notion of Mdm2 as an oncogene is quite reasonable. Nevertheless, there is a growing body of evidence to suggest that Mdm2 may also exert effects that suppress cell proliferation (see Deb 2003). Targeting Mdm2 to the mouse mammary gland did not lead to tumor formation, but rather a hyperplasia that was associated with multiple rounds of DNA synthesis in the absence of cell division (Lundgren et al. 1997). This finding is consistent with the idea that Mdm2 somehow suppresses mitotic progression. Studies in cell culture have indicated a growth-suppressing function of Mdm2, but the molecular basis for this remains unclear (Brown et al. 1998; Dang et al. 2002). In one study, overexpression of Mdm2 altered the percent of cells arrested in the G1 versus the G2/M phases of the cell cycle in response to p53 (Ohkubo et al. 2006). A requirement for Mdm2 in p53-dependent cell cycle arrest via the cyclin-dependent kinase inhibitor p21 has also been suggested (Giono and Manfredi 2007). Recently, it was shown that Mdm2 targets Slug for proteolytic degradation (Wang et al. 2009). As Slug has been implicated as a key transcriptional regulator in the epithelial-to-mesenchymal transition (see Polyak and Weinberg 2009; Thiery et al. 2009), this is more consistent with a tumor suppressor role for Mdm2. Although full-length Mdm2 was used for expression in certain transgenic models, examination of Mdm2 in resulting tumors showed that the Mdm2 can be truncated (Steinman et al. 2004). This is consistent with the notion that there may be selective pressure to lose some function of Mdm2 during tumorigenesis. Remarkably, in some cases, these forms of Mdm2 lack the p53-binding region at the N terminus.
Recently, the Lozano laboratory (Lang et al. 2004) described the generation of a mutant mouse model in which there is expression of the mouse equivalent of a tumor-derived mutant p53. Previous studies had argued that mutant p53 is stabilized and expressed at higher levels than the wild-type p53 protein (see Prives and White 2008). In this mouse model, it was found that only in tumor tissue was the mutant p53 overexpressed, whereas, in the normal tissues, its level was kept low (Lang et al. 2004). To determine the molecular basis for this finding, a role for Mdm2 was explored, and it was shown that loss of Mdm2 leads to stabilization of mutant p53 (Terzian et al. 2008). Intriguingly, loss of Mdm2 enhanced tumor formation in these mice. This clearly demonstrates that, at least under these conditions, Mdm2 functions as a tumor suppressor, and this is likely due to reduced mutant p53 expression. Whether similar situations can be identified in human cancers remains to be seen.
Several laboratories have argued that Mdm2 also plays a role in nuclear export of p53 (Boyd et al. 2000; Geyer et al. 2000). The resulting inhibition of p53 transcriptional activity is consistent with an oncogenic role for Mdm2. More recently, a cytoplasmic function for p53 has been established (see Manfredi 2003; Vaseva and Moll 2009). It has been shown that p53 can interact functionally with the Bcl-2 family of proteins in the cytoplasm, thereby activating the mitochondrial apoptotic pathway, leading to cell death (Dumont et al. 2003; Mihara et al. 2003; Chipuk et al. 2004, 2005). In this context, the ability of Mdm2 to facilitate nuclear exclusion of p53, in part through effects on ubiquitination, facilitates an apoptotic response and would contribute to the tumor suppressor activity of p53 (Dumont et al. 2003; Marchenko et al. 2007).
Future directions
While it is clear that Mdm2 plays an important role in oncogenesis, the ability to translate these laboratory findings into clinically relevant outcomes will require that a full understanding of the underlying molecular mechanisms be elucidated. The notion that Mdm2 may act as an oncogene or tumor suppressor, depending on the experimental conditions, is intriguing from a research perspective. However, the benefit of this knowledge to prognosis and treatment of human disease will be limited without clear determination of its basis. Given what is currently known, there are several avenues of study that are likely to shed further light. These include an appreciation of the possibly distinct biology of various isoforms of Mdm2 (see Bartel et al. 2002; Harris 2005); its interplay with its closely related partner, MdmX (see Marine et al. 2007; Marine and Lozano 2010; Wade et al. 2010); and a more detailed analysis of the alterations in Mdm2 that are found in human disease (see Rayburn et al. 2005).
To date, most transgenic models have examined ectopic expression of Mdm2 that relies on the use of a cDNA that encodes the full-length protein. It will be important to examine the relative effects of targeting distinct splice variants of Mdm2 to specific tissues to elucidate their respective abilities to affect tumorigenesis. Recent studies have indicated that Mdm2 is likely to have different effects, depending on the tissue and cell type that is examined (Grier et al. 2006; Xiong et al. 2006; Maetens et al. 2007). While adding to the complexity of the analysis, these findings argue that comparison of splice forms needs to be done with care under similar conditions and cell types. The role of splicing of Mdm2 itself during oncogenesis remains an intriguing area as well. This is especially true given the detection in human tumors of Mdm2 variants that fail to interact with p53 (Sigalas et al. 1996; Sanchez-Aguilera et al. 2006). In vivo models in which Mdm2 is expressed from vectors that preserve genomic structure will likewise be key.
The interplay between MdmX and Mdm2 remains a fascinating and important area of study. There is growing evidence, summarized elsewhere in several excellent recent reviews, that Mdm2 homo-oligomers are likely to have distinct biochemical functions from those that involve Mdm2–MdmX heterodimers (see Kruse and Gu 2009; Vousden and Prives 2009; Wade et al. 2010). Comparison of mouse models in which either Mdm2 or MdmX has been deleted reveal both overlapping and nonoverlapping roles in tumorigenesis, as well as in normal development (see Marine and Lozano 2010). A complete understanding of Mdm2 biology and its role in cancer certainly will need to be fully integrated with MdmX.
Although Mdm2 has been shown to be overexpressed in human tumors through either gene amplification or other means, a more careful analysis is needed. Reliance on the use of immunohistochemistry or even determination of mRNA levels is of limited utility given the existence of distinct isoforms of Mdm2 in human tumors. Reagents to address this important question in clinical samples will need to be developed so that it can be rigorously determined which isoforms—and in what relative amounts—are being expressed in specific cancer types. There have been limited reports on the frequency and nature of missense mutations in Mdm2 in human tumors (Schlott et al. 1997). Importantly, characterization of some of these mutants has revealed quite interesting insights into Mdm2 function involving the zinc finger (Lindstrom et al. 2007). These mutants retain the ability to inhibit the transcriptional activity of p53, but are defective at directing the nuclear export and proteolytic degradation of p53. This suggests the need for further genomic analysis of Mdm2 in human tumors as well. Whether overexpression of Mdm2 is associated with better or worse prognosis appears to vary depending on the tumor type (Onel and Cordon-Cardo 2004). Many of these studies relied on the use of assay methods that would not have distinguished between splice forms. It will be important to revisit this in the future to gain molecular insight into the basis for the varying prognostic outcomes.
As Mdm2 has been characterized extensively for its interaction with p53, it is not surprising that an emphasis has been placed on its role in tumorigenesis in that context. Nevertheless, the increasing numbers of p53-independent effects that have been ascribed to Mdm2 suggest that it has relevance in human cancer on its own. Given the exciting and sometimes confusing findings that are emerging from laboratory studies, placing these recent developments in an integrated context will be critical. This is especially true as Mdm2 emerges from the shadow of its relationship with p53. Whether Mdm2 proves to be an oncogene or tumor suppressor, and under which conditions, remains to be determined. Either way, this pursuit will certainly have an impact on human disease.
Acknowledgments
Discussions with Carol Prives, Jiandong Chen, Wei Gu, Gigi Lozano, Lois Resnick-Silverman, Lubo Vassilev, and Jim Xiao are gratefully appreciated, and were invaluable in developing the ideas presented here. I am supported by grants from the National Cancer Institute (P01 CA080058, R01 CA125741, and R01 CA086001).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1941710.
References
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2: E139 doi: 10.1371/journal.pbio.0020139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki S, Eitel JA, Batuello CN, Bijangi-Vishehsaraei K, Xie XJ, Danielpour D, Pollok KE, Boothman DA, Mayo LD 2010. TGF-β1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J Clin Invest 120: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M 1993. mdm2 expression is induced by wild type p53 activity. EMBO J 12: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Gottlieb E, Juven-Gershon T, Oren M 1994. Regulation of mdm2 expression by p53: Alternative promoters produce transcripts with nonidentical translation potential. Genes Dev 8: 1739–1749 [DOI] [PubMed] [Google Scholar]
- Bartel F, Taubert H, Harris LC 2002. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2: 9–15 [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591–602 [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Levine A 2005. A single nucleotide polymorphism in the MDM2 gene: From a molecular and cellular explanation to clinical effect. Cancer Res 65: 5481–5484 [DOI] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2: 563–568 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W 2006. p53 ubiquitination: Mdm2 and beyond. Mol Cell 21: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Thomas CA, Deb SP 1998. The human oncoprotein MDM2 arrests the cell cycle: Elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J 17: 2513–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CY, Mize GJ, Pineda M, George DL, Morris DR 1999. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene 18: 5631–5637 [DOI] [PubMed] [Google Scholar]
- Burns TF, Bernhard EJ, El-Deiry WS 2001. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 20: 4601–4612 [DOI] [PubMed] [Google Scholar]
- Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, Bourougaa K, Calvo F, Fahraeus R 2008. p53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol 10: 1098–1105 [DOI] [PubMed] [Google Scholar]
- Cheng TH, Cohen SN 2007. Human MDM2 isoforms translated differentially on constitutive versus p53-regulated transcripts have distinct functions in the p53/MDM2 and TSG101/MDM2 feedback control loops. Mol Cell Biol 27: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Chen L, Li Z, Lane WS, Chen J 2009. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J 28: 3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR 2004. Direct activation of bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303: 1010–1014 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1735 [DOI] [PubMed] [Google Scholar]
- Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol 4: 529–533 [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, Marechal V, Chen J, Brennan MF, Levine AJ 1994. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res 54: 794–799 [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H 2004. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol 24: 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H 2008. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol 28: 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF 2002. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res 62: 1222–1230 [PubMed] [Google Scholar]
- Deb SP 2003. Cell cycle regulatory functions of the human oncoprotein MDM2. Mol Cancer Res 1: 1009–1016 [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC 3rd, George DL, Murphy M 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365 [DOI] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL 1999. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 13: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 10: 1565–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch RA, Donoviel DB, Potter D, Shi M, Fan A, Freed DD, Wang CY, Zambrowicz BP, Ramirez-Solis R, Sands AT, et al. 2002. mdmx is a negative regulator of p53 activity in vivo. Cancer Res 62: 3221–3225 [PubMed] [Google Scholar]
- Freeman-Anderson NE, Zheng Y, McCalla-Martin AC, Treanor LM, Zhao YD, Garfin PM, He TC, Mary MN, Thornton JD, Anderson C, et al. 2009. Expression of the Arf tumor suppressor gene is controlled by Tgfβ2 during development. Development 136: 2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW 2003. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res 63: 5703–5706 [PubMed] [Google Scholar]
- Fu W, Ma Q, Chen L, Li P, Zhang M, Ramamoorthy S, Nawaz Z, Shimojima T, Wang H, Yang Y, et al. 2009. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem 284: 13987–14000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli G, Wasylyk B 2003. p53-independent functions of MDM2. Mol Cancer Res 1: 1027–1035 [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol 2: 569–573 [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G 2006. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nat Rev Mol Cell Biol 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Giono LE, Manfredi JJ 2007. Mdm2 is required for inhibition of Cdk2 activity by p21, thereby contributing to p53-dependent cell cycle arrest. Mol Cell Biol 27: 4166–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G 2006. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 26: 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M 2009. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell 15: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Harris LC 2005. MDM2 splice variants and their therapeutic implications. Curr Cancer Drug Targets 5: 21–26 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27 [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Teresky AK, Levine AJ 2007. p53 regulates maternal reproduction through LIF. Nature 450: 721–724 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208 [DOI] [PubMed] [Google Scholar]
- Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A 1998. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci 95: 15608–15612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8: 3411–3416 [PubMed] [Google Scholar]
- Kruse JP, Gu W 2009. Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Lahav G 2008. Oscillations by the p53–Mdm2 feedback loop. Adv Exp Med Biol 641: 28–38 [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119: 861–872 [DOI] [PubMed] [Google Scholar]
- Leveillard T, Gorry P, Niederreither K, Wasylyk B 1998. MDM2 expression during mouse embryogenesis and the requirement of p53. Mech Dev 74: 189–193 [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y 2007. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol 27: 1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587 [DOI] [PubMed] [Google Scholar]
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP, Finlay CA 1997. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev 11: 714–725 [DOI] [PubMed] [Google Scholar]
- MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA 2008. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci 105: 10408–10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T 1995. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev 9: 935–944 [DOI] [PubMed] [Google Scholar]
- Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, Klingmuller U, Lozano G, Marine JC 2007. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood 109: 2630–2633 [DOI] [PubMed] [Google Scholar]
- Manfredi JJ 2003. p53 and apoptosis: It's not just in the nucleus anymore. Mol Cell 11: 552–554 [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM 2007. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 26: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G 2010. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ 17: 93–102 [DOI] [PubMed] [Google Scholar]
- Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G 2006. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ 13: 927–934 [DOI] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG 2007. MDMX: From bench to bedside. J Cell Sci 120: 371–378 [DOI] [PubMed] [Google Scholar]
- McDonnell TJ, Montes de Oca Luna R, Cho S, Amelse LL, Chavez-Reyes A, Lozano G 1999. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J Pathol 188: 322–328 [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, Perry ME 2000. The p53 tumor suppressor protein does not regulate expression of its own inhibitor, MDM2, except under conditions of stress. Mol Cell Biol 20: 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM 2003. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590 [DOI] [PubMed] [Google Scholar]
- Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC 1994. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9: 1799–1805 [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206 [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M 2008. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 32: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C 2006. Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem 281: 16943–16950 [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83 [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362: 857–860 [DOI] [PubMed] [Google Scholar]
- Onel K, Cordon-Cardo C 2004. MDM2 and prognosis. Mol Cancer Res 2: 1–8 [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29: 92–95 [DOI] [PubMed] [Google Scholar]
- Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267: 1024–1027 [DOI] [PubMed] [Google Scholar]
- Perry ME, Piette J, Zawadzki JA, Harvey D, Levine AJ 1993. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc Natl Acad Sci 90: 11623–11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps M, Darley M, Primrose JN, Blaydes JP 2003. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor α-positive breast cancer cells. Cancer Res 63: 2616–2623 [PubMed] [Google Scholar]
- Polyak K, Weinberg RA 2009. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer 9: 265–273 [DOI] [PubMed] [Google Scholar]
- Prives C, White E 2008. Does control of mutant p53 by Mdm2 complicate cancer therapy? Genes Dev 22: 1259–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn E, Zhang R, He J, Wang H 2005. MDM2 and human malignancies: Expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets 5: 27–41 [DOI] [PubMed] [Google Scholar]
- Ries S, Biederer C, Woods D, Shifman O, Shirasawa S, Sasazuki T, McMahon M, Oren M, McCormick F 2000. Opposing effects of Ras on p53: Transcriptional activation of mdm2 and induction of p19ARF. Cell 103: 321–330 [DOI] [PubMed] [Google Scholar]
- Sanchez-Aguilera A, Garcia JF, Sanchez-Beato M, Piris MA 2006. Hodgkin's lymphoma cells express alternatively spliced forms of HDM2 with multiple effects on cell cycle control. Oncogene 25: 2565–2574 [DOI] [PubMed] [Google Scholar]
- Schlott T, Reimer S, Jahns A, Ohlenbusch A, Ruschenburg I, Nagel H, Droese M 1997. Point mutations and nucleotide insertions in the MDM2 zinc finger structure of human tumours. J Pathol 182: 54–61 [DOI] [PubMed] [Google Scholar]
- Sherr CJ 2006. Divorcing ARF and p53: An unsettled case. Nat Rev Cancer 6: 663–673 [DOI] [PubMed] [Google Scholar]
- Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J 1996. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: Transforming ability and frequent detection in human cancer. Nat Med 2: 912–917 [DOI] [PubMed] [Google Scholar]
- Steinman HA, Burstein E, Lengner C, Gosselin J, Pihan G, Duckett CS, Jones SN 2004. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem 279: 4877–4886 [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB 2005. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123: 49–63 [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W 2008. Acetylation is indispensable for p53 activation. Cell 133: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 447: 5–9 [DOI] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G 2008. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 22: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, Wahl GM, Heimbrook DC, Vassilev LT 2004. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem 279: 53015–53022 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2007. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol 39: 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM 2009. The mitochondrial p53 pathway. Biochim Biophys Acta 1787: 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM 2010. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 20: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, et al. 2009. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol 11: 694–704 [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G 2006. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci 103: 3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar SC, Abramson DH, Cobrinik D 2009. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137: 1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, Broglio K, Berry DA, Hung MC 2006. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol 26: 7269–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al. 2008. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 10: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M 1995. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res 23: 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H 2009. Signaling to p53: Ribosomal proteins find their way. Cancer Cell 16: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang R 2005. p53-independent activities of MDM2 and their relevance to cancer therapy. Curr Cancer Drug Targets 5: 9–20 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23: 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 3: 973–982 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C 2009. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35: 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]