Abstract
Oncogene-mediated transformation of hematopoietic cells has been studied extensively, but little is known about the molecular basis for restriction of oncogenes to certain target cells and differential cellular context-specific requirements for oncogenic transformation between infant and adult leukemias. Understanding cell type-specific interplay of signaling pathways and oncogenes is essential for developing targeted cancer therapies. Here, we address the vexing issue of how developmental restriction is achieved in Down syndrome acute megakaryoblastic leukemia (DS-AMKL), characterized by the triad of fetal origin, mutated GATA1 (GATA1s), and trisomy 21. We demonstrate overactivity of insulin-like growth factor (IGF) signaling in authentic human DS-AMKL and in a DS-AMKL mouse model generated through retroviral insertional mutagenesis. Fetal but not adult megakaryocytic progenitors are dependent on this pathway. GATA1 restricts IGF-mediated activation of the E2F transcription network to coordinate proliferation and differentiation. Failure of a direct GATA1–E2F interaction in mutated GATA1s converges with overactive IGF signaling to promote cellular transformation of DS fetal progenitors, revealing a complex, fetal stage-specific regulatory network. Our study underscores context-dependent requirements during oncogenesis, and explains resistance to transformation of ostensibly similar adult progenitors.
Keywords: DS-AMKL, Down syndrome, GATA1, IGF signaling, IGF1R, megakaryoblastic leukemia
Leukemias in infants or young children—e.g., acute megakaryoblastic leukemia (AMKL) in Down syndrome (DS-AMKL; also known as myeloid leukemia in DS [ML-DS]), or juvenile myelomonocytic leukemia (JMML)—are distinct from adult leukemias. Growing evidence supports that these leukemias originate in utero from fetal hematopoietic cells (Chou et al. 2008; Klusmann et al. 2008; Tunstall-Pedoe et al. 2008). Thus, pathogenic and phenotypic differences may arise from intrinsic genetic programs in fetal hematopoietic stem and progenitor cells (HSPCs) and their response to fetal environmental stimuli, both of which may differ from those in adult HSPCs. However, little is understood regarding the specific dependency on intrinsic and extrinsic signaling pathways supporting self-renewal and expansion of fetal HSPCs, and how these pathways intersect with lineage-specific transcription factors (TFs) to coordinate proliferation and differentiation. Moreover, how these pathways provide a unique background for malignant transformation, specifically in infant leukemia, and why ostensibly similar adult HSPCs are resistant to transformation by certain oncogenes remains elusive. Better understanding of the cell type-specific interplay of signaling pathways and oncogenes forms a basis for developing novel targeted cancer therapies.
DS-AMKL is strictly restricted to neonates, infants, or young children with DS or trisomy 21 (Hasle 2001; Klusmann et al. 2008), thus providing a unique context in which to address these issues. Five percent to 10% of DS infants develop the related, antecedent transient leukemia (DS-TL, also known as transient myeloproliferative disorder [TMD]) (Pine et al. 2007). Although DS-TL resolves spontaneously in the majority of cases, 20%–30% of patients progress to DS-AMKL within the first 4 years of life (Klusmann et al. 2008). Acquired mutations in the hematopoietic TF GATA1, leading to expression of a shorter GATA1 variant (referred to as GATA1s) truncated at its N terminus, are consistently present in the affected cells of children with DS-AMKL and DS-TL (Wechsler et al. 2002; Klusmann et al. 2008).
GATA1 is essential for terminal erythroid and megakaryocytic differentiation (Orkin 1992). Using knock-in mice (Gata1ΔN and Gata1Δe2) that express Gata1s (referred to as Gata1s mice) exclusively, we previously identified a unique transient embryonic/fetal progenitor population present only from embryonic days 9.5–16.5 (E9.5–E16.5) of mouse development (Li et al. 2005). In these megakaryocytic progenitors (MPs), Gata1s dominantly induces their hyperproliferation, while allowing them to attain a relatively mature stage of differentiation. Adult Gata1s mice exhibit normal hematopoiesis (Li et al. 2005), an observation that underscores differences in the requirement for Gata1-mediated regulation of proliferation in fetal and adult megakaryopoiesis. However, in both humans and mice, germline GATA1s mutation alone is not associated with leukemia in the absence of trisomy 21 (Hollanda et al. 2006), consistent with a stringent requirement for trisomy 21 in cellular transformation. We hypothesize that the trisomy 21 genetic background may perturb the normal signaling network in affected cells, which contributes to leukemogenesis in cooperation with GATA1s. Identification of pathways that synergize with GATA1s should provide unique insights into the regulation of normal and malignant fetal hematopoiesis, and those pathways aberrantly controlled in the trisomy 21 cells.
Intriguingly, Zhang et al. (Zhang and Lodish 2004; Chou and Lodish 2010) showed recently that hepatic stromal cells, uniquely found in the fetal liver (FL), support hematopoietic stem cell (HSC) expansion by secreting insulin-like growth factor 2 (Igf2). IGFs (IGF1 and IGF2) are critical regulators of energy metabolism, cell growth, proliferation, and apoptosis (for review, see Pollak 2008). Maternally imprinted IGF2 is essential for embryonic and fetal but not adult growth and development (DeChiara et al. 1990, 1991; Baker et al. 1993). The mitogenic and anti-apoptotic activity of IGF1 and IGF2 is mediated principally by the IGF1 receptor (IGF1R), a cell surface tyrosine kinase signaling molecule, which plays a role in diverse human cancers, including acute myeloid leukemia (AML) (for review, see Pollak 2008). This suggests that the FL niche may support fetal hematopoiesis through IGF/IGF1R signaling. However, whether fetal HSPCs and infant leukemias arising from fetal HSPCs are specifically dependent on IGF2 and IGF/IGF1R signaling remains elusive.
Here, we demonstrate that IGF signaling is constitutively activated in DS-AMKL and DS-TL with a fetal origin. We further show that dependency on this pathway distinguishes fetal MPs from their adult counterparts. Full-length GATA1 serves as a molecular “brake” to intersect with the mitogenic IGF/IGF1R/mTOR pathway, and likely restricts proliferation of fetal MPs through a direct inhibitory interaction with E2F TFs, which are activated by IGF signaling. GATA1s mutation contributes to leukemogenesis by perturbing this function of GATA1, which may be aggravated further by excessive IGF signaling under the trisomy 21 genetic background. Our findings reveal a unique, developmental stage-specific interplay of a lineage-specific TF and a major mitogenic pathway, and how perturbation of both players in fetal HSPCs converges in leukemia. Our findings also explain how ostensibly similar adult cells are resistant to transformation, and provide an entry into deciphering the complex regulatory network between cell type-specific signaling pathways and oncogenes in cancer cells.
Results
IGF/IGF1R signaling is critical for proliferation and survival of GATA1s mutant AMKL cells
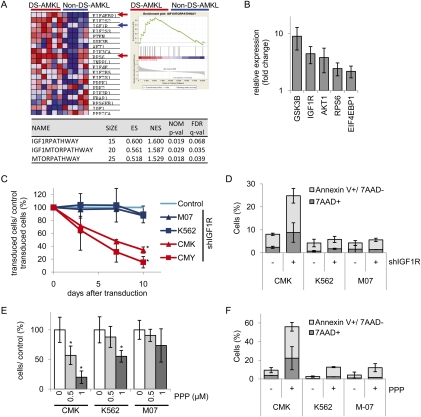
Since DS-TL and DS-AMKL arise from fetal progenitors, we reasoned that the FL microenvironment might be necessary to support development of this type of leukemia. The FL niche secrets Igf2, which supports fetal HSC expansion through the IGF/IGF1R pathway (Zhang and Lodish 2004; Chou and Lodish 2010). To assess the role of IGF signaling in the pathogenesis of DS-AMKL arising from fetal progenitors (Li et al. 2005; Klusmann et al. 2008), we first compared mRNA expression levels of IGF1R in sorted primary human DS-AMKL blasts (n = 6) in comparison with non-DS-AMKL blasts (n = 5; cytogenetic data were available for DS-AMKL [n = 4] and non-DS-AMKL [n = 3] patients) (Supplemental Table S1). We observed that IGF1R expression was significantly greater in DS-AMKL blasts compared with non-DS-AMKL blasts (Fig. 1A,B), and in the DS-AMKL cell line CMK compared with the non-DS-AMKL cell line (M-07) and the erythroleukemia cell line K562 (Supplemental Fig. S1A). By Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005), we found that gene sets related to IGF/IGF1R signaling (IGF1RPATHWAY, IGF1MTORPATHWAY, and MTORPATHWAY) from the c2 collection (curated gene sets) of the Molecular Signatures Database (MSigDB) (Subramanian et al. 2005) are enriched in DS-AMKL blasts compared with non-DS-AMKL blasts (Fig. 1A). The negative regulator of PI3K signaling, PTEN, is also up-regulated in DS-AMKL cells compared with non-DS-AMKL cells, consistent with the previously shown requirement of PTEN expression for the sensitivity of some cancer cells to upstream IGF and insulin signaling (Lackey et al. 2007). The up-regulation of key genes within the IGF1–mTOR pathway in primary DS-AMKL blasts was validated by quantitative RT–PCR (qRT–PCR) (Fig. 1B).
Figure 1.
IGF/IGF1R signaling is critical for proliferation and survival of human DS-AMKL cell lines. (A) GSEA showing enrichment of IGF1RPATHWAY, IGF1MTORPATHWAY, and MTORPATHWAY gene sets from the c2 collection of the MSigDB in leukemic blasts from patients with DS-AMKL (n = 6) in comparison with those with non-DS-AMKL (n = 5). The GSEA enrichment plot (right) and heat map (left) of the IGF1MTORPATHWAY gene set are shown. (Blue arrow) Showing higher levels of IGF1R expression in DS-AMKL samples; (red arrows) key genes involved in mTOR signaling. (Bottom) Statistics for all gene sets. (ES) Enrichment score; (NES) normalized enrichment score; (NOM p-val) nominal P-value; (FDR) false discovery rate. Gene sets with P < 0.05 and FDR q < 0.25 are considered as significant (Subramanian et al. 2005). (B) Relative expression of key genes in the IGF1MTORPATHWAY gene set in DS-AMKL blasts (n = 3) and non-DS-AMKL blasts (n = 3) analyzed by qRT–PCR. The expression in DS-AMKL is presented in relation to the expression in non-DS-AMKL (1). (C) Growth curves of shIGF1R and empty vector (LMPIG or Tripz)-transduced cell lines (CMK, CMY, M-07, and K562) grown in liquid culture. GFP or RFP expression (i.e., transduced cells) was assessed by flow cytometric analysis. The percentage of positive cells in relation to empty vector controls and the starting point (100%) from pooled data of two independent experiments are shown. (B,C) Error bars represent ±standard deviation (SD); (*) P < 0.05. (D) Percentage of Annexin V+/7-AAD− apoptotic and 7-AAD+ dead CMK, K562, and M-07 cells 72 h after induction of shIGF1R expression using Dox (500 ng/mL) in comparison with noninduced control cells. Data from n = 2 independent experiments are shown as means ±SD. (E) Number of CMK, M-07, and K562 cells 48 h after addition of the indicated concentrations of the IGF1R inhibitor PPP in relation to the untreated control (DMSO; 100%). Error bars represent ±SD of triplicates; (*) P < 0.05. (F) Percentage of Annexin V+/7-AAD− apoptotic and 7-AAD+ dead CMK, K562, and M-07 cells 48 h after addition of PPP (0.5 μM; +) in comparison with the untreated control cells (DMSO; −). Data from n = 2 independent experiments are shown as means ± SD.
To test dependency of DS-AMKL cells on IGF/IGF1R signaling for their proliferation and survival, we used shRNA (shIGF1R) to knock down IGF1R expression in the human DS-AMKL cell lines CMK and CMY. shRNA-mediated IGF1R knockdown, confirmed by qRT–PCR and Western blot (Supplemental Fig. S1B,C), led to a dramatic reduction of the percentage of transduced CMK and CMY cells, while the percentage of empty vector-transduced control cells remained constant (Fig. 1C). Additionally, we noted a larger number of annexin V+/7-AAD− apoptotic and 7-AAD+ dead CMK-shIGF1R cells and a reduction of actively cycling cells compared with the control CMK cells (Fig. 1D; Supplemental Fig. S1D). In contrast, only a modest effect of shIGF1R was observed in control cell lines (K562 and M-07) (Fig. 1C,D; Supplemental Fig. S1D).
Upon treatment with a specific IGF1R inhibitor, Picropodophyllin (PPP), we observed a remarkable reduction in cell growth and a high percentage of apoptotic and dead cells for the DS-AMKL cell lines CMK and CMY (Fig. 1E,F; Supplemental Fig. S1E,F), excluding the possibility of shRNA off-target effects. In contrast, the control cell lines K562 and M-07 showed substantially less responses to PPP treatment (Fig. 1E,F). In K562 cells, the observed level of response is consistent with previous findings using a different IGF1R inhibitor (AG1024) (Lakshmikuttyamma et al. 2008).
Taken together, these experiments indicate that IGF/IGF1R signaling is activated in human primary DS-AMKL blasts, and is critical for the survival and proliferation of DS-AMKL cell lines.
Mitogenic activity of IGF/IGF1R signaling is mediated through mTOR
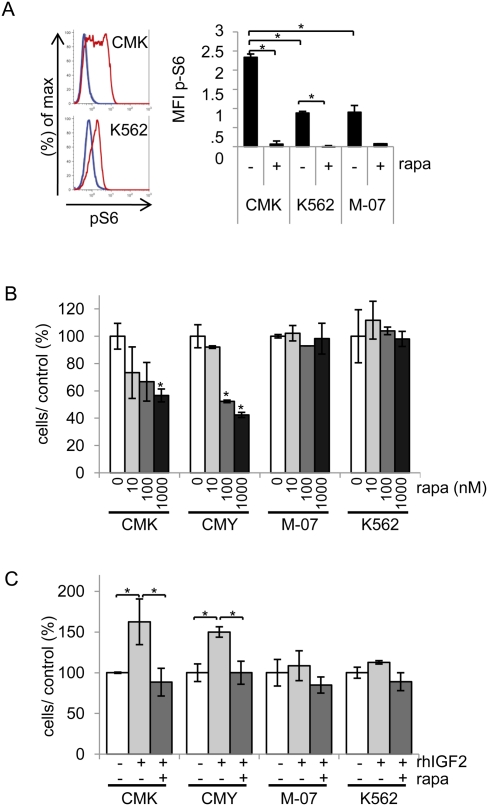
Downstream intracellular signaling molecules of IGF1R include IRS proteins, PI3K, AKT, mTOR, S6 kinase, and MAPK (for review, see Pollak 2008). In patient samples, GSEA identified IGF/mTOR and mTOR pathway gene sets to be activated in primary DS-AMKL blasts compared with non-DS-AMKL cells (Fig. 1A), suggesting that IGF/IGF1R signaling may employ the mTOR pathway to induce proliferation and survival of DS-AMKL blasts. Consistent with this hypothesis, we found that CMK cells exhibited high levels (higher than in K562 and M-07 cells) of intracellular phosphorylated S6 protein (p-S6), a downstream target of mTOR, whereas mTOR inhibition by treatment with rapamycin led to a marked reduction of p-S6 in these cells (Fig. 2A), confirming activation of the mTOR/p70S6K pathway. Concomitantly, the numbers of CMK and CMY cells were reduced gradually with increasing concentrations of rapamycin, whereas growth of control cell lines (M-07 and K562) was less sensitive (Fig. 2B; Supplemental Fig. S1G). The weaker sensitivity of K562 cells to rapamycin observed here is consistent with a previous study (Mohi et al. 2004). Conversely, treatment of CMK and CMY with IGF2 significantly enhanced their growth, whereas simultaneous addition of rapamycin to the culture medium inhibited this proproliferative effect (Fig. 2C). In contrast, IGF2 alone or IGF2 plus rapamycin had less effect on control M-07 and K562 cells. Thus, the IGF/IGF1R/mTOR signaling pathway is activated in DS-AMKL cell lines, which are hypersensitive to both further activation and inhibition of this pathway.
Figure 2.
IGF/IGF1R signaling promotes growth of DS-AMKL cells via the mTOR pathway. (A) p-S6 expression in CMK, M-07, and K562 cells with (+) or without (−) addition of rapamycin (rapa, 100 nM). (Left) Representative flow cytometry profiles for CMK and K562 cells from one of two independent experiments. (Gray line) IgG control; (red line) p-S6-stained untreated cells; (blue line) p-S6-stained rapamycin-treated cells. (Right) Diagram and statistics showing the difference of the mean fluorescence intensity (MFI) of p-S6 expression in comparison with the IgG control. Error bars represent ±SD of pooled data from two independent experiments. (*) P < 0.05. (B,C) Number of CMK, CMY, M-07, and K562 cells 96 h after addition of the indicated concentrations of rapamycin (in nanomolar) (B), or 48 h after addition of IGF2 (rhIGF: recombinant human IGF, 500 ng/mL) or IGF2 (500 ng/mL) plus rapamycin (rapa; 100 nM) (C), in relation to the untreated control (DMSO; 100%). Error bars represent ±SD of replicates. (*) P < 0.05.
Primary human DS-TL and DS-AMKL blasts are hypersensitive to IGF signaling activation and inhibition
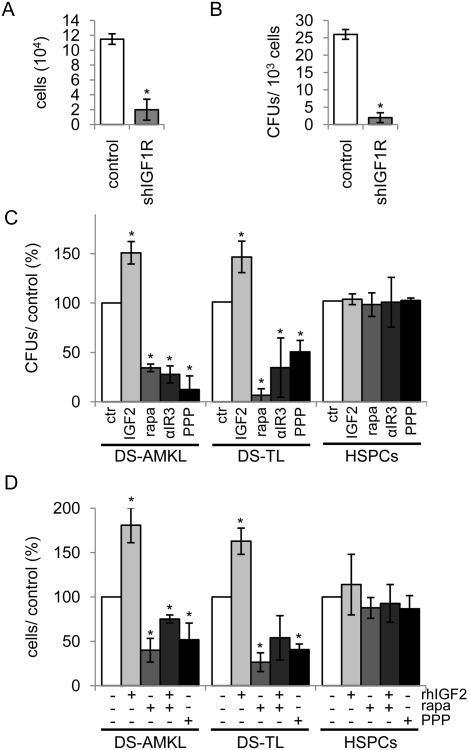
Although studies in primary DS-AMKL/DS-TL blasts are particularly challenging, they are crucial in the validation of effects observed in cell lines. To confirm the role of IGF signaling in primary human leukemic blasts, we took advantage of DS-TL blasts from one patient, which were obtained in high numbers after leukapheresis, and transduced them with shIGF1R. Since DS-TL antecedes DS-AMKL, and both originate from the same fetal target cell, active signaling pathways of their cells of origin should be shared in both entities. Colony formation and proliferation of DS-TL blasts were greatly impaired upon IGF1R knockdown in comparison with the empty vector-transduced cells (Fig. 3A,B). In a larger set of primary human DS-AMKL and DS-TL samples, IGF2 significantly enhanced their colony formation (Fig. 3C) and in vitro proliferation (Fig. 3D), whereas addition of rapamycin, IGF1R inhibitor PPP, or IGF1R antibody αIR3 exhibited the opposite effect. Simultaneous addition of rapamycin and IGF2 blocked the proproliferative effect of IGF2 (Fig. 3D). In contrast, adult CD34+ HSPCs showed only a modest response to IGF2, PPP, rapamycin, and αIR3 treatments (Fig. 3C,D). The response of normal CD34+ HSPCs to rapamycin and IGF1R inhibition is consistent with previous reports (Ratajczak et al. 1994; Recher et al. 2005). As another set of the control, we found that leukemic blasts from patients with non-DS-AMKL, AML FAB M2, and M5 did not exhibit a similar response to IGF2 and rapamycin (Supplemental Fig. S2A). Lastly, inhibition of the IGF1R activity by PPP also led to an enhanced rate of apoptosis of DS-AMKL and DS-TL cells (similar to DS-AMKL cell lines), compared with CD34+ HSPCs (Supplemental Fig. S2B).
Figure 3.
Primary human DS-AMKL/DS-TL cells from patients are dependent on IGF/IGF1R/mTOR signaling for their growth. (A) Number of shIGF1R and empty vector (LMPIG; control)-transduced DS-TL leukemic blasts after 96 h grown in liquid culture. (B) Number of CFUs formed in the colony-forming assay of shIGF1R and empty vector (LMPIG; control)-transduced DS-TL leukemic blasts. (A,B) Error bars represent ±SD of two independent experiments. (*) P < 0.05. (C) Number of CFUs formed in the colony-forming assay of DS-AMKL (n = 2) and DS-TL (n = 3) blasts, as well as HSPCs (n = 2) in the presence of rapamycin (rapa; 100 nM), IGF1R inhibitor PPP (0.5 μM), IGF2 (200 ng/mL), or IGF1R antibody αIR3 (1 μg/mL) in relation to the untreated control cells (ctr, 100%). (D) Number of blasts from patients with DS-AMKL (n = 3), DS-TL (n = 4), and of HSPCs from healthy individuals (n = 2) grown in liquid culture (all in triplicate) 72 h after addition of IGF1R inhibitor PPP (0.5 μM), rapamycin (rapa; 100 nM), and/or IGF2 (200 ng/mL) in relation to the untreated control (100%). (C,D) Error bars represent ±SD. (*) P < 0.05.
In summary, we established a central role of IGF/IGF1R/mTOR signaling in the growth and survival of primary human DS-AMKL and DS-TL cells.
A DS-AMKL mouse model shows sensitivity to IGF1R inhibition
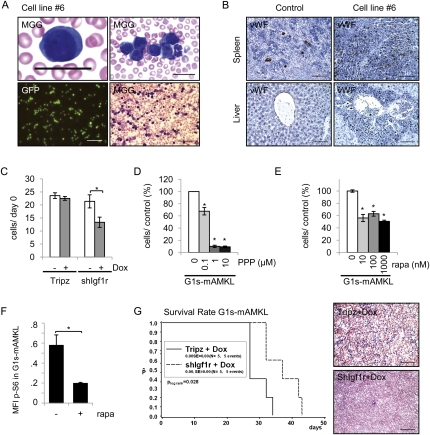
To validate the oncogenic mechanism implicated from in vitro experiments, we tested the role of IGF signaling in a DS-AMKL mouse model in vivo. Previously, we found that three existing DS mouse models—Tc1, Ts65Dn, and Ts1Cje—do not provide a cellular context in which Gata1s cooperates with trisomy 21 for generation of DS-TL or DS-AMKL (Supplemental Fig. S3; Alford et al. 2010; data not shown). To overcome this obstacle, we used a different strategy by conducting retroviral insertional mutagenesis in Gata1s mutant FL MPs using the MSCV-ires-Gfp (MIGR1) retrovirus, and by selecting for immortalized cell lines in vitro in the presence of thrombopoietin (Tpo), followed by transplantation into Rag2−/− mice (J-H Klusmann, SH Orkin, and Z Li, unpubl.). Although the majority of the in vitro generated cell lines were not leukemogenic in vivo, cell line #6—which contains retroviral insertion sites (RISs) at the Evi1, Plag1, and Il15 loci (Supplemental Fig. S4A [up-regulation of Evi1 and Plag1 were confirmed by RT–PCR], B)—gave rise to vWF+ AMKL in all recipient mice (n = 6) (Fig. 4A,B; Supplemental Fig. S4C).
Figure 4.
A DS-AMKL mouse model generated by retroviral insertional mutagenesis shows sensitivity to Igf1r inhibition. (A) May-Grünwald-Giemsa (MGG)-stained blood smears from one representative Rag2−/− recipient mouse transplanted with cell line #6 (G1s-mAMKL) (bars: top panels, 20 μm; bottom right panel, 50 μm). The bottom left panel shows GFP expression from the leukemic cells (bar, 200 μm). (B) Immunohistochemical staining of liver and spleen sections showing infiltration of von Willebrand factor (vWF)-positive (brown) leukemic blasts in one representative Rag2−/− mouse transplanted with cell line #6 in comparison with a nontransplanted Rag2−/− mouse (control). Sections were counterstained with hematoxylin. Bars, 100 μm. (C) Number of G1s-mAMKL cells transduced with shRNA against Igf1r (shIgf1r) under the control of a Dox-inducible promoter (or the empty vector Tripz) with and without addition of Dox (100 ng/mL). The data were collected 7 d after addition of Dox, and are in relation to day 0. Error bars represent ±SD of pooled data from two independent experiments. (*) P < 0.05. (D,E) Number of G1s-mAMKL cells 48 h after addition of the indicated concentrations of Igf1r inhibitor PPP (D), or 96 h after addition of the indicated concentrations of rapamycin (rapa) (E) in relation to the control (100%). Error bars represent ±SD of replicates. (*) P < 0.05. (F) p-S6 expression in the G1s-mAMKL blasts with (+) and without (−) addition of rapamycin (rapa; 100 nM). Diagram and statistics showing the difference of the mean fluorescence intensity (MFI) in comparison with the IgG control. Error bars represent ±SD of pooled data from two independent experiments. (*) P < 0.05. (G) Kaplan-Meier survival analysis (left) and MGG staining of representative blood smears (right, at day 28) from Dox-treated Rag2−/− mice transplanted with G1s-mAMKL blasts transduced with shIgf1r (n = 5) or the empty vector (Tripz; n = 5). Plog-rank = 0.028. Bars, 100 μm.
Since these murine AMKL cells [referred to as T6(6) cells] have a fetal origin and carry the Gata1s mutation, two hallmarks of human DS-AMKL, we investigated whether their transcriptome recapitulates that of human DS-AMKL globally. We analyzed the gene expression profile of flow-sorted GFP+ T6(6) leukemic blasts. Megakaryocytic and erythroid genes were up-regulated in the T6(6) cells (Supplemental Fig. S4D), compared with FL MPs (Li et al. 2005), a finding typical of DS-AMKL (Bourquin et al. 2006; Ge et al. 2006). Next, we compared the gene expression signature of murine T6(6) AMKL to that of human DS-AMKL. We first identified the top 500 up-regulated and down-regulated genes in DS-AMKL compared with non-DS-AMKL (ranked ordered gene list using a signal-to-noise statistic) (Subramanian et al. 2005). Most importantly, GSEA demonstrated strong enrichment of the top 500 up-regulated human DS-AMKL genes in T6(6) blasts, whereas the top 500 down-regulated genes were negatively enriched (Supplemental Fig. S5A,B [enrichment plot, heat map, and statistics], C,D [qRT–PCR validation of selected genes]). These data demonstrate that T6(6) leukemic blasts are murine Gata1s AMKL cells that resemble human DS-AMKL morphologically (Fig. 4A) and recapitulate gene expression programs found in human DS-AMKL. Hereafter, these T6(6) cells are referred to as G1s-mAMKL cells.
Knockdown of Igf1r (Fig. 4C), as confirmed by qRT–PCR and Western blot (Supplemental Fig. S6A,B), and treatment with the Igf1r inhibitor PPP (Fig. 4D) or rapamycin (Fig. 4E) markedly reduced proliferation of G1s-mAMKL cells in vitro. This indicates a similar in vitro response of these murine blasts to IGF1R/mTOR inhibition as human DS-AMKL and DS-TL cells. Concomitantly, S6 protein was phosphorylated in these cells, and was dephosphorylated upon rapamycin treatment (Fig. 4F). In contrast, murine leukemic blasts from the spleen and bone marrow (BM) of recipient mice transplanted with Mll-Af9-transduced BM progenitors (Krivtsov et al. 2006) were insensitive to PPP treatment in vitro (Supplemental Fig. S6C). These Mll-Af9 blasts are of an adult origin, and do not carry the Gata1s mutation.
Next, we tested the requirement of Igf/Igf1r signaling for leukemogenesis of G1s-mAMKL cells in vivo using a doxycycline (Dox)-inducible shRNA expression system (Tripz). Dox-treated mice transplanted with G1s-mAMKL cells with activated shIgf1r exhibited a significant delay in the onset of leukemia and improved survival, as estimated by Kaplan-Meier analysis, in comparison with Dox-treated control mice (transplanted with the empty vector [Tripz]-transduced G1s-mAMKL cells) (Fig. 4G). Thus, the proliferation of G1s-mAMKL cells in vitro, as well as their engraftment and establishment of leukemia in vivo, is dependent on the Igf/Igf1r signaling pathway. Further studies are required to elucidate the mechanism by which overexpression of Plag1 and Evi1, in concert with mutated Gata1s, induces an expression signature similar to that of human DS-AMKL.
Mitogenic activity of IGF/IGF1R signaling is mediated through activation of E2F target genes
To this point, our data identify the IGF/IGF1R/mTOR pathway as a major mitogenic pathway for human DS-AMKL/DS-TL and G1s-mAMKL cells. To further define IGF-regulated gene sets downstream from mTOR, we performed global gene expression profiling of G1s-mAMKL cells with and without Igf1r knockdown, and performed GSEA. Intriguingly, compared with G1s-mAMKL cells with Igf1r knockdown, all of the top 20 gene sets of the c3 collection (MSigDB; motifs) enriched in the control G1s-mAMKL cells (i.e., no Igf1r knockdown) were related to targets of E2F TFs (Supplemental Table S2).
The result was further validated with another gene set (REN_E2F1_TARGETS) from the c2 collection (MSigDB; curated gene sets) that was generated by a chromatin immunoprecipiationa (ChIP)–chip experiment (Supplemental Fig. S7A; Ren et al. 2002). Down-regulation of selected E2F target genes in G1s-mAMKL blasts upon Igf1r knockdown, as well as in CMK cells treated with the IGF1R inhibitor PPP, was further confirmed by qRT–PCR (Supplemental Fig. S7B,C). Thus, these studies demonstrate that, in both murine and human AMKL cells expressing GATA1s, the highly activated mitogenic IGF/IGF1R/mTOR pathway is exerted at the transcriptional level by activation of E2F target genes.
GATA1s mutant protein fails to repress E2F target genes
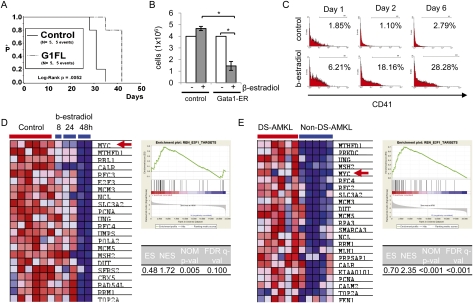
Next, we sought to determine the role of GATA1s in disease pathogenesis in concert with activated IGF signaling. Expression of full-length Gata1 in G1s-mAMKL cells led to delayed leukemia development in vivo (Fig. 5A). Concomitantly, upon synchronized restoration of full-length Gata1 activity using a β-estradiol-inducible system (full-length Gata1 fused to the estrogen receptor [G1ER]) (Rylski et al. 2003), G1s-mAMKL cells proliferated much slower (Fig. 5B) and differentiated along the megakaryocytic lineage (Fig. 5C). These results demonstrate that the G1s-mAMKL phenotype is strongly dependent on exclusive Gata1s expression, similar to GATA1s dependency of human DS-AMKL cells (Supplemental Fig. S8A,B; Xu et al. 2003).
Figure 5.
GATA1s mutant protein contributes to megakaryocytic leukemia through derepression of E2F target genes. (A) Kaplan-Meier survival analysis of Rag2−/− mice transplanted with G1s-mAMKL cells transduced with full-length Gata1 (G1FL; n = 5) or the empty vector (control; n = 5). (B) Cell counts after 48 h of β-estradiol-treated and untreated G1-ER-transduced and empty vector (MSCVpuro, control)-transduced G1s-mAMKL cells, respectively. Error bars represent ±SD of replicates. (*) P < 0.05. (C) Flow cytometric analysis for CD41 expression of β-estradiol-treated and untreated (control) G1ER-transduced G1s-mAMKL cells 1 d, 2 d, and 6 d after treatment. (D,E) GSEAs of E2F1 target genes previously identified by ChIP–chip analysis (Ren et al. 2002), showing their significant down-regulation in a time-dependent manner after full-length Gata1 induction in G1s-mAMKL cells (D), as well as significant up-regulation in DS-AMKL patient samples (n = 6) in comparison with non-DS-AMKL samples (n = 5) (E). For each panel a heat map showing the leading edge subset is on the left, and enrichment plot and statistics are on the right. MYC is indicated by a red arrow.
Next, we generated global gene expression profiles of G1ER-transduced G1s-mAMKL cells with or without β-estradiol treatment after 0, 8, 24, and 48 h (Supplemental Fig. S9A). Interestingly, among the top 20 gene sets from the c3 collection (MSigDB; motifs) enriched in the control cells (i.e., without full-length Gata1 induction), 19 gene sets were related to targets of E2F factors (Supplemental Fig. S9A; Supplemental Table S3). The E2F1 gene set generated by a ChIP–chip experiment (REN_E2F1_TARGETS) was also significantly enriched (Fig. 5D; Ren et al. 2002). The down-regulation of selected target genes within this gene set was validated by qRT–PCR (Supplemental Fig. S9B).
We validated the repression of E2F target genes as a normal function of full-length GATA1 by analyzing three independent global gene expression data sets using GSEA: Gata1s in comparison with wild-type FL-derived megakaryocytes (Supplemental Fig. S9D; Supplemental Table S4; Li et al. 2005), leukemic blasts from DS-AMKL patients in comparison with those from non-DS-AMKL (Fig. 5E; Supplemental Fig. S10A,B; Supplemental Table S5), and AMKL cell lines with (CMK) and without (UT-7) GATA1s mutation (Supplemental Fig. S11). In all cases, E2F targets were significantly enriched in GATA1s mutant cells.
Taken together, these data strongly suggest that full-length GATA1 protein is both necessary and sufficient to repress E2F target genes.
GATA1 interacts physically with E2F1 in megakaryoblastic cells
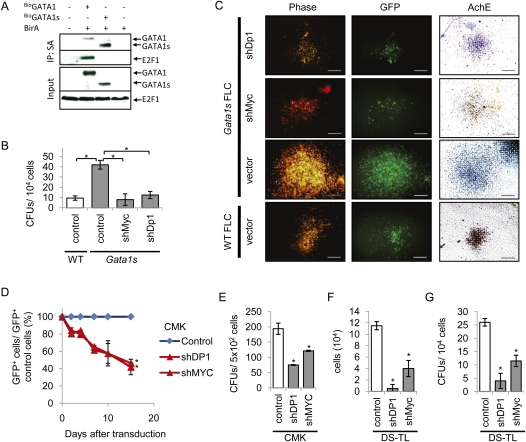
As the mRNA levels of activating E2F TFs (E2F1–3) were not significantly down-regulated in G1ER-transduced G1s-mAMKL cells treated with β-estradiol (Supplemental Fig. S9C), or up-regulated in DS-AMKL cells compared with non-DS-AMKL cells (Supplemental Fig. S10C), we conclude that GATA1 does not negatively regulate E2Fs through a direct transcriptional repression mechanism. Repression of E2F activity by GATA1 can also be caused by a direct inhibitory protein–protein interaction between GATA1 and E2F TFs. We tested this possibility in CMK cells. E2F1 is highly expressed in this cell line (Fig. 6A). We analyzed a potential direct protein–protein interaction between GATA1 and E2F1, by performing coimmunoprecipitation (co-IP) using a biotinylation approach (Wang et al. 2006), in CMK cells. GATA1 fused to a short peptide tag that serves as a substrate for biotinylation (BioGATA1) was ectopically expressed in CMK cells, along with the Escherichia coli biotin ligase BirA, through a retroviral expression system. Using an anti-E2F1-antibody, we found that GATA1, but not GATA1s, interacts physically with E2F1 (Fig. 6A), suggesting that the failure of GATA1s to repress E2F activity may be caused by this failure in direct interaction.
Figure 6.
The GATA1–E2F1 interaction is lost in GATA1s cells, and knockdown of DP1 and MYC reverts the GATA1s-mediated hyperproliferative phenotype. (A) Co-IP of bioGATA1/bioGATA1s and E2F1 in CMK cells ectopically expressing bioGATA1/bioGATA1s and BirA using Streptavidin (SA) beads and an anti-E2F1 antibody. (B,C) Number of CFU-MKs (B) and representative microscopic images (C) of colony-forming assays of shDp1-transduced, shMyc-transduced, and empty vector (LMPIG; control)-transduced wild-type (WT) and Gata1s FL progenitors. (B) Error bars represent ±SD of replicates. (*) P < 0.05. (C) Bars, 200 μm. (D) Growth curves of shDP1-transduced, shMYC-transduced, and empty vector (LMPIG, control)-transduced CMK cells grown in liquid culture. The percentage of GFP+ cells was calculated in relation to empty vector controls and the starting point (100%). Data from n = 2 independent experiments are shown as means ±SD. (E) Number of CFUs formed in the colony-forming assay of shDP1-transduced, shMYC-transduced, and empty vector (LMPIG; control)-transduced CMK cells. Error bars represent ±SD of replicates. (*) P < 0.05 (in comparison with empty vector control). (F,G) Number of cells after 96 h grown in liquid culture (F), and number of CFUs formed in the colony-forming assay (G) of shDP1-transduced, shMYC-transduced, and empty vector (LMPIG; control)-transduced primary DS-TL leukemic blasts. Error bars represent ±SD of replicates. (*) P < 0.05 (in comparison with empty vector control).
Knockdown of Dp1 and Myc reverts the Gata1s hyperproliferative phenotype
We next determined in functional genetic studies if failure of Gata1s to repress E2f function contributes to the hyperproliferative phenotype of Gata1s FL MPs. To inhibit E2f function broadly, we used shRNA-mediated knockdown of Dp1 (shDp1) (Supplemental Fig. S12A, Western blot confirming knockdown), the essential dimerization partner of E2f1–6 for DNA binding and activation of gene expression (for review, see Rowland and Bernards 2006). The number and size of megakaryocyte colony-forming units (CFU-MKs) from shDp1-transduced Gata1s FL progenitors were greatly reduced to the level seen with wild-type MPs (Fig. 6B,C). In addition, Gata1s FL MPs cultured in the presence of Tpo ceased to proliferate upon Dp1 knockdown (Supplemental Fig. S12B). In contrast, wild-type FL MPs were less sensitive to knockdown of Dp1 (Supplemental Fig. S12B). The colony assay was validated by another shRNA in a lentiviral system (LKO.1) (Supplemental Fig. S12A,C). Consistent with this, proliferation and colony-forming capacity of human DS-AMKL CMK cells were also arrested upon knockdown of DP1 (shDP1) (Fig. 6D,E; Supplemental Fig. S12D, Western blot), thereby underscoring the role of E2F factors in the hyperproliferation of GATA1s mutant cells.
The Myc (c-myc) proto-oncogene was among the top down-regulated genes upon expression of full-length Gata1 (Fig. 5D), as well as in DS-AMKL blasts compared with non-DS-AMKL blasts (Fig. 5E), and is included in most E2F-related gene sets used. shRNA-mediated depletion of MYC (LMPIG-shMyc, knockdown confirmed by Western blot) (Supplemental Fig. S12A,D) in Gata1s FL MPs, as well as in CMK cells, resulted in a similar inhibition of the colony-forming capacity and proliferation as those from DP1 knockdown, whereas wild-type FL MPs were largely unaffected upon Myc knockdown (Fig. 6B–E; Supplemental Fig. S12B,C).
In accordance with these findings, the proliferation and colony-forming capacity of primary human DS-TL blasts were markedly reduced in vitro upon DP1 or MYC knockdown (Fig. 6F,G). Thus, our experiments establish that repression of E2F activity (through DP1 knockdown) or knockdown of MYC abrogates GATA1s-mediated hyperproliferation of FL MPs, the DS-AMKL cell line, and primary DS-TL blasts, and therefore rescues the GATA1s phenotype.
GATA1 intersects with IGF/IGF1R signaling by restricting IGF-stimulated proliferation of fetal progenitors
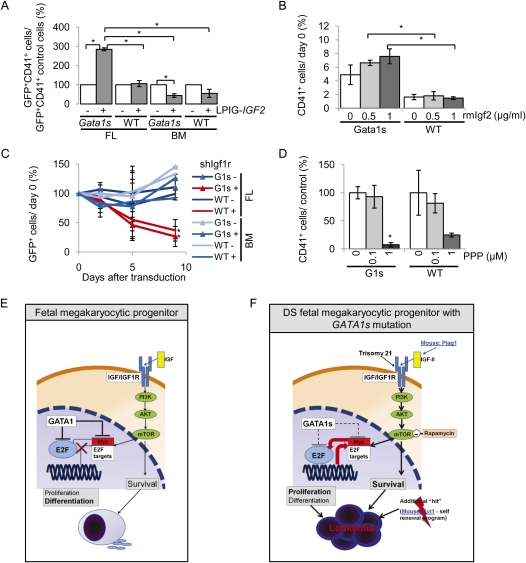
We showed that IGF/IGF1R signaling exerts its mitogenic effect by activating E2F target genes, whereas GATA1s is insufficient to repress these genes. To functionally explore the intersection between GATA1 and mitogenic IGF/IGF1R signaling, we constitutively activated IGF/IGF1R signaling through retroviral expression of the human IGF2 gene in primary FL or BM MPs derived from Gata1s or wild-type animals. This led to a marked expansion of CD41+ Gata1s fetal MPs, but had negligible effects on wild-type fetal MPs or adult MPs (both Gata1s and wild type) (Fig. 7A). To further validate this finding, we cultured Gata1s or wild-type FL progenitors in the presence of Tpo with or without addition of purified mouse Igf2. Gata1s CD41+ fetal MPs exhibited an even higher level of hyperproliferation with increasing concentrations of Igf2 (Fig. 7B). In contrast, addition of the same amounts of Igf2 to the culture also had a negligible effect on wild-type fetal MPs. These data suggest that full-length GATA1 intersects with IGF signaling to restrict IGF signaling-stimulated proliferation of fetal MPs, whereas GATA1s is defective in this function.
Figure 7.
GATA1 full-length protein restricts IGF signaling-induced proliferation of fetal megakaryocytic progenitors in a developmental stage-dependent manner. (A) Relative number of GFP+/CD41+ wild-type and Gata1s FL as well as BM-derived megakaryocytic cells grown in liquid culture in the presence of Tpo 96 h after transduction with human IGF2-expressing retrovirus (LPIG-IGF2; +) or the empty vector as the control (LPIG; −). Cell numbers are in relation to those of the control of each subgroup (control = 100%). Error bars represent ±SD of two independent experiments. (*) P < 0.05. (B) Fold increase (at day 2) of GFP+/CD41+ wild-type and Gata1s FL-derived megakaryocytic cells in liquid culture in the presence of Tpo, treated with indicated concentrations of recombinant mouse Igf2 (rmIgf2), in comparison with the starting point (at day 0) of each cell type. Error bars represent ±SD of replicates. (*) P < 0.05. (C) Growth curves of GFP+ shIgf1r (+)-transduced or empty vector (LMPIG, −)-transduced wild-type and Gata1s (G1s) FL-derived as well as BM-derived megakaryocytic cells grown in liquid culture in the presence of Tpo. The percentage of GFP+ cells was calculated in relation to the starting point (day 0 = 100%). Data from n = 3 independent experiments are shown as means ± SD. (*) P < 0.05 (in comparison with empty vector). (D) Number of Gata1s (G1s) and wild-type CD41+ FL-derived megakaryocytic cells 48 h after addition of indicated concentrations of Igf1r inhibitor PPP in relation to the untreated control (DMSO; 100%). Error bars represent ±SD from two independent samples. (*) P < 0.05. (E,F) Proposed model for the development of DS-TL and DS-AMKL. (E) Full-length GATA1 coordinates terminal megakaryocytic differentiation and proliferation of IGF1/IGF1R signaling-dependent fetal progenitors by repressing E2F activity. (F) Failure to repress E2F activity by GATA1s, along with a possible perturbation of IGF/IGF1R signaling by trisomy 21 in fetal progenitors, leads to their uncontrolled proliferation and increased survival, ultimately leading to leukemia in conjunction with additional mutations. In G1s-mAMKL, Plag1 overexpression may mimic the effect of trisomy 21 to constitutively activate IGF/IGF1R signaling, leading to leukemia in cooperation with Gata1s mutation and Evi1 overexpression.
Dependency on IGF/IGF1R signaling distinguishes fetal megakaryocytic progenitors from their adult counterparts
We showed previously that Gata1s has a developmental stage-selective effect, and fetal progenitors are specifically sensitive to this mutant protein (Li et al. 2005). To determine if the interplay of Gata1 and Igf/Igf1r signaling is also developmental stage-dependent, we examined the dependency of fetal and adult megakaryocytic cell proliferation on Igf/Igf1r signaling. We transduced FL and BM MPs from wild-type and Gata1s mice with the LMPIG-shIgf1r retrovirus. Knockdown of Igf/Igf1r signaling specifically impaired the growth of GFP+ FL MPs from both Gata1s and wild-type animals (Fig. 7C). In contrast, the percentage of GFP+ LMPIG-shIgf1r BM MPs was largely unaffected compared with empty vector-transduced controls. Similarly, when we treated fetal MPs with the IGF1R inhibitor PPP, we observed reduction of CD41+ FL MPs from both Gata1s and wild-type animals (Fig. 7D). These data thus demonstrated differential sensitivity of fetal and adult MPs, particularly Gata1s mutant fetal MPs, to inhibition of IGF/IGF1R signaling, and are consistent with our previous data showing greater sensitivity of leukemia cell lines with fetal origin and GATA1s mutation (e.g., CMK and CMY) to IGF1R knockdown than those with adult origin and wild-type GATA1 (e.g., K562 and M07) (Fig. 1C).
Discussion
Our study of a unique population of transient fetal progenitors, and the related fetal hematopoietic disorder DS-TL/DS-AMKL, has provided an entry into the largely unknown signaling network regulating proliferation and differentiation of normal FL progenitors and its role in the pathogenesis of infant leukemia. We address the vexing issue of how a specific oncogene may transform one cell type (i.e., fetal MPs), while another ostensibly similar cell type (i.e., adult MPs) remains unaffected, and discovered the specific requirement for IGF signaling in expansion of fetal, but not adult, MPs. Cross-species comparison of global gene expression data from murine models and primary human DS-TL and DS-AMKL samples revealed the convergence of IGF/IGF1R signaling, and consequences of GATA1s mutation in deregulation of the E2F transcription network. The IGF/IGF1R signaling pathway activates E2F target genes to exert its mitogenic potential in fetal MPs. Normally, proliferation is regulated and restricted by GATA1 by interacting directly with E2F. In contrast, failure of this direct GATA1–E2F interaction in mutated GATA1s leads to a failure to limit proliferation during fetal megakaryocytic differentiation. Thus, our findings point to a unique developmental stage-specific interplay of an activated signaling pathway and a lineage-determining TF during normal and malignant fetal hematopoiesis (Fig. 7E,F), and explain the resistance to transformation of ostensibly similar adult MPs. Deciphering the complex regulatory network between cell type-specific pathways and oncogenes in cancer cells is essential for understanding disease pathogenesis, and will guide us to develop more target cell-specific cancer therapies.
Dependency on IGF signaling distinguishes fetal from adult megakaryopoiesis
Pathogenic and phenotypic differences between leukemias in infants or young children (e.g., DS-AMKL or JMML) and adults may arise from phenotypic and functional differences between fetal and adult hematopoiesis. Differences in the immunophenotype, gene expression profile, and gene regulation between fetal and adult HSPCs have been described (Morrison et al. 1995; Ivanova et al. 2002; Hock et al. 2004; Kim et al. 2007). However, little is known regarding the specific dependency on extrinsic and intrinsic signaling pathways supporting self-renewal and expansion of fetal HSCs compared with adult HSCs. We demonstrated the specific dependency of a transient population of fetal progenitors on IGF/IGF1R signaling. SCF+DLK+ hepatic stromal cells, uniquely found in the FL, secrete Igf2 (Zhang and Lodish 2004; Chou and Lodish 2010), suggesting that the FL niche supports fetal hematopoiesis through the Igf/Igf1r pathway. In our retroviral insertional mutagenesis study, only immortalized cell lines with Plag1 overexpression engrafted in adult BM and gave rise to leukemia. As Igf2 is an established, direct target of Plag1 (Voz et al. 2004), we suspect that up-regulation of Igf2 by Plag1 overexpression constitutively activates the Igf/Igf1r pathway by providing an autocrine stimulation of the immortalized cells. One might speculate that this provides FL-derived cells, which are typically restricted to the FL microenvironment, the required stimuli to engraft in the BM microenvironment. We hypothesize that the spontaneous remission of DS-TL that coincides with the transition of hematopoiesis from FL to BM may also be explained by the dependency of DS-TL cells on FL-secreted IGF2. In addition, whether more advanced DS-AMKL blasts acquire additional mutation(s) within the IGF/IGF1R/mTOR cascade to support their growth in the BM microenvironment remains to be determined. Although our data only address developmental stage-specific dependency on IGF/IGF1R signaling at the progenitor level, it is possible that fetal and adult HSCs may also exhibit differential sensitivity to IGF/IGF1R signaling.
Interplay of GATA1 and mitogenic IGF signaling regulates fetal megakaryopoiesis
We showed that IGF/IGF1R signals through the mTOR pathway in human DS-AMKL blasts and in the murine model. It has been shown that activated mTOR phosphorylates 4E-BP1, thereby releasing eIF4E and activating cap-dependent translation of key cell cycle regulators, such as Myc, Cyclin D1, and p27kip1 (West et al. 1998). These cell cycle regulators in turn activate the E2F transcription network, explaining our observed down-regulation of E2F target genes upon Igf1r knockdown in G1s-mAMKL cells (Supplemental Fig. S7), and establishing the E2F network as the crucial downstream effector of mitogenic IGF/IGF1R/mTOR signaling.
Globally, our data demonstrate both activation of E2F target genes by IGF signaling and their repression by the full-length GATA1 protein in fetal megakaryoblastic cells. The core enrichment genes of the examined E2F target gene sets activated by IGF signaling overlap with those controlled by full-length Gata1 (Fig. 5D; Supplemental Fig. S7), thus pointing to conflicting control of the E2F transcription network by these two players. This finding is further supported by enhanced proliferation of Gata1s FL, but not wild-type FL, MPs upon exposure to Igf2 (Fig. 7A,B). Furthermore, our data suggest that repression of E2F targets by GATA1 is mediated by a direct interaction with E2F1, and not through transcriptional repression of activating E2F TFs. This interaction is attenuated in the absence of the N-terminal domain in truncated GATA1s. In erythropoiesis, Kadri et al. (2009) showed recently the interaction of GATA1 and E2F2 in a tricomplex with RB, leading to the inactivation of E2F2. This interaction is abrogated by mutated GATA1s (Kadri et al. 2009). Similar results were demonstrated for C/EBPα and E2F1 during granulopoiesis (Porse et al. 2001), suggesting that coordination of proliferation and terminal differentiation through repression of E2F activity appears to be a conserved feature of lineage-determining TFs.
MYC is a critical target gene controlled by GATA1 and E2F TFs, whereas inappropriate Myc expression is largely responsible for Gata1s-mediated uncoupling of proliferation and differentiation (Fig. 6). MYC can activate E2Fs and their target genes by inducing E2F gene expression (Sears et al. 1997) or by inducing CDK4 and CDK6, augmenting the phosphorylation and inactivation of RB (Mateyak et al. 1999). Myc itself is a target gene of E2Fs and GATA1 (Thalmeier et al. 1989; Rylski et al. 2003). It remains to be determined whether GATA1-mediated repression of E2F targets is caused by the inhibitory direct interaction between GATA1 and E2F TFs or indirectly by repressing MYC expression, or both.
Our data are best accommodated by a model in which wild-type GATA1 serves as a “brake” to restrict IGF signaling (“engine”)-stimulated proliferation in fetal MPs by repressing E2F target genes for cell cycle exit during megakaryocytic differentiation (Fig. 7E). Fetal HSPCs have high proliferation rates and are heavily dependent on IGF signaling (i.e., they require a larger “engine”). As a result, they also require a stronger “brake” to coordinate proliferation and cellular differentiation. Wild-type GATA1 protein functions as this strong “brake” during megakaryocyte differentiation. Even with high levels of Igf2, wild-type Gata1 is sufficient to restrict proliferation. In Gata1s fetal progenitors, the “brake” is impaired, and Gata1s mutant protein cannot induce cell cycle arrest due to derepression of E2F targets (Fig. 7F). As a result, elevated IGF signaling exhibits a stronger effect to enhance proliferation of Gata1s fetal MPs, possibly through increased activation of the E2F transcription network (i.e., a larger “engine” with an impaired “brake”). During pathogenesis, deregulation of these two players eventually converges at E2F targets, leading to development of leukemia (Fig. 7F). The importance of E2f1 repression during megakaryopoiesis is underscored by a previous in vivo study, in which megakaryocyte-specific, transgenic expression of E2f1 blocked terminal differentiation and caused proliferation of megakaryocytes (Guy et al. 1996). This phenotype partially resembles the Gata1s phenotype during fetal development.
Trisomy 21 may deregulate IGF signaling during fetal hematopoiesis
Our global gene expression analysis of primary leukemic cells demonstrated that gene sets related to IGF signaling are more enriched in pediatric DS-AMKL samples than pediatric non-DS-AMKL samples (Fig. 1A). Therefore, we speculate that trisomy 21 may further enhance IGF signaling in fetal hematopoietic progenitors, and perhaps more widely. Overactivation of the IGF/IGF1R pathway might result from increased expression of a set of genes on human Hsa21; e.g., GABPA. Its protein product, GABPα, is involved in both induction of cell cycle entry by activation of E2F and inhibition of CDK inhibitors, and insulin signaling (Ouyang et al. 1996; Yang et al. 2007). Interestingly, shRNA-mediated knockdown of GABPα impairs proliferation of the DS-AMKL cell line CMK and abrogates the mitogenic effect of IGF2 on these cells (Supplemental Fig. S13). In adults, trisomy 21 does not predispose DS patients to AMKL, possibly due to lesser dependency of adult HSPCs on IGF/IGF1R signaling and, therefore, a weaker requirement of lineage-determining TFs to restrict the E2F transcription network in slow-cycling adult HSPCs during terminal differentiation (Bowie et al. 2006). The specific requirement of GATA1, essential for terminal megakaryocytic differentiation, in the context of trisomy 21, can be reconciled with preferential differentiation toward megakaryopoiesis of fetal HSPCs, as observed in FLs of healthy fetuses with DS (Chou et al. 2008; Tunstall-Pedoe et al. 2008). Overexpression of several megakaryopoiesis-related genes encoded on Hsa21—e.g., RUNX1, ETS2, ERG, and miR-125b-2 (Elagib et al. 2003; Ge et al. 2008; Klusmann et al. 2010)—may cooperate with excessive IGF signaling to perturb baseline hematopoiesis in DS.
In summary, we discovered a novel developmental stage-specific interplay of a central mitogenic pathway (IGF signaling) and a lineage-determining TF, GATA1, during fetal MP expansion and differentiation (Fig. 7E). Perturbation of this conserved feature of lineage-determining TFs, in conjunction with overactivation of IGF/IGF1R signaling, jointly deregulates cell cycle control and contributes to malignant transformation (Fig. 7F). Our study underscores the necessity to study these context-dependent requirements during oncogenesis. Understanding the cell type-specific signaling pathways and their intersection with oncogenes during malignant transformation will ultimately guide us to develop more target cell-specific cancer therapies.
Materials and methods
Cell lines and patient samples
All human leukemic cell lines were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) and maintained according to the supplier's instructions. CMY cells were a kind gift from Dr. J.P. Bourquin (Zurich). For global gene expression analysis, BM and peripheral blood samples were obtained from six patients with DS-AMKL and five patients with non-DS-AMKL, provided by the AML-‘Berlin-Frankfurt-Münster’ Study Group (AML-BFM-SG, Münster, Germany). For in vitro studies, BM and peripheral blood samples were obtained from 18 patients, including three patients with DS-AMKL, four patients with DS-TL, three patients with non-DS-AMKL, three patients with AML FAB M2, and three patients with AML FAB M5. Mobilized adult CD34+-HSPCs from donors were positively selected by immunomagnetic labeling with corresponding magnetic cell-sorting beads (Miltenyi Biotech). AML-BFM-SG performed a central review of the diagnosis, classification, and clinical follow-up of the patients. All investigations had been approved by the Ethics Committee of the Hannover Medical School.
Mice
Gata1s mice were generated as described (Li et al. 2005). Ts65Dn and Ts1Cje mice were acquired from the Jackson Laboratory Cytogenetic Models Resource. Rag2−/− immunodeficient mice are maintained in our mouse colony. All studies involving mice were approved by the Children's Hospital (Boston) Institutional Animal Care and Use Committee, and were performed in accordance with the relevant protocol.
Retroviral insertional mutagenesis
Retroviral insertional mutagenesis and the subsequent identification of retroviral integration sites were performed according to Du et al. (2005). Detailed procedures are provided in the Supplemental Material.
Pathology, immunostaining, and flow cytometry
Standard protocols were followed. Detailed procedures are provided in the Supplemental Material.
Viral vectors and transduction
cDNAs were subcloned into retroviral expression vectors, including pMSCV-Puro (Clontech) and pMSCV-Puro-IRES-GFP (LPIG) (Dickins et al. 2005). shRNAs were subcloned in the miR-30 backbone of the pTRIPZ lentiviral vector (Open Biosytems) or the LMPIG retroviral vector (Dickins et al. 2005) using standard protocols, or were purchased from Open Biosystems (LKO.1-based lentiviral system). Both LPIG and LMPIG were generous gifts from Dr. Scott Lowe. For production of retroviral and lentiviral supernatant and for viral infection, standard protocols were followed. Detailed procedures and shRNA sequences are provided in the Supplemental Material.
Microarray data collection and analysis
Microarray expression profiles were collected using Affymetrix chips, and were analyzed using dChip (Li and Wong 2001) and GSEA (Subramanian et al. 2005) as described. For murine cells and human flow-sorted leukemic blasts, Affymetrix Mouse Genome 430 2.0 and Human Genome U133 Plus 2.0 arrays were used, respectively. All microarray data have been deposited in NCBI's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) with GEO Series accession number GSE16655.
Acknowledgments
We thank Dr. H. Hock, J. Schindler, J. Schöning, T.V. Raghavan, K. Böhmer, M. Wackerhahn, T. Pushpanathan, Dr. J. Skokowa, K. Gupta, Dr. B. Thakur, and Dr. Y. Fujiwara for suggestions and/or technical assistance; M. Hamblen for general laboratory support; G. Losyev and Dr. M. Ballmaier for cell sorting; Dr. M. Zimmermann for statistical assistance; Drs. A. Schambach and C. Baum for providing plasmids and assistance with lentiviral transduction; Dr. N. von Neuhoff for providing support and assistance with protein analyses; Drs. K. Bernt and S. Armstrong for Mll-Af9 leukemic cells; and Dr. A. Cantor for the BioGata1/BioGata1s constructs and for sharing unpublished data. Z.L. was a Fellow of the Leukemia and Lymphoma Society, and was supported by a K99 Pathway-to-Independence award from NCI. This work was supported by a grant to J.H.K. and D.R. from the German National Academic Foundation (KL-2374/1-1), and a grant to S.H.O. from the NIH. S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1903410.
Supplemental material is available at http://www.genesdev.org.
References
- Alford KA, Slender A, Vanes L, Li Z, Fisher EM, Nizetic D, Orkin SH, Roberts I, Tybulewicz VL 2010. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood 115: 2928–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J, Liu JP, Robertson EJ, Efstratiadis A 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75: 73–82 [PubMed] [Google Scholar]
- Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, Baruchel A, Cave H, Dastugue N, Hasle H, et al. 2006. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci 103: 3339–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ 2006. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest 116: 2808–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Lodish HF 2010. Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc Natl Acad Sci 107: 7799–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Opalinska JB, Yao Y, Fernandes MA, Kalota A, Brooks JS, Choi JK, Gewirtz AM, Danet-Desnoyers GA, Nemiroff RL, et al. 2008. Trisomy 21 enhances human fetal erythro-megakaryocytic development. Blood 112: 4503–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara TM, Efstratiadis A, Robertson EJ 1990. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345: 78–80 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859 [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Du Y, Jenkins NA, Copeland NG 2005. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood 106: 3932–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN 2003. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101: 4333–4341 [DOI] [PubMed] [Google Scholar]
- Ge Y, Dombkowski AA, LaFiura KM, Tatman D, Yedidi RS, Stout ML, Buck SA, Massey G, Becton DL, Weinstein HJ, et al. 2006. Differential gene expression, GATA1 target genes, and the chemotherapy sensitivity of Down syndrome megakaryocytic leukemia. Blood 107: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, LaFiura KM, Dombkowski AA, Chen Q, Payton SG, Buck SA, Salagrama S, Diakiw AE, Matherly LH, Taub JW 2008. The role of the proto-oncogene ETS2 in acute megakaryocytic leukemia biology and therapy. Leukemia 22: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Zhou W, Kaufman S, Robinson MO 1996. E2F-1 blocks terminal differentiation and causes proliferation in transgenic megakaryocytes. Mol Cell Biol 16: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasle H 2001. Pattern of malignant disorders in individuals with Down's syndrome. Lancet Oncol 2: 429–436 [DOI] [PubMed] [Google Scholar]
- Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, Orkin SH 2004. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev 18: 2336–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollanda LM, Lima CS, Cunha AF, Albuquerque DM, Vassallo J, Ozelo MC, Joazeiro PP, Saad ST, Costa FF 2006. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet 38: 807–812 [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR 2002. A stem cell molecular signature. Science 298: 601–604 [DOI] [PubMed] [Google Scholar]
- Kadri Z, Shimizu R, Ohneda O, Maouche-Chretien L, Gisselbrecht S, Yamamoto M, Romeo PH, Leboulch P, Chretien S 2009. Direct binding of pRb/E2F-2 to GATA-1 regulates maturation and terminal cell division during erythropoiesis. PLoS Biol 7: e1000123 doi: 10.1371/journal.pbio.1000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ 2007. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130: 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann JH, Creutzig U, Zimmermann M, Dworzak M, Jorch N, Langebrake C, Pekrun A, Macakova-Reinhardt K, Reinhardt D 2008. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood 111: 2991–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D 2010. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev 24: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Lackey J, Barnett J, Davidson L, Batty IH, Leslie NR, Downes CP 2007. Loss of PTEN selectively desensitizes upstream IGF1 and insulin signaling. Oncogene 26: 7132–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmikuttyamma A, Pastural E, Takahashi N, Sawada K, Sheridan DP, DeCoteau JF, Geyer CR 2008. Bcr-Abl induces autocrine IGF-1 signaling. Oncogene 27: 3831–3844 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH 2001. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH 2005. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet 37: 613–619 [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM 1999. c-Myc regulates cyclin D–Cdk4 and –Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol 19: 4672–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohi MG, Boulton C, Gu TL, Sternberg DW, Neuberg D, Griffin JD, Gilliland DG, Neel BG 2004. Combination of rapamycin and protein tyrosine kinase (PTK) inhibitors for the treatment of leukemias caused by oncogenic PTKs. Proc Natl Acad Sci 101: 3130–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL 1995. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci 92: 10302–10306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH 1992. GATA-binding transcription factors in hematopoietic cells. Blood 80: 575–581 [PubMed] [Google Scholar]
- Ouyang L, Jacob KK, Stanley FM 1996. GABP mediates insulin-increased prolactin gene transcription. J Biol Chem 271: 10425–10428 [DOI] [PubMed] [Google Scholar]
- Pine SR, Guo Q, Yin C, Jayabose S, Druschel CM, Sandoval C 2007. Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood 110: 2128–2131 [DOI] [PubMed] [Google Scholar]
- Pollak M 2008. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 8: 915–928 [DOI] [PubMed] [Google Scholar]
- Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, Nerlov C 2001. E2F repression by C/EBPα is required for adipogenesis and granulopoiesis in vivo. Cell 107: 247–258 [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Kuczynski WI, Onodera K, Moore J, Ratajczak J, Kregenow DA, DeRiel K, Gewirtz AM 1994. A reappraisal of the role of insulin-like growth factor I in the regulation of human hematopoiesis. J Clin Invest 94: 320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recher C, Beyne-Rauzy O, Demur C, Chicanne G, Dos SC, Mas VM, Benzaquen D, Laurent G, Huguet F, Payrastre B 2005. Antileukemic activity of rapamycin in acute myeloid leukemia. Blood 105: 2527–2534 [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, Bernards R 2006. Re-evaluating cell-cycle regulation by E2Fs. Cell 127: 871–874 [DOI] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ 2003. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol 23: 5031–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol 17: 5227–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmeier K, Synovzik H, Mertz R, Winnacker EL, Lipp M 1989. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes Dev 3: 527–536 [DOI] [PubMed] [Google Scholar]
- Tunstall-Pedoe O, Roy A, Karadimitris A, de la Fuente J, Fisk NM, Bennett P, Norton A, Vyas P, Roberts I 2008. Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations. Blood 112: 4507–4511 [DOI] [PubMed] [Google Scholar]
- Voz ML, Mathys J, Hensen K, Pendeville H, Van Valckenborgh I, Van Huffel C, Chavez M, Van Damme B, De Moor B, Moreau Y, et al. 2004. Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene 23: 179–191 [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444: 364–368 [DOI] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD 2002. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet 32: 148–152 [DOI] [PubMed] [Google Scholar]
- West MJ, Stoneley M, Willis AE 1998. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17: 769–780 [DOI] [PubMed] [Google Scholar]
- Xu G, Nagano M, Kanezaki R, Toki T, Hayashi Y, Taketani T, Taki T, Mitui T, Koike K, Kato K, et al. 2003. Frequent mutations in the GATA-1 gene in the transient myeloproliferative disorder of Down syndrome. Blood 102: 2960–2968 [DOI] [PubMed] [Google Scholar]
- Yang ZF, Mott S, Rosmarin AG 2007. The Ets transcription factor GABP is required for cell-cycle progression. Nat Cell Biol 9: 339–346 [DOI] [PubMed] [Google Scholar]
- Zhang CC, Lodish HF 2004. Insulin-like growth factor 2 expressed in a novel fetal liver cell population is a growth factor for hematopoietic stem cells. Blood 103: 2513–2521 [DOI] [PubMed] [Google Scholar]