Abstract
Spliced leader (SL) trans-splicing in Caenorhabditis elegans attaches a 22-nucleotide (nt) exon onto the 5′ end of many mRNAs. A particular class of SL, SL2, splices mRNAs of downstream operon genes. Here we use an embryonic extract-based in vitro splicing system to show that SL2 specificity information is encoded within the polycistronic pre-mRNA, and that trans-splicing specificity is recapitulated in vitro. We define an RNA sequence required for SL2 trans-splicing, the U-rich (Ur) element, through mutational analysis and bioinformatics as a short stem–loop followed by a sequence motif, UAYYUU, located ∼50 nt upstream of the trans-splice site. Furthermore, this element is predicted in intercistronic regions of numerous operons of C. elegans and other species that use SL2 trans-splicing. We propose that the UAYYUU motif hybridizes with the 5′ splice site on the SL2 RNA to recruit the SL to the pre-mRNA. In this way, the UAYYUU motif in the pre-mRNA would serve an analogous function to the similar sequence in the U1 snRNA, which binds to the 5′ splice site of introns, effectively reversing the roles of snRNP and pre-mRNA in trans-splicing.
Keywords: Operon, spliced leader, SL, trans-splice
In spliced leader (SL) trans-splicing, a short leader sequence is transferred from the 5′ end of an SL RNA onto the first exon of a pre-mRNA (for review, see Nilsen 1993; Blumenthal 2005; Hastings 2005). This joins two discontinuous RNA sequences, and provides a common 5′ sequence to many mRNAs. SL trans-splicing was identified first in trypanosomes and subsequently in numerous other organisms, ranging from protists to primitive chordates, and including roundworms, flatworms, arthropods, and cnidarians. However, it is not found in plants, fungi, or vertebrates (Hastings 2005; Marlétaz and Le Parco 2008; Douris et al. 2010). It has a number of proposed functions relating to handling of mRNA and translation (see Lall et al. 2004 and references therein).
A different form of trans-splicing has been reported in flies (Mongelard et al. 2002), and also recently in worms (Fischer et al. 2008). In these instances, exons of separate pre-mRNAs, neither one an SL RNA, are spliced together to form a mature mRNA containing portions of both pre-mRNAs. Additionally, there have been numerous reports of a similar process in mammalian cells (Akiva et al. 2006) and plants (Zhang et al. 2010), where normally separate mRNAs are “accidentally” trans-spliced together. This sort of splicing has been implicated recently in cancers associated with translocations (Li et al. 2008).
SL trans-splicing and cis-splicing (intron removal) are similar, and trans-splicing likely evolved from cis-splicing (Blumenthal 2004, 2005). The splice site on the SL RNA closely matches the 5′ splice site (5′ss) consensus sequence, and the trans-splice site of the pre-mRNA has the same consensus sequence as intronic 3′ss (Kent and Zahler 2000; Blumenthal 2005). In Caenorhabditis elegans, trans-splicing is signaled by a 3′ss without an upstream 5′ss. Artificial trans-splice sites can be created from cis-splice sites by removing an upstream 5′ss (Conrad et al. 1991, 1995; Maroney et al. 2000; Boukis and Bruzik 2001), and a trans-splice site can be made to cis-splice instead by insertion of a 5′ss into the outron, the intron-like sequence upstream of the trans-splice site (Conrad et al. 1993).
Mechanistically, SL trans-splicing occurs like cis-splicing, and requires most of the same spliceosomal components. The one notable exception is U1 snRNP, which is not needed for trans-splicing (Hannon et al. 1991; Nilsen 1993; Maroney et al. 1996). In cis-splicing, U1 snRNP recognizes the pre-mRNA by hybridization between the U1 snRNA and the 5′ss (for review, see Brow 2002). It was proposed that, in trans-splicing, the first stem of the SL RNA itself hybridizes across the SL splice site and performs this role (Bruzik et al. 1988; Bruzik and Steitz 1990). However, this base-pairing interaction is not required in vitro (Maroney et al. 1991), implying that a different mechanism may identify the 5′ss.
One key purpose of SL trans-splicing in C. elegans is the resolution of operons, or polycistronic transcription units (Spieth et al. 1993)—clusters of two to eight genes organized end to end and transcribed by a single upstream promoter. Each gene pair within an operon is separated by an intercistronic region (ICR) of typically ∼110 nucleotides (nt) (Blumenthal 2005; MA Allen, unpubl.). The mRNAs from genes within the operon are processed by cleavage and polyadenylation at each 3′ end, and trans-splicing at the 5′ end of each downstream gene. While some nonoperon genes and some first genes in operons are trans-spliced, all downstream genes in operons are necessarily trans-spliced. In C. elegans, this category includes at least 1733 genes (http://www.wormbase.org, release WS195). For downstream genes in operons, SL trans-splicing separates the individual mRNAs and provides the 5′ TMG (trimethylguanosine) cap on the SL exon to the mRNA. Since downstream mRNA genes are not 5′ end-capped cotranscriptionally as first genes are, trans-splicing supplies the cap needed for protection for the mRNA 5′ end and for translation.
In most SL trans-splicing organisms, a single SL RNA (or variants) is used for all trans-splicing (Hastings 2005; Guiliano and Blaxter 2006; Douris et al. 2010), but Rhabditid nematodes have a specialized class of SL RNA (SL2) devoted to processing downstream operon genes (Huang and Hirsh 1989; Evans et al. 1997; Lee and Sommer 2003). SL1 and SL2 RNAs both donate a 22-nt exon with a 5′ TMG cap, fold into a three stem–loop structure, and are bound by Sm proteins. Despite these similarities, SL1 is used to process outrons, while SL2 is used to splice downstream genes in operons (Spieth et al. 1993; Blumenthal 2005).
SL2 interacts with the 3′ end processing factor CstF-64 (Evans et al. 2001). This likely provides some of the SL2 (vs. SL1) specificity, since only downstream gene trans-splice sites would be expected to be in proximity to a 3′ end formation site. In addition, a sequence element, named Ur because it is U-rich, within the ICR of the gpd-3 operon was required for downstream SL2 trans-splicing (Huang et al. 2001). Computational analyses extended the definition of the Ur element to include position +40 to +60 relative to the 3′ end cleavage site, and an adenosine preceding the uracil run (Graber et al. 2007).
Yeast and mammalian in vitro splicing systems have been used extensively to analyze mechanisms of cis-splicing. The ability to manipulate substrates has facilitated the characterization of primary splicing recognition signals, enhancer and silencer elements, and spliceosomal complexes and reaction intermediates (Lindsey and Garcia-Blanco 1999; Jurica and Moore 2002). In nematodes, an Ascaris in vitro splicing system has allowed for the comparison of cis-splicing with SL trans-splicing (Hannon et al. 1990, 1991). Currently, however, there is no published C. elegans in vitro splicing system and no system that has the ability to correctly specify SL1 and SL2 trans-splicing.
In this study, we report the use of a C. elegans embryonic extract in vitro trans-splicing system that can splice SL1 and SL2 with specificity approaching that seen in vivo. We identify a Ur element experimentally, demonstrate that it is essential for SL2 trans-splicing in vitro, and analyze the prevalence of this element within operons. We demonstrate that the element comprises a short stem–loop followed by one or more copies of the sequence UAYYUU. This pre-mRNA sequence may anneal to the SL splice site. We propose that it functions in a manner analogous to the U1 snRNA in cis-splicing, hybridizing with—and thereby defining—the 5′ss on the SL RNA in a role reversal of the snRNP and pre-mRNA.
Results
The in vitro trans-splicing system recapitulates in vivo SL specificity
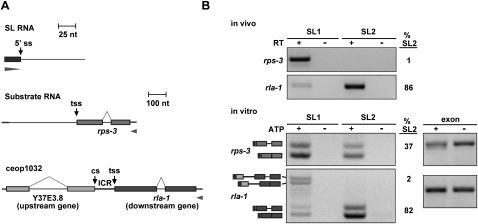
In order to study the mechanism of trans-splicing, as well as the requirements for SL specificity, we developed an in vitro trans-splicing system using C. elegans crude embryonic extracts. Unlabeled T7 transcripts were trans-spliced to SL RNAs present in the extract, and SL specificity was detected by RT–PCR specific for SL1 or SL2. Reverse primers specific for vector-derived portions of the substrates ensured detection of the T7 transcript rather than endogenous mRNA (Fig. 1A). The presence of all trans-spliced products was dependent on ATP, confirming that trans-splicing occurred in vitro (Fig. 1B; data not shown). (Reactions without ATP are not shown in subsequent figures, although all bands are ATP-dependent.) Although some substrates were not studied further due to failure to splice, unexpected trans-splicing at 3′ cis-splice sites, or failure to show SL specificity (data not shown), in the majority of cases, we found that single gene RNAs that splice to SL1 in vivo likewise splice to SL1 in vitro. The reason for the lack of specificity of some templates is unknown. Importantly, substrates derived from operons and containing portions of an upstream and downstream gene predominantly splice SL2 at the downstream gene trans-splice site, just as in vivo. These substrates (including rps-3, tct-1, nuo-4, and rla-1) are from highly expressed C. elegans genes and operons that showed robust SL specificity in vivo and were consistently cis-spliced and trans-spliced accurately in vitro (Fig. 1; Supplemental Fig. S1A). Since SL2 snRNP is far less abundant than SL1 snRNP, SL2 is clearly specifically chosen over SL1 for operon substrates. The level of product correlated well with the level of substrate added, but the ratio of SL1 to SL2 trans-splicing remained constant. Additionally, neither the temperature nor time of the reaction altered the SL1/SL2 ratio (data not shown). Examples of one SL1 and one SL2 splicing substrate are shown in Figure 1, and these substrates were used throughout these experiments. The rps-3 substrate was spliced in vitro predominantly by SL1, as it is in vivo (Fig. 1). Furthermore, the substrate derived from rla-1, a downstream operon gene, was spliced predominantly to SL2, as it is in vivo. Thus, the in vitro trans-splicing system recapitulated the SL specificity seen for these genes in vivo, and indicated that, in these cases, SL2 specificity is a property of the RNA sequences.
Figure 1.
The in vitro trans-splicing system recapitulates in vivo SL specificity. (A) Diagram of in vitro splicing RNA substrates. The SL1 and SL2 RNAs are present in the extract. rps-3 is trans-spliced to SL1 in vivo. rla-1 is a downstream gene in operon ceop1032, and is trans-spliced to SL2 in vivo. (tss) Trans-splice site; (cs) cleavage site; (ICR) ICR between upstream gene Y37E3.8 and downstream gene rla-1. Bars: for SL RNAs, 25 nt; for substrate RNAs, 100 nt. Boxes represent exons, and arrowheads represent the locations of RT and PCR primers. Forward primers are specific for the SL sequences of the endogenous SL1 or SL2, and reverse primers are specific to the substrate RNA. (B, top) RT–PCR with RNA from whole worms. Forward primers are specific for SL1 or SL2, and reverse primers are specific for rps-3 and rla-1. Lanes 2 and 4 lacked reverse transcriptase. Percent SL2 (%SL2; SL2 divided by the sum of SL1 and SL2) is given for the +RT reactions. (Bottom) RT–PCR of in vitro splicing reactions. Lanes 2 and 4 lacked ATP, creatine phosphate, and creatine phosphokinase. Reverse primers are specific for the splicing substrate RNA. Spliced products for the rps-3 and rla-1 substrates are shown (determined by sequencing the PCR products). The top bands in the rla-1 panel arise from splicing of SL1 to the last cis-splice site in the upstream operon gene. (Dark gray) 22-nt SL; (lighter gray) rps-3 and rla-1 spliced substrates. Percent SL2 is given for the +ATP reactions. Exon lanes are RT–PCR products using forward and reverse primers specific for the substrate, and are used as a measure of input RNA.
The two in vitro trans-spliced PCR products from each substrate correspond to the trans-spliced product with and without the removal of the intron (cis-splicing) (Fig. 1B, see the diagram at the side of the gel), as verified by sequencing (data not shown). Intron-containing products were not detected in vivo (Fig. 1B, top) due to efficient cis-splicing.
The two top bands in the bottom panel of Figure 1B, representing the rla-1 SL1 PCR, result from splicing of SL1 to the 3′ss of the last intron of the upstream Y37E3.8 gene (see the diagram at the side of gel). This intron is much larger than most C. elegans introns (214 nt as compared with 47 nt) (Blumenthal 2005), possibly resulting in the intron being mistaken for an outron. Equivalent bands are not seen in the SL2 PCR, indicating that the 3′ cis-splice site lacks information for SL2 specificity.
As SL1 is spliced only at a low level at the rla-1 trans-splice site in vitro, and this work deals primarily with operon processing and SL2 trans-splicing, only the SL2 data at the trans-splice site is presented for most figures. Analysis of SL1 in vitro trans-splicing was carried out in parallel, and is shown in Supplemental Figure S2. Notably, in no instance did the level of SL1 trans-splicing at the expected rla-1 trans-splice site increase when SL2 trans-splicing was reduced by mutation, indicating that SL1 is not able to substitute when SL2 trans-splicing is lost.
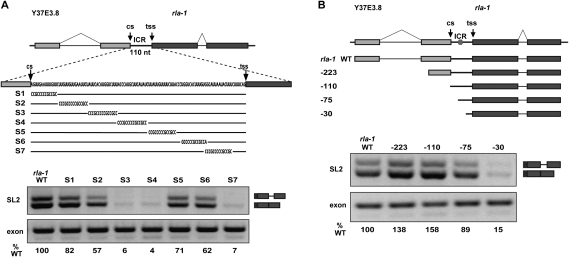
A pre-mRNA region required for SL2 trans-splicing
We wanted to investigate the sequence requirements for the strong SL2 specificity of the rla-1 substrate. We targeted the ICR with blocks of substitution mutations throughout the 110-nt ICR (Fig. 2A; Supplemental Fig. S1B). This identified a region in the middle of the rla-1 ICR required for SL2 trans-splicing. The reduction of SL2 trans-splicing with constructs S3 and S4 is comparable with construct S7, which eliminates the presumed branchpoint. We also shortened the rla-1 substrate from the 5′ end (Fig. 2B). The 5′ end of the genomic region of the wild-type rla-1 construct is −604 upstream of the trans-splice site. We created constructs that begin in the terminal exon of Y37E3.8 (−223 nt upstream of the rla-1 trans-splice site), immediately after the upstream gene 3′ end cleavage site (−110), or within the ICR (−75 and −30). In each case, an additional upstream 22 nt of RNA is provided by the vector-derived portion of the substrate. Robust SL2 trans-splicing is clearly maintained even when upstream rla-1 RNA is limited to only −75 nt (97 nt total), which contains the region of the ICR implicated by the scanning substitutions. In contrast, the −30 construct is not efficiently trans-spliced.
Figure 2.
A pre-mRNA region required for SL2 trans-splicing. (A, top) Diagram of in vitro splicing substrates and substitution mutations within the ICR of the rla-1 substrate RNA. (Bottom) RT–PCR of in vitro splicing reactions. SL2 trans-spliced products are indicated in the diagram at the right. (%WT) Ratio of the values of the experimental to the wild-type SL2 PCR products, each normalized to their respective exon PCR products. (B, top) Diagram of in vitro splicing substrates. The 5′ ends of each progressively shortened construct are indicated. Nucleotide distances upstream of the trans-splice site are −223, −110, −75, and −30 (wild type is −604). (Hexagon) Approximate location of S3/S4 element from A. (Bottom) RT–PCR of in vitro splicing reactions.
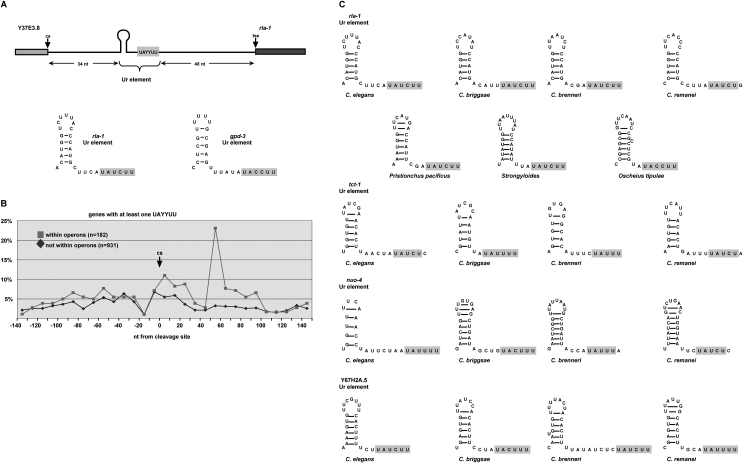
The Ur element comprises a short stem–loop and a consensus UAYYUU motif
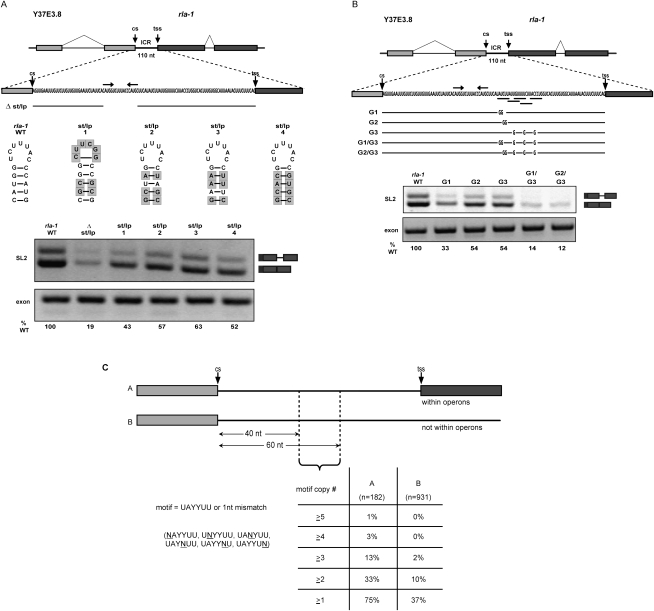
To determine whether different ICRs shared any sequence, we aligned those upstream of rla-1, tct-1, and nuo-4, each of which was robustly SL2 trans-spliced both in vivo and in vitro (Supplemental Fig. S1). This alignment, annotated with the rla-1 substitution mutations of Figure 2A, reveals a short motif of identical sequence corresponding to the last 4 nt of rla-1 substitution S4 and the next few nucleotides. A close examination of the entire region defined by constructs S3 and S4, and the corresponding regions in the other operons, led to the observation that, although the sequences were different, a short stem–loop with five or six stem base pairs could be formed in each case just upstream of the identical sequence motif (Fig. 3A). The mutations S3 and S4 disrupt this two-part RNA element, and it corresponds to the short Ur element required for in vivo SL2 trans-splicing in the gpd-3 operon (Huang et al. 2001); that element can also be divided into two parts, each required for trans-splicing, and separated by a short spacer region. Our analysis revealed that the first part of the gpd-3 Ur element can form a 5-base-pair (bp) stem (although with different sequences), and the second part has the sequence motif observed in the rla-1, tct-1, and nuo-4 ICRs (Fig. 3A,C; Supplemental Fig. S1B). In each of these operons, the sequence element is located near the middle of the ICR, with the stem just upstream. Since mutations in this region of the gpd-3 operon lost trans-splicing activity in vivo, and in rla-1 lost trans-splicing activity in vitro, we concluded that we had identified the rla-1 Ur element. The existence of a stem of different sequences explained the previous difficulty in finding the particular sequence of the gpd-3 Ur element in other operons.
Figure 3.
The Ur element comprises a short stem–loop and a consensus UAYYUU motif. (A) Diagram of the ICR between Y37E3.8 and rla-1 indicating the two components of the Ur element. Shown below is the nucleotide sequence of this Ur element and that in the previously described gpd-3 operon (Huang et al. 2001). (B) Bioinformatic analysis comparing the percentages of genes containing UAYYUU upstream of and downstream from the 3′ end cleavage site (cs) within (gray squares) or not within (black diamonds) operons. (C) Nucleotide sequences of presumptive Ur elements of different nematode species shown for four operons. The trans-spliced downstream gene name is given.
When we examined the identical sequence motif of the second part of the Ur element, we observed that it is complementary to the 5′ss of the SL RNA, suggesting that it could anneal in much the same way as U1 snRNA anneals to and defines the 5′ss in cis-splicing. The consensus sequence for a base-pairing interaction across the splice site (nucleotide positions −3 to +3) of the SL RNA is UAYYUU (where Y = C/U), and this sequence occurs within the same region of each of the four ICRs initially examined. Figure 3A shows the two-part Ur element and its location within the rla-1 operon, as well as the nucleotide sequences of the rla-1 and gpd-3 stem–loop and UAYYUU.
Bioinformatic analysis of the Ur element
In order to analyze the prevalence of the UAYYUU motif in other ICRs, we compared the incidence of different nucleotide sequences surrounding the 3′ cleavage sites found within operons (ICRs) compared with those not within operons (terminal genes in operons and nonoperon genes). The percentage of genes containing UAYYUU (Fig. 3B) or the 1-nt-shorter UAYYU (Supplemental Fig. S1C) was plotted in 10-nt windows for 140 nt upstream of and downstream from the 3′ end cleavage sites. There is a sharp peak of this sequence occurring 50–70 nt downstream from the cleavage site. The peak is present within ICRs (Fig. 3B, gray line), but not downstream from terminal cleavage sites (Fig. 3B, black line). These data support the involvement of this sequence in trans-splicing, since only ICRs end in a trans-splice site. The 50- to 70-nt location is consistent with the UAYYUU position of the experimentally studied operons. In the rla-1 operon, the 3′ nt of the UAYYUU is located at +62 nt from the cleavage site, and in the gpd-3 operon, it is +51 nt. The majority of C. elegans ICRs have ∼110 nt between the 3′ cleavage site of the upstream gene and the trans-splice site of the downstream gene (Blumenthal 2005), so the Ur element is positioned in the middle of the ICR, ∼50 nt from the splice site.
Using the evidence from the C. elegans in vitro splicing mutagenesis, we looked for similar rla-1 Ur elements in seven different nematode species. In each case, the rla-1 gene was downstream in a predicted operon, and the presumptive Ur element was in the middle of the ICR. As shown in Figure 3C, each Ur element comprises a short stem–loop, followed closely by UAYYUU (except Caenorhabditis remanei, where it is UAYYUG). While these features of the Ur elements are conserved, the primary sequence is not. The nucleotides within the stem, within the loop, and within the spacer sequence all vary; even the pyrimidine nucleotides of the consensus motif can vary. In addition, the length of the stem, the number of bases in the loop, and the length of the spacer are variable. Based on this definition, we identified a predicted Ur element in the middle of numerous C. elegans ICRs (examples are shown in Fig. 3C). Indeed, upon extending these predictions to include various species (Fig. 3C), we found conservation of location and predicted stem and UAYYUU (or occasionally UAYYU), but rarely of particular sequence (Fig. 3C; data not shown).
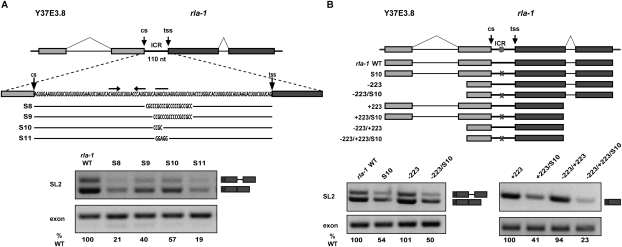
The Ur element is required for SL2 trans-splicing in a variety of contexts
In order to examine the boundaries of the rla-1 Ur element, we designed mutations that targeted the downstream part (Fig. 4A). SL2 trans-splicing was reduced in all cases, showing that substituting as few as 4 nt (S10 mutation) could cause a decrease in SL2 trans-splicing. To determine whether SL2 trans-splicing is dependent on upstream and/or downstream introns and in a Ur-dependent way, we tested constructs shortened from the 5′ end, the 3′ end, or both that remove introns. Figure 4B shows that neither intron was required for SL2 trans-splicing. In fact, the construct consisting of only a portion of the upstream exon, the ICR, and most of the downstream exon is capable of splicing specifically to SL2 (Fig. 4B, −223/+223). Furthermore, the S10 substitution (Fig. 4A, see diagram) leads to loss of SL2 trans-splicing in all four substrates: wild type, −223, +223, and −223/+223. Since, in each case, S10 shows reduced SL2 trans-splicing, we concluded that the Ur element is involved in SL2 trans-splicing in a variety of contexts, and that this involvement does not require cis-splicing.
Figure 4.
The Ur element is required for SL2 trans-splicing in a variety of contexts. (A, top) Diagram of in vitro splicing substrates and substitution mutations within the Ur element of the rla-1 ICR. Horizontal end-to-end arrows and the bar above the sequence indicate the nucleotides of the Ur element stem and UAYYUU, respectively. (Bottom) RT–PCR of in vitro splicing reactions. (B, top) Diagram of in vitro splicing substrates. Substrates have been shortened from either the 5′ end (−223 upstream of the trans-splice site; wild type is −604), the 3′ end (+223 downstream of the trans-splice site; wild type is +449), or both. X indicates the Ur element substitution S10 shown in A. (Bottom) RT–PCR of in vitro splicing reactions. (Right panel) The +223 constructs have smaller PCR products.
The stem–loop region is needed for SL2 trans-splicing
In order to examine the roles of the two parts of the Ur element, we created additional targeted substitution and deletion mutation constructs. As shown in Figure 5A, a precise deletion of the predicted stem results in loss of SL2 trans-splicing (19% of wild type). This may indicate that the stem is necessary for some aspect of trans-splicing, or that it is important for the integrity of the downstream RNA following upstream gene 3′ end cleavage. We also replaced the wild-type stem–loop with different heterologous stem–loops, including one that is predicted to be quite stable (Fig. 5A, st/lp 1) and three that maintain the wild-type loop sequence, altering only 3 or 4 bp within the stem. Substitution with any of these stem–loops increases SL2 trans-splicing compared with the deletion, but wild-type levels are never completely restored. This is consistent with the idea that a stem–loop is required (for trans-splicing or RNA integrity), but some aspect of the actual Ur element sequence also plays a role, since heterologous stems can only partially substitute.
Figure 5.
A stem–loop and a UAYYUU are necessary for SL2 trans-splicing. (A, top) Diagram of Ur element stem mutation in vitro splicing substrates. Δ st/lp is a deletion of the 17 nt in the stem–loop. The sequence of each stem–loop substitution is shown next to the wild-type stem–loop, with shaded boxes indicating changed nucleotides. (Bottom) RT–PCR of in vitro splicing reactions. (B, top) Diagram of Ur element UAYYUU mutation in vitro splicing substrates. Horizontal bars below the sequence indicate the nucleotides of the Ur element UAYYUU and the four overlapping downstream mismatched copies. G1 and G2 contain substitutions in the first full UAYYUU motif, G3 contains substitutions in the four downstream mismatched UAYYUU copies, and G1/G3 and G2/G3 contain substitutions in all of them. (Bottom) RT–PCR of in vitro splicing reactions. (C) Bioinformatic analysis comparing the percentages of genes within (column A) or not within (column B) operons that contain multiple copies of the sequence motif UAYYUU (allowing a 1-nt mismatch), within a 20-nt window located 40–60 nt downstream from 3′ end cleavage sites (cs). Motif copy # indicates the number of instances of the motif occurring within the window. Motif copies may overlap.
Multiple copies of UAYYUU play a role in SL2 trans-splicing
The rla-1 ICR contains multiple, overlapping copies of UAYYUU, some with a single nucleotide mismatch (underlined in Fig. 5B, top). In fact, the majority of Ur elements in other operons have multiple copies of UAYYUU in this small region (Fig. 5C; data not shown). To assess the contribution of these “extra” copies, we created constructs that contained substitutions within the first full UAYYUU (Fig. 5B, G1 and G2), within the four downstream mismatched UAYYUU copies (Fig. 5B, G3), or within all the copies combined. G1 and G2 demonstrate that 2-nt substitutions within the most 5′ UAYYUU reduce, but do not eliminate, SL2 trans-splicing. The same is true where all downstream copies are mutated (Fig. 5B, G3). Importantly, when the mutations are combined (Fig. 5B, G1/G3 or G2/G3), SL2 trans-splicing is almost completely lost (14% and 12% wild type). This demonstrates that the additional mismatched copies act additively in SL2 trans-splicing, and are partially redundant.
To ask whether partial redundancy was widespread among ICRs, we searched for the UAYYUU motif or any single nucleotide mismatch within a specified region (40–60 nt downstream from 3′ end cleavage sites). Figure 5C shows that 75% of operon ICRs (column A) have at least one UAYYUU (allowing a single mismatch) within that limited region, while only 37% of nonoperon genes do (column B). Furthermore, operons are far more likely to have multiple copies. The chart shows the percentages of genes with at least one, two, three, four, or even five copies. Whereas only 2% of nonoperon genes have three or more copies within this region, 13% of ICRs within operons do. It is also notable that nearly half (44%) of the operon genes that have the motif or a mismatched copy have more than one within this very limited region. This shows that the multiple copies of UAYYUU, which we showed to be important to rla-1 trans-splicing, frequently occur in other operon ICRs.
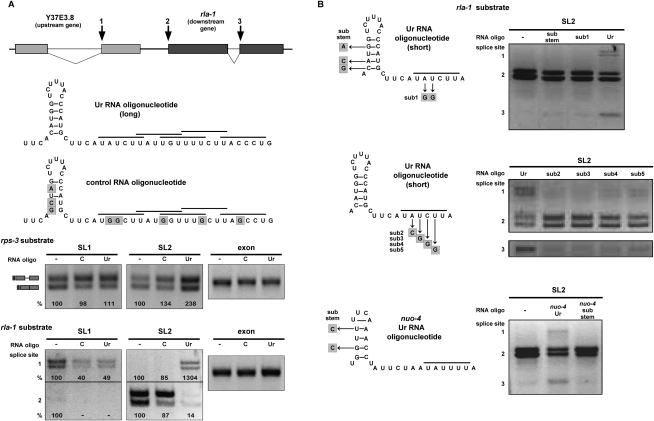
A Ur element RNA oligonucleotide alters SL trans-splicing specificity
In order to address the mechanism of the Ur element and its effect on trans-splicing, we asked whether addition of an RNA oligonucleotide spanning the element could act as a competitive inhibitor of SL2 trans-splicing, leaving SL1 trans-splicing unaffected. Instead, we got unexpected, but quite interesting, results. The long Ur element RNA oligonucleotide is depicted in Figure 6A. It is 49 nt long and contains the stem and all copies of the UAYYUU sequence. An oligonucleotide containing substitutions within the predicted stem and each of the UAYYUU copies (the G1/G3 substitutions in Fig. 5B) was used as a control. In each splicing reaction, the RNA oligonucleotide was added immediately prior to addition of the RNA substrate. We first tested a range of oligonucleotide concentrations (0–333 ng/μL), and determined that nonspecific total splicing inhibition occurred at the highest concentrations (data not shown). However, at lower concentrations (optimally 33 ng/μL, which is a 300–500 times molar excess over the substrate, depending on the substrate used), we observed a consistent and surprising effect. As Figure 6A shows, the Ur element RNA oligonucleotide, but not the control, alters the SL trans-splicing specificity on both the rps-3 and rla-1 substrates. The rps-3 substrate, which is normally spliced predominantly by SL1, now shows a distinct increase in SL2 splicing. The most intriguing change is seen in the two top bands of the rla-1 substrate that correspond to products trans-spliced at the upstream gene 3′ cis-splice site (Fig. 6A, splice site designated 1). That splice site, which was only SL1 trans-spliced in the wild-type or mutant rla-1 constructs (Fig. 1B; Supplemental Fig. S2), is trans-spliced by SL2 in the presence of the Ur oligonucleotide, but not the control oligonucleotide. In addition, there is a decrease of SL2 trans-splicing at the expected trans-splice site. This same change in SL specificity was seen with other splicing substrates (data not shown), where SL2 trans-splicing is increased at sites that normally splice SL1 in vitro.
Figure 6.
A Ur element RNA oligonucleotide alters SL trans-splicing specificity. (A, top) Diagram of wild-type rla-1 substrate, as in Figure 1. Numbers 1, 2, and 3 indicate sites of SL2 trans-splicing in the presence of a Ur element RNA oligo. (Middle) Diagram of long RNA oligos added to in vitro splicing reactions. The long Ur element oligo (Ur) includes the stem and all of the UAYYUU motifs (each indicated by a horizontal bar above the sequence). The control RNA oligo (C) contains substitutions, indicated by shaded boxes. (Bottom) SL1, SL2, and exon RT–PCR of in vitro splicing reactions of the wild-type rps-3 and rla-1 RNA substrates (shown in Fig. 1 and in the top panel) in the absence (−) or presence of an excess amount of control or long Ur element RNA oligo. Percent is calculated relative to the no oligo lanes. (B, left) Diagrams of short RNA oligos added to in vitro splicing reactions. The wild-type short Ur element sequence contains the stem–loop and one UAYYUU motif (horizontal bar above the sequence). Substitutions in other oligos are indicated by shaded boxes. (Right) SL2 RT–PCR of in vitro splicing reactions of the wild-type rla-1 substrate in the absence (−) or presence of indicated RNA oligos. Spliced products 1, 2, and 3 are SL2 trans-spliced at the sites indicated in the diagram in A.
Next, we analyzed the requirements for this novel activity of the Ur element RNA oligo. We began by testing a shorter oligonucleotide containing the stem and the perfect UAYYUU (Fig. 6B). This oligo had the same effect as the long oligo, increasing SL2 trans-splicing at both rla-1 substrate cis-splice sites (Fig. 6B, designated 1 and 3). Both the stem and the UAYYUU were required for this activity (Fig. 6B, top). Furthermore, changing any of four bases in the UAYYUU to bases that would interfere with base-pairing to the SL2 snRNA abrogated the effect of the oligo (Fig. 6B, middle), strongly supporting the SL2 RNA/Ur element base-pairing model. Finally, we showed that an unrelated oligo with the sequence of the Ur element from a different operon showed a similar activity on the rla-1 in vitro splicing substrate, and this activity was also dependent on the ability to form a stem (Fig. 6B, bottom gel).
While it is not clear how the Ur element RNA oligonucleotides act, there is a distinct effect on SL2 trans-splicing, which supports the idea that the Ur element is involved in SL2 specificity. When a Ur element oligo is added, SL1 trans-splicing is barely affected, relative to the control. SL2, however, promiscuously trans-splices at many sites that normally splice introns or SL1, and also shows reduced use at downstream operon trans-splice sites. These data are consistent with the idea that the SL2 snRNP itself is altered by the Ur oligo and loses its specificity, supporting the idea of a direct interaction between the SL2 snRNP and the Ur element.
Discussion
Here we show that a C. elegans extract both cis-splices and trans-splices several pre-mRNA substrates, recapitulating in vivo SL specificity. The Ur element upstream of the trans-splice site and required for SL2 trans-splicing comprises a short variable stem–loop, followed by one or more UAYYUU motifs. We show that this element is prevalent between genes in C. elegans operons. Additionally, a Ur element oligonucleotide alters the specificity of endogenous SL2, consistent with a direct Ur/SL2 snRNP interaction. We propose a model for SL2 trans-splicing involving base-pairing between the UAYYUU and the SL2 snRNP. This interaction is analogous to the 5′ss/U1 snRNP interaction, but in trans-splicing the snRNP contains the 5′ss and the pre-mRNA contains the site to which it anneals.
Correspondence between in vivo and in vitro results
In vivo mutagenesis studies of the gpd-2/gpd-3 operon showed the Ur element is required for downstream gene trans-splicing (Huang et al. 2001). A 20-nt U-rich region located in the middle of the gpd-3 ICR contained two required parts. Even a 1-nt substitution severely reduced gpd-3 mRNA accumulation. Similarly, in vitro, we show that a bipartite Ur region in the middle of a different operon ICR is required for SL2 trans-splicing to the downstream gene, and that a 2-nt substitution can dramatically reduce SL2 trans-splicing. Our re-examination of the gpd-2/gpd-3 ICR sequences indicated that the 5′ part can form a 5-bp stem–loop, and the 3′ part contains UACCUU. The in vivo mutagenesis results on gpd-2/gpd-3 are in excellent agreement with the in vitro results on the rla-1 operon reported here. Furthermore, the in vitro trans-splicing system not only recapitulates the in vivo SL specificity, but it depends on the same cis-acting sequence elements. The fact that such similar results were obtained validates both systems, and indicates that in vitro trans-splicing can be used to dissect operon trans-splicing.
Concurrent transcription is not required for SL2 specificity
Downstream gene trans-splicing might have required concurrent transcription, making this sort of in vitro splicing system relatively uninformative (Maniatis and Reed 2002; Kornblihtt et al. 2004; de Almeida and Carmo-Fonseca 2008). In vivo 3′ end formation and trans-splicing appear to occur concurrently with transcription, since the unprocessed precursor is rarely detected (T Blumenthal, unpubl.). Fortunately, the trans-splicing signals and the SL specificity information must be encoded within the RNA transcript, since trans-splicing occurs with the correct SL specificity in vitro in the absence of concurrent transcription. This is intriguing for both types of substrates. It appears that, for SL1 specificity, proximity to promoter elements, RNA polymerase II, and 5′ end-capping machinery is not necessary. Similarly, for SL2 specificity, transcription rate and RNA polymerase II and associated proteins are not required. However, coupling of trans-splicing to transcription in vivo could still increase trans-splicing efficiency or specificity.
Possible linkage between 3′ end formation and trans-splicing
3′ end formation of an upstream operon gene may be linked to SL2 trans-splicing of a downstream gene. Significantly, the distance between the two RNA processing sites appears to be constrained, with a median length of 112 nt (MA Allen, LW Hillier, RH Waterston, T Blumenthal, unpubl.). In addition, 3′ end formation factor CstF-64 coimmunoprecipitated SL2 RNA but not SL1 RNA. Mutant SL2 RNA sequences that had lost this interaction were unable to trans-splice at downstream operon sites (Evans and Blumenthal 2000; Evans et al. 2001). Furthermore, there is a strong rationale for a connection between 3′ end formation and trans-splicing. Downstream operon genes are not cotranscriptionally capped; therefore, upstream gene 3′ end cleavage would leave an unprotected 5′ end on the downstream pre-mRNA. The uncapped mRNA would likely be rapidly degraded 5′ to 3′ (Liu et al. 2003). Linking the cleavage that generates the uncapped 5′ RNA end with trans-splicing that provides a 5′ TMG cap would ensure protection of the downstream mRNA, as well as prevent transcription termination that is associated with 3′ end formation. The distance constraint may be explained by the interaction of the 3′ end formation and trans-splicing machineries, or by the need to coordinate the two RNA processing events.
Nonetheless, there is evidence that 3′ end cleavage and trans-splicing are not absolutely dependent on one another. Mutation of an operon gene AAUAAA abolished 3′ end formation without leading to loss of downstream SL2 trans-splicing, although SL1 trans-splicing was increased (Kuersten et al. 1997; Huang et al. 2001; data not shown). Furthermore, in vitro substrates that contained no AAUAAA because they were shortened from the 5′ end were still able to splice SL2. In addition, SL2 trans-splicing at downstream genes was not affected by mutating the predicted CstF-binding region (J Morton and E Lasda, unpubl.), or by adding an RNA oligonucleotide containing that same region. Since 3′ end processing and SL2 trans-splicing occur close to one another, and 3′ end factor CstF-64 is associated with SL2 snRNP, it seems likely that the 3′ end machinery, or 3′ end formation itself, is part of the signal for SL2 trans-splicing. However, at least in all of the contexts tested, this interaction is not required, as SL2 trans-splicing can occur in the absence of any known polyA signal.
Similarly, trans-splicing is not required for upstream 3′ end formation, since mutation of neither the trans-splice site nor the Ur element had an effect on upstream gene 3′ end formation in vivo, although these mutations abolished the downstream mRNA (Kuersten et al. 1997; Huang et al. 2001). This may mean that the two processes are capable of occurring independently, but that linking them serves to increase the specificity and/or efficiency, as has been shown for other RNA processing events (Maniatis and Reed 2002; Kornblihtt et al. 2004; de Almeida and Carmo-Fonseca 2008).
The Ur element: a required SL2 trans-splicing signal
In contrast to the 3′ end formation signal, the Ur element is clearly required for downstream gene SL2 trans-splicing, both in vivo and in vitro. Constructs that do not have an intact Ur element splice SL2 poorly, if at all. Furthermore, shortened constructs that lack the introns still robustly trans-splice SL2, if the Ur element is intact. Thus, the introns are not required for the Ur element to function in SL2 trans-splicing in vitro. Mutation of either part of the Ur element leads to a severe reduction in trans-splicing. Although this element was originally described in the gpd-2/gpd-3 operon (Huang et al. 2001), predictions of sequences that might constitute the Ur element in other operon ICRs, based solely on sequence, proved inaccurate. Computational analysis revealed two Ur regions in ICRs: the CstF-binding site just downstream from the 3′ cleavage site, and the Ur element (Graber et al. 2007). This study indicated that position within the ICR is a key component of the Ur element definition. Here we show that the Ur element can be predicted in different species as well as in different operons based on a combination of sequence, position, and structure.
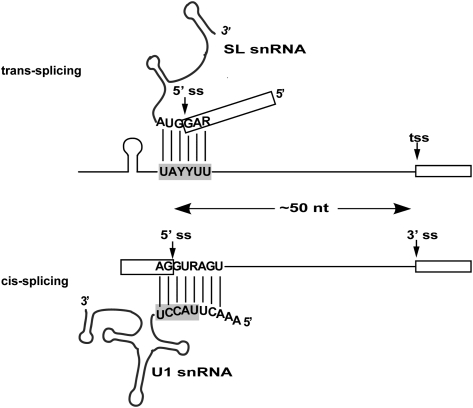
We propose that the Ur element interacts directly with the SL2 snRNP (Fig. 7). We hypothesize that the UAYYUU transiently base-pairs across the SL RNA splice site analogous to the U1 snRNA interaction in cis-splicing, and that the Ur element stem–loop provides additional specificity or stability to the binding interaction. This model is attractive in three ways. First, it provides for direct recruitment of the SL RNA containing the 5′ss by the pre-mRNA. Second, it positions the 5′ss at a location 50 nt upstream of the pre-mRNA trans-splice site. This spacing is the same as the typical C. elegans intron. Third, it proposes a parallel for a U1-like 5′ss recognition in SL2 trans-splicing. In cis-splicing, U1 inactivation inhibits splicing. Similarly, in SL2 trans-splicing, Ur element inactivation inhibits splicing.
Figure 7.
The Ur element may hybridize with the 5′ss of the SL RNA. The UAYYUU of the Ur element on the trans-splicing pre-mRNA is predicted to hybridize with the 5′ss located on the SL RNA. This is analogous to the U1 role in cis-splicing, in which the U1 snRNA recognizes and base pairs with the pre-mRNA 5′ss, except in trans-splicing these roles would be inverted: The 5′ss is on the snRNP, while the proposed hybridizing sequence is on the pre-mRNA. This would position the 5′ss of the SL RNA ∼50 nt upstream of the 3′ss (trans-splice site), the same distance as the majority of C. elegans introns. Exons are indicated as boxes. Proposed Ur or known U1 base-pairing interactions are depicted as solid lines.
The stem–loop of the Ur element
The existence of a short stem–loop at the 5′ end of the Ur element is supported by three facts. First, all predicted Ur elements of different operons share this feature. Second, variations in primary sequence among species still maintain stem-forming potential. Third, the in vivo mutagenesis of gpd-3 and the in vitro experiments of rla-1 demonstrate that different stem–loop sequences recovered partial SL2 trans-splicing compared with the construct in which the stem–loop was deleted and substitutions were not predicted to form stems (Fig. 5A; data not shown). Lack of full recovery by heterologous stem–loops may indicate that subtle structural or sequence aspects of the region are playing a role. Perhaps a stem–loop supplies a binding surface, but some feature of the sequence provides the ideal context. We propose that one purpose of the stem is to add a discriminating interaction with the SL2 snRNP, contributing to the SL specificity of downstream operon genes. Additionally, the stem or anything bound to it may block 5′-to-3′ exonucleolytic RNA degradation following cleavage at the 3′ end of the upstream gene. Supporting this, the laboratory (Liu et al. 2003) has reported previously the existence of a “Ur RNA” from the gpd-2/gpd-3 transgenic operon with a 5′ end 4 nt upstream of the Ur element stem, a location dependent on the position of the Ur element.
The UAYYUU motif of the Ur element
Previous in vivo results (Huang et al. 2001) as well as our in vitro results demonstrate that the UAYYUU is required for SL2 trans-splicing. Furthermore, UAYYUU is overrepresented in a 20-nt window between operon genes. The consensus sequence agrees well with the overrepresented pentamer sequences (Graber et al. 2007) within ICRs 40–60 nt downstream from the 3′ end cleavage site, (UACUU, UAUCU, and UAUUU). These sequences are well conserved among Caenorhabditid species. Interestingly, the identity of the pyrimidines (U or C) within the UAYYUU of a particular ICR are often not the same among different species, lending support to the base-pairing idea, since the two guanines of the SL splice site could pair with either U or C.
Many operons contain clusters of UAYYUU repeats, and these repeats are functionally important in vitro. During cis-splicing, the U1:5′ss duplex must be exchanged for a 5′ss interaction with U6 and U5. Hyperstabilization of the U1 interaction inhibits this switch (Staley and Guthrie 1999). In the trans-splicing parallel, the SL RNA 5′ss base-pairing interaction would need to be unwound in order for splicing to occur. Perhaps the Ur element contains multiple UAYYUU repeats to increase the possibility of base-pairing without increasing its stability.
Since UAYYUU could hybridize to the same relative positions across the 5′ss of either SL1 or SL2 RNA, it cannot be the sole determinant of SL1 versus SL2 specificity. Perhaps the SL2:CstF interaction is sufficient to bring SL2 into proximity with the Ur element, and hence with the nearest downstream trans-splice site. Alternatively, the specificity may be provided by some surface of the stem–loop part of the Ur element.
In Ascaris extracts, in vitro transcribed SL RNA can be trans-spliced to substrate RNAs (Maroney et al. 1990). In order to test the base-pairing model, we attempted to restore the splicing defects of UAYYUU substitutions through compensatory base changes in exogenously added SL2 RNA. However, our extracts did not splice in vitro transcribed SL RNA (data not shown). Nonetheless, we did show that four different single-base mutations in the UAYYUU predicted to interfere with base-pairing with the SL2 RNA 5′ss each prevented the oligonucelotide from altering SL2 snRNP specificity in vitro (Fig. 6B), providing strong support for the base-pairing model of Figure 7.
As an alternative to the base-pairing idea, the Ur element could bind some other trans-acting factor responsible for recruiting the SL2 snRNP. Indeed, the sequence looks like the AU-rich motif bound by ARE-binding proteins. However, to date, no SL2-specific proteins have been identified, and the homologs of the trans-splicing-specific proteins identified in Ascaris (Denker et al. 2002) have been found on SL1 but not SL2 in C. elegans (MacMorris et al. 2007). Finally, many mutations that fortuitously reduced the proposed base-pairing interfered with SL2 trans-splicing (Huang et al. 2001).
A Ur element oligonucleotide acts specifically on SL2
In vivo, C. elegans trans-splicing substrates splice almost exclusively either SL1 or SL2, with very few trans-splice sites allowing splicing to both SLs (MA Allen, LW Hillier, RH Waterston, T Blumenthal, unpubl.). This specificity is at least partially determined by gene context; the 5′-most trans-splice sites receive SL1, whereas downstream operon genes receive SL2. In addition to this, or perhaps alternatively, there may be sequence elements that establish SL bias. The Ur element RNA oligo specifically affected SL2 activity, either directly or indirectly: SL2 became capable of trans-splicing to sites that normally splice only SL1 in vitro, such as the cis-splice site of Y37E3.8 and the SL1 splice site of rps-3. We propose that, in the presence of the Ur oligo, SL2 either splices indiscriminately at any splice site, or acts explicitly with SL1-like specificity. The oligo could be decreasing the specificity of the SL2 snRNP for downstream operon genes, perhaps by titrating away some specificity factor (for example, a factor that bridges the SL2 snRNP with 3′ end processing proteins). In this case, an SL2 snRNP would be able to trans-splice at any splice site either indiscriminately or like SL1. Alternatively, SL2 snRNP may normally be in an inactive state, and is only activated when it interacts with a Ur element, perhaps through 5′ss definition. In this case, the oligo could activate the SL2 RNA for splicing. However, the SL2 activated by a Ur element located on an oligo instead of on the pre-mRNA is not tethered to a particular downstream trans-splice site, like in an operon ICR, and therefore can splice indiscriminately.
Implications for trans-splicing mechanism
The Ur element may point toward a general mechanism for SL trans-splicing (Fig. 7). Does a corresponding element function in SL1 trans-splicing? Interestingly, a UC-rich element has been identified ∼50 nt upstream of SL1 trans-splice sites: the Ou (for outron) element (Graber et al. 2007). Although we do not know the precise sequence or whether there is a structural component to this element, we suggest this element may hybridize across the SL1 5′ss (GAG/guaaaca). Furthermore, this mechanism of SL splice site recognition could be used by other trans-splicing phyla. Although trans-splicing likely arose many times independently (Douris et al. 2010), in each lineage it would require SL recruitment, appropriate positioning relative to the pre-mRNA splice site, and splice site definition. A search for a pre-mRNA structure/sequence element in other phyla would be worthwhile. Even chimeric mRNAs created by trans-splicing of separate pre-mRNAs at low levels in mammals or constitutively in flies could require a related interaction between a 5′ss on the donor mRNA and its antisense that is fortuitously present upstream of the 3′ss. If so, this kind of trans-splicing event could even be U1 independent.
Materials and methods
Endogenous RNA and RT–PCR
C. elegans N2 mixed-stage populations were harvested from plates and broken by freeze/thaw in TRIzol reagent (GIBCO-BRL). RNA was isolated by TRIzol extraction and DNase treatment. First strand cDNA was prepared using SuperScript II RT (Invitrogen) and gene-specific primers for either rps-3 or rla-1 (Supplemental Table S1). PCR for SL1 and SL2 was as described below, except that reverse PCR primers anneal within exon 2 of each gene (Supplemental Table S1).
Splicing substrates
Genomic regions surrounding the trans-splice sites of rps-3 (669 nt) and rla-1 (1053 nt from operon ceop1032 containing portions of Y37E3.8 and rla-1) were PCR-amplified and cloned ino the ApaI and EcoRV sites of pBluescript SK+ (Invitrogen). Deletion and substitution mutations were made with standard PCR cloning techniques. Genomic cloning primer sequences are in Supplemental Table S1, and mutation cloning primer sequences are available on request. In vitro transcription with T7 RNA polymerase adds 22 nt of vector-derived sequence to the 5′ ends of constructs and 37 nt to the 3′ ends of constructs when linearized with XbaI.
Plasmid DNA was linearized with XbaI and purified by Qiagen MinElute PCR Purification kit (#28004), and 1 μg was transcribed in vitro with T7 RNA polymerase using the mMessage mMachine kit with cap analog (Ambion/Applied Biosystems #AM1344). Samples were DNase-treated, then polyadenylated by the Poly(A) Tailing kit (Ambion/Applied Biosystems #AM1350). RNA was isolated by phenol/chloroform extraction and isopropanol precipitation, resuspended, and then column-purified by either Micro Bio-Spin columns (Bio-Rad #732-6250) or NucAway Spin columns (Ambion/Applied Biosystems #AM10070).
Embryonic extract
Synchronous liquid cultures of C. elegans N2 worms were grown at 20°C, shaking at 180 rpm for 3 d, collected by centrifugation, and washed three times with cold water. Embryos were isolated by hypochlorite treatment and a 1 M sucrose float, then washed three times with cold water and two times with cold homogenization buffer (10 mM TrisHCl at pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 50 mM sucrose, 0.05% NP-40, 1× complete protease inhibitor tablet, EDTA-free [Roche #1873560], 1 mM PMSF). Embryos were resuspended in an equal volume of cold homogenization buffer, and were broken using a tight metal dounce homogenizer 30 times. Homogenate was sedimented in 1.5-mL tubes for 5 min at 6000 rpm (3100g) at 4°C. The supernatant was dialyzed for 1 h at 4°C in dialysis buffer (20 mM Tris-HCl at pH 8.0, 50 mM KCl, 1 mM DTT, 10% glycerol, 0.5 mM EDTA, 1 mM PMSF). Aliquots were quick-frozen in liquid N2 and stored at −70°C. Final concentration was ∼25 μg/μL.
In vitro splicing
Twenty-five nanograms of splicing substrate RNA was added to a 15-μL final volume reaction containing 50% embryonic extract (final reaction concentrations: 60 mM KCl, 4 mM MgCl2, 2 mM ATP, 20 mM creatine phosphate, 50 μg/mL creatine phosphokinase, 2 mM DTT, 3% PEG 8000, 0.25 mM EDTA, 5% glycerol, 10 mM Tris-HCl at pH 8.0, 0.5 mM PMSF, 40 U/μL RNase inhibitor). Reactions designated −ATP omitted the ATP, creatine phosphate, and creatine phosphokinase. Reactions with RNA oligonucleotides (synthesized by IDT) added 500 ng of the RNA oligo (sequences listed in Supplemental Table S2) to the reaction immediately preceding substrate RNA addition. All reactions were incubated for 2 h at 15°C, then stopped by the addition of STOP buffer (10 mM Tris-HCl at pH 7.4, 250 mM NaOAc, 1 mM EDTA, 0.25% SDS, 2 μg/mL glycogen).
In vitro splicing analysis
Total splicing reactions were digested by proteinase K for 15 min at 37°C. RNA was isolated by phenol/chloroform extraction and ethanol precipitation. Twenty percent of recovered RNA was used in a 10-μL RT reaction with SuperScript II RT (Invitrogen) and a reverse primer corresponding to a pBluescript SK+ region. −RT reactions omitted the enzyme. Two microliters of the RT reactions was used in 50-μL PCR reactions. Initial substrate amount and PCR cycle number were optimized for linear range. Forward PCR primers were specific for SL1, SL2, or an exon, and reverse PCR primers were specific for a pBluescript SK+ region. RT and PCR primers are in Supplemental Table S1. Twenty microliters of each PCR was analyzed on an agarose gel with ethidium bromide. Volume analysis (cis-spliced ± bands combined) with local background correction was done using Quantity One software version 4.6.1 (Bio-Rad). Percent SL2 was calculated as the volume of the SL2 bands divided by the sum of the volume of SL1 and SL2 bands. Percent wild type was calculated as the ratio of the experimental to the wild-type (or no oligo for Fig. 6) volume of SL2 bands, each normalized to the volume of their respective exon bands. Gels and numbers shown are representative of at least two different splicing experiments.
Bioinformatics
Collection of sequences
The data set of sequences within operons are the 182 putative internal 3′ processing sites, and sequences not within operons are the 931 putative terminal 3′ processing sites determined and used in our previous publication (Graber et al. 2007). Sequences were represented in the standard International Union of Biochemistry/International Union of Pure and Applied Chemistry nucleic acid codes.
Computational searches for UAYYUU and UAYYU
Sequences were aligned by the 3′ end processing site and divided into 10-nt blocks. Regular expressions were used to search for the words TAYYTT and TAYYT within the sequence (T was used instead of U because DNA sequences were searched). The word was counted as present in a 10-nt block of sequence if the 3′-most nucleotide of the word was within the block. The percentage of sequences with at least one word present was plotted for each 10-nt block relative to the 3′ cleavage site.
Computational searches for UAYYUU and all 1-nt variants
All 1-nt variants of the word TAYYTT were determined (for example, VAYYTT is a 1-nt variant of TAYYTT). Each of the data sets was searched for the word and each of its variants. Locations were determined based on the 5′-most nucleotide of the word or variant. The number of copies of each word or variant located 40–60 nt downstream from the cleavage site was determined for each sequence. The percentage of genes in each data set with x copies of the word or variant was calculated (x = 0–5).
Acknowledgments
We thank past and present members of the Blumenthal laboratory, especially Scott Kuersten, Peg MacMorris, and Maria Pagratis, for both technical and intellectual contributions. We also thank Richard E. Davis for helpful discussions, and David Bentley for discussions and comments on the manuscript. This work was supported by National Institute of General Medical Science Grant NIH R01 GM42432.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1940010.
Supplemental material is available at http://www.genesdev.org.
References
- Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R 2006. Transcription-mediated gene fusion in the human genome. Genome Res 16: 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T 2004. Operons in eukaryotes. Brief Funct Genomics Proteomics 3: 199–211 [DOI] [PubMed] [Google Scholar]
- Blumenthal T 2005. Trans-splicing and operons. In WormBook (ed The C. elegans Research Community), WormBook, doi: 10.1895/wormbook.1.5.1, http://www.wormbook.org
- Boukis LA, Bruzik JP 2001. Functional selection of splicing enhancers that stimulate trans-splicing in vitro. RNA 7: 793–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Steitz JA 1990. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell 62: 889–899 [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Van Doren K, Hirsh D, Steitz JA 1988. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature 335: 559–562 [DOI] [PubMed] [Google Scholar]
- Conrad R, Thomas J, Spieth J, Blumenthal T 1991. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol 11: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Liou RF, Blumenthal T 1993. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J 12: 1249–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Lea K, Blumenthal T 1995. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA 1: 164–170 [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Carmo-Fonseca M 2008. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett 582: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Denker JA, Zuckerman DM, Maroney PA, Nilsen TW 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417: 667–670 [DOI] [PubMed] [Google Scholar]
- Douris V, Telford MJ, Averof M 2010. Evidence for multiple independent origins of trans-splicing in Metazoa. Mol Biol Evol 27: 684–693 [DOI] [PubMed] [Google Scholar]
- Evans D, Blumenthal T 2000. trans splicing of polycistronic Caenorhabditis elegans pre-mRNAs: Analysis of the SL2 RNA. Mol Cell Biol 20: 6659–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Zorio D, MacMorris M, Winter CE, Lea K, Blumenthal T 1997. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc Natl Acad Sci 94: 9751–9756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Perez I, MacMorris M, Leake D, Wilusz CJ, Blumenthal T 2001. A complex containing CstF-64 and the SL2 snRNP connects mRNA 3′ end formation and trans-splicing in C. elegans operons. Genes Dev 15: 2562–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SEJ, Butler MD, Pan Q, Ruvkun G 2008. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 455: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, Salisbury J, Hutchins LN, Blumenthal T 2007. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA 13: 1409–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiliano DB, Blaxter ML 2006. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. PLoS Genet 2: e198 doi: 10.1371/journal.pgen.0020198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Denker JA, Nilsen TW 1990. Trans splicing of nematode pre-messenger RNA in vitro. Cell 61: 1247–1255 [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Maroney PA, Nilsen TW 1991. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J Biol Chem 266: 22792–22795 [PubMed] [Google Scholar]
- Hastings KEM 2005. SL trans-splicing: Easy come or easy go? Trends Genet 21: 240–247 [DOI] [PubMed] [Google Scholar]
- Huang XY, Hirsh D 1989. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc Natl Acad Sci 86: 8640–8644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Kuersten S, Deshpande AM, Spieth J, MacMorris M, Blumenthal T 2001. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol Cell Biol 21: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ 2002. Capturing splicing complexes to study structure and mechanism. Methods 28: 336–345 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res 10: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Lea K, MacMorris M, Spieth J, Blumenthal T 1997. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA 3: 269–278 [PMC free article] [PubMed] [Google Scholar]
- Lall S, Friedman CC, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE 2004. Contribution of trans-splicing, 5′-leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. J Biol Chem 279: 45573–45585 [DOI] [PubMed] [Google Scholar]
- Lee K, Sommer RJ 2003. Operon structure and trans-splicing in the nematode Pristionchus pacificus. Mol Biol Evol 20: 2097–2103 [DOI] [PubMed] [Google Scholar]
- Li H, Wang J, Mor G, Sklar J 2008. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 321: 1357–1361 [DOI] [PubMed] [Google Scholar]
- Lindsey LA, Garcia-Blanco MA 1999. Prespliceosome and spliceosome isolation and analysis. Methods Mol Biol 118: 351–364 [DOI] [PubMed] [Google Scholar]
- Liu Y, Kuersten S, Huang T, Larsen A, MacMorris M, Blumenthal T 2003. An uncapped RNA suggests a model for Caenorhabditis elegans polycistronic pre-mRNA processing. RNA 9: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMorris M, Kumar M, Lasda E, Larsen A, Kraemer B, Blumenthal T 2007. A novel family of C. elegans snRNPs contains proteins associated with trans-splicing. RNA 13: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Marlétaz F, Le Parco Y 2008. Careful with understudied phyla: The case of chaetognath. BMC Evol Biol 8: 251 doi: 10.1186/1471-2148-8-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Hannon GJ, Denker JA, Nilsen TW 1990. The nematode spliced leader RNA participates in trans-splicing as an Sm snRNP. EMBO J 9: 3667–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Hannon GJ, Shambaugh JD, Nilsen TW 1991. Intramolecular base pairing between the nematode spliced leader and its 5′ splice site is not essential for trans-splicing in vitro. EMBO J 10: 3869–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu YT, Jankowska M, Nilsen TW 1996. Direct analysis of nematode cis- and trans-spliceosomes: A functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA 2: 735–745 [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW 2000. Functional recognition of the 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol Cell 6: 317–328 [DOI] [PubMed] [Google Scholar]
- Mongelard F, Labrador M, Baxter EM, Gerasimova TI, Corces VG 2002. Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160: 1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW 1993. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol 47: 413–440 [DOI] [PubMed] [Google Scholar]
- Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T 1993. Operons in C. elegans: Polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73: 521–532 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64 [DOI] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. 2010. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20: 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]