Abstract
Anopheles arabiensis is a major vector of Plasmodium falciparum in southern Zambia. This study aimed to determine the rate of multiple human blood meals taken by An. arabiensis to more accurately estimate entomologic inoculation rates (EIRs). Mosquitoes were collected in four village areas over two seasons. DNA from human blood meals was extracted and amplified at four microsatellite loci. Using the three-allele method, which counts ≥ 3 alleles at any microsatellite locus as a multiple blood meal, we determined that the overall frequency of multiple blood meals was 18.9%, which was higher than rates reported for An. gambiae in Kenya and An. funestus in Tanzania. Computer simulations showed that the three-allele method underestimates the true multiple blood meal proportion by 3–5%. Although P. falciparum infection status was not shown to influence the frequency of multiple blood feeding, the high multiple feeding rate found in this study increased predicted malaria risk by increasing EIR.
Introduction
Anopheles arabiensis Patton is one of the major malaria vectors in Africa.1–7 This species is the primary malaria vector in large regions of southern Zambia.8 Unlike its sibling species An. gambiae Giles sensu stricto, which is predominantly anthropophilic, endophilic, and endophagic, An. arabiensis displays a large degree of variation in foraging behavior.9 This species varies in anthropophily, with estimates of the human blood index from 0.2 to 1.0.8 Data from indoor spray catches and window traps show that this species may rest indoors but often exits after feeding there.10 It is also known to feed indoors and outdoors.10,11
The Southern Province of Zambia has historically had hyperendemic transmission of Plasmodium falciparum,12 although there has been a significant decrease in malaria cases since 2003 (Thuma P, unpublished data). Until recently, the two primary malaria vectors in the Macha region were An. arabiensis and An. funestus.8 No specimens of An. funestus have been captured since a drought in 2004–2005, making An. arabiensis the only recognized malaria vector in the region. The entomologic inoculation rate (EIR) for An. arabiensis in the region has ranged from 1.6 to 18.3 infective bites per person per season.8 These estimates assumed that every blood fed mosquito captured fed on only one human host. However, if infected female mosquitoes take more than one human blood meal per gonotrophic cycle, the effective biting rate is increased, increasing the EIR and malaria transmission risk to humans.13 Therefore, accurate estimation of the multiple human feeding rate is critical to determining and understanding the intensity of Plasmodium transmission.
Our specific aim was to determine the proportion of female An. arabiensis mosquitoes that take multiple human blood meals in the Southern Province of Zambia to better understand An. arabiesis feeding behavior and to more accurately estimate EIR. Previous DNA fingerprinting studies have estimated the multiple blood meal frequency in An. gambiae (0–11%),13,14 An. funestus (3–14%),15 and Aedes aegypti (18%).16 Although An. gambiae and An. arabiensis are closely related, behavioral plasticity among populations of An. arabiensis suggests that this trait will also vary.9
Additionally, P. falciparum infection has been shown to increase multiple feeding in An. gambiae possibly by increasing the blood meal volume at which a mosquito stops host-seeking behavior, or by decreasing apyrase activity.13 We therefore investigated whether P. falciparum infection correlates with multiple blood feeding in An. arabiensis.
Materials and Methods
Study area.
The Johns Hopkins Malaria Research Institute's field station in Macha, Zambia is located in the Southern Province (16.39292°S, 26.79061°E) at an elevation of approximately 1,000 meters above sea level. The habitat around the field station, the Malaria Institute at Macha (MIAM), is characterized as Miombo woodland. Annual rainfall is variable, but averages 600–1,000 mm in the Southern Province.2 There is a single rainy season each year that lasts from approximately November to April, followed by cool dry (April–August) and hot dry (August–November) seasons. Mosquitoes were collected in four village areas: Chidakwa, Lupata, Namwalinda, and Chilumbwe. Chidakwa and Lupata are 3 km from the research facility and Macha Mission Hospital, and Namwalinda and Chilumbwe are 10 km away.
Mosquito collection and handling.
Field specimens of An. arabiensis were collected during the rainy season in both years by pyrethrum spray catch or CDC light traps hung next to persons sleeping under bed nets.17–19 Collections were conducted in Chidakwa and Lupata (65 households) during November 2005–May 2006 and Chidakwa, Lupata, Namwalinda, and Chilumbwe (107 households) during November 2006–May 2007. Specimens were collected in each village area multiple times each month throughout the season. After collection, specimens were killed by freezing, identified morphologically,1,20 packed individually in tubes containing silica gel desiccant and cotton, and stored at room temperature until processing.
Preparation of DNA and polymerase chain reaction.
Mosquito heads/thoraces were separated from abdomens and DNA was extracted from each by a modified salt procedure.21 Specimens morphologically identified as An. gambiae sensu lato were confirmed as An. arabiensis by polymerase chain reaction (PCR).22 Specimens were screened for P. falciparum infection by nested PCR of the head/thorax DNA extractions.23 The host animals that contributed to the An. arabiensis blood meals were identified by PCR by using 2 μL of the abdomen DNA extraction.21,24
Microsatellite analysis.
All human blood meals were analyzed to determine the minimum number of humans that contributed during the gonotrophic cycle. Primers fluorescently tagged with HEX and FAM were used to amplify DNA at the CTT (CSF1PO and TPOX) and Silver STR (D7S and D13S) loci.13,25 Each 20-μL reaction contained 10 mM Tris, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 0.8 mM dNTPs, 3.2 units Taq polymerase, 25 pmol each forward and reverse primer, and 1 μL template DNA. PCR conditions consisted of a two-minute initial denaturation at 96°C, followed by 30 cycles for 1 minute at 94°C, 1 minute at 60°C, 1.5 minutes at 70°C, and a 45-minute final extension at 70°C. One microliter of each PCR product was multiplexed (CSF with TPOX, D7S with D13S) with 15 μL of deionized formamide and 0.5 μL of GeneScan-500 Rox size standard (Applied Biosystems, Foster City, CA), and incubated at 95°C for 5 minutes before running on an ABI 3100 Avant Genetic Analyzer (Applied Biosystems). Data were analyzed with GeneScan Analysis 3.7 and Genotyper (Applied Biosystems).
Time course analysis.
Human whole blood preserved with K3EDTA anticoagulant was used for a time course experiment to determine the effect of blood meal digestion on microsatellite amplification. Colonized An. gambiae s.s. (Keele strain) maintained at the Johns Hopkins Bloomberg School of Public Health (Baltimore, MD) were fed with an artificial feeding apparatus until fully engorged and held in an insectary facility at 27°C. Six mosquitoes were killed at each time point (0, 6, 12, 18, 24, 30, 36, and 48 hours) by freezing and stored at −20°C until DNA extraction and microsatellite PCR. Microsatellite PCRs were performed by the same methods used for field samples.
Statistical analysis.
The proportion of blood meals with contributions from multiple humans was estimated by using the three-allele method. Because humans are diploid and should have at most two alleles at all microsatellite loci, a blood meal is assumed to be from multiple human hosts if there are three or more alleles at any microsatellite locus. However, some samples that contain multiple blood meals may be scored incorrectly as single blood meals because of alleles shared between hosts or because of homozygosity. To account for this bias, statistical analysis and computer simulations were performed. All samples with at least one detected microsatellite loci were included in the statistical analysis (n = 323). Data were entered into a MySQL relational database (http://www.mysql.com) and subsequently analyzed with the R statistical analysis package (http://cran.r-project.org/). 95% confidence intervals were calculated using the Agresti-Coull method26 as implemented in the binom.confint R-function. Significance testing is performed by using a two-sided Fisher exact test for proportions using the fisher.test R-function.
Computer simulation.
Computer simulation was used to estimate the magnitude of bias caused by shared alleles. Briefly, a population of h simulated diploid human genomes containing l simulated microsatellite loci were generated as follows. 1) We define four classes of microsatellite loci corresponding to each of the four microsatellite loci for which we collected data (CSF1, TPOX, D7S, and D13S). 2) Each of the l simulated loci, L1,…, Ll in each simulated genome is assigned to one of the four previously defined classes (drawn at random with replacement). 3) The two alleles, in each simulated locus, in each chromosome, are drawn at random from the set of alleles of the corresponding class. This procedure yields a population of h simulated diploid genomes in Hardy-Weinberg equilibrium. Inbreeding is accounted for, by randomly converting heterozygous genomes into homozygous genomes so as to achieve an inbreeding factor f, equal to ((exp. frequency of heterozygotes) – (observed frequency of heterozygotes))/(expected frequency of heterozygoes). A factor of f = 0 is equivalent to Hardy-Weinberg equilibrium, and f = 1 is equivalent to a clonal population. 4) The alleles that are present in b simulated blood meals are determined by drawing genomes from the population simulated diploid genomes. For each blood meal, a single genome is drawn with probability 1 – p or two (different) genomes are drawn with probability p. 5) A simulated PCR experiment, with perfect detection, is carried out on each simulated blood meal by simply counting the number of unique alleles present in each simulated blood meal. 6) The proportion, 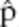 , of simulated PCR experiments that included at least one microsatellite locus with three or more alleles is calculated and reported. 7) The simulation parameters are P = 0.19, h = 1000, l = {1,2,3,4,5}, b=324, and f = {0,0.2, 0.5}. The simulated experiment is repeated 100 times for each of the 15 sets of simulation parameters. For each parameter set, the difference between the mean
, of simulated PCR experiments that included at least one microsatellite locus with three or more alleles is calculated and reported. 7) The simulation parameters are P = 0.19, h = 1000, l = {1,2,3,4,5}, b=324, and f = {0,0.2, 0.5}. The simulated experiment is repeated 100 times for each of the 15 sets of simulation parameters. For each parameter set, the difference between the mean 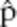 and the “true” p (i.e., the parameter p) is an estimate of the bias in the three-allele-method (Figure 1).
and the “true” p (i.e., the parameter p) is an estimate of the bias in the three-allele-method (Figure 1).
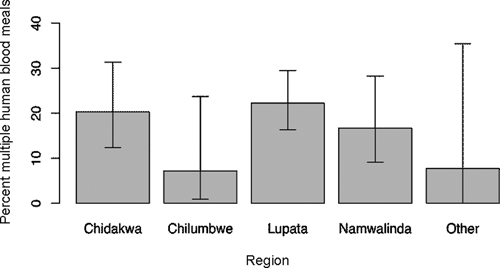
Figure 3.
Proportion of mosquitoes that took multiple human blood meals in each village area, Zambia. Namwalinda and Chilumbwe include data from 2006–2007 only. Error bars indicate 95% confidence intervals. Regional differences are not statistically significant (P = 0.1).
Results
A total of 1,284 An. arabiensis were collected over two seasons. Of these, 324 had human blood meals and were screened for multiple human host contributions, 175 samples from the 2005–2006 season and 149 samples from the 2006–2007 season. In the whole sample set, 13 distinct alleles were detected at the CSF1PO locus, 7 at the TPOX locus, 8 at the D7S locus, and 8 at the D13S locus. In both seasons, a substantial number of samples (32% and 30%, respectively) had PCRs that failed to yield a product for one or more microsatellite loci after two PCR attempts. The D7S locus consistently had the largest number of failed PCRs, and TPOX had the least number of failed PCRs. Only one sample failed to produce PCR products for all four loci over two attempts. Thus, a total of 323 samples yielded usable PCR detections of microsatellite loci. The failed PCRs at some loci could be caused by null alleles, which do not amplify because of mutations in primer binding sites, digestion of the blood meal by the mosquito, or because DNA samples were archived at −20°C for up to a year before screening. The time course experiment demonstrated that, despite blood meal digestion, complete microsatellite profiles were detectable up to 30 hours post-feeding at 27°C, and no microsatellites were detectable by 48 hours (Figure 2). At heterozygous loci, both alleles were consistently amplified at 0 and 6 hour time points. After 12 hours, only one allele was amplified at heterozygous loci in some samples, but both alleles were detectable in at least one sample at each time point up to 30 hours.
Figure 2.
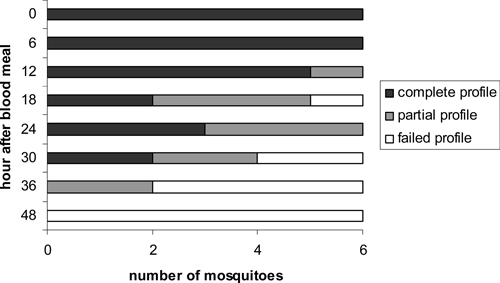
Microsatellite detection time course in Anopheles arabiensis, Zambia. Mosquitoes were held at 27°C after blood feeding and killed at various time points for microsatellite detection. Samples where all four microsatellite loci were dectected are identified as complete, samples where at least one locus was detected are partial, and samples where no loci were detected are failed.
During the 2005–2006 season, we obtained 174 samples with one or more microsatellite detections. We estimated that 19.0% (95% confidence interval [CI] = 14–25%) of these samples contained multiple blood meals based on the three-allele method. During the 2006–2007 season, we obtained 149 samples with one or more microsatellite detections. We estimated that 19.8% (95% CI = 13–26%) of these samples contained multiple blood meals. There was no statistically significant difference (P = 0.97) in the multiple blood meal proportions between the two seasons. We estimated that the proportion of samples with multiple blood meals over both seasons was 18.9% (95% CI = 15–23%). Comparison of the village areas showed no significant difference in multiple blood meal proportions (P = 0.1;Figure 3). This finding was the case for each season, as well as for the total data set. In addition, there was no significant difference in multiple blood meal proportions between mosquitoes collected by spray catch and those collected by CDC light trap (P = 1.0). Our data provided no evidence that mosquitoes infected with P. falciparum are more likely to take meals from multiple humans than uninfected mosquitoes (P = 0.3). The proportion of infected mosquitoes with meals from multiple humans was 11% (n = 28, 95% CI = 3–28%), and the proportion of uninfected mosquitoes with meals from multiple humans was 20% (n = 295, 95% CI = 15–25%).
Figure 1.
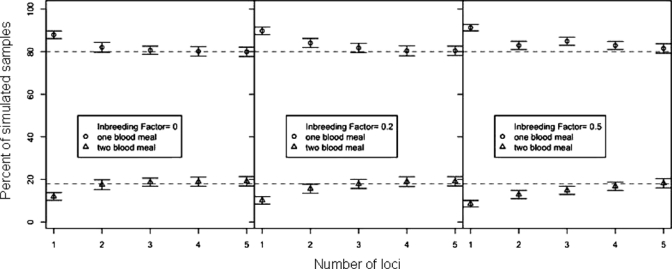
Simulated results of percentage of single and multiple blood meals estimated by the three-allele method, as a function of the number of microsatellite loci (1–5) used in the experiment with inbreeding factors of f = 0, 0.2, and 0.5, Zambia. Perfect detection is assumed. The upper dashed line indicates the true proportion of single blood meals, and the lower dashed line indicates the true proportion of multiple blood meals. Each point represents the mean percentage of single and multiple blood meals returned by the simulation for a given number of loci. Error bars represent standard error of the mean. Detection bias is equal to the difference between each point and the true proportion (dashed line).
Discussion
Over the combined 2005–2006 and 2006–2007 malaria seasons, An. arabiensis in the Macha region had an average multiple human feeding rate of 18.9% (95% CI = 15–23%). This rate is much higher than the 0–11% rate reported for An. gambiae s.s. in Kenya,13 or the 14% rate reported for An. funestus in Tanzania.15 Both of these studies directly identified the components of individual humans present in the blood meals by their DNA fingerprint. It is reasonable to ask whether the differences in the measured rates can be attributed to differences in experimental protocols. The study in Kenya used a six-locus fingerprint and the study in Tanzania used an eight-locus fingerprint. This study uses only four loci and eschews DNA fingerprinting of the human population in favor of the three-allele method. We note that the three-allele method produces a systematically low estimate of the true proportion of meals from multiple humans.
This bias has not been characterized in previous studies.13–16,27 The three-allele method is biased for two reasons. First, inbreeding in the human population reduces the number of unique alleles that are likely to be present in a given blood meal (e.g., in a clonal human population there can never be more than two alleles per locus, thereby making it impossible to detect multiple blood meals). Second, failed PCRs exacerbate the bias because they result in an undercount of the number of unique alleles, thereby reducing the probability of detecting three or more alleles at a given locus. The magnitude of the bias in the three-allele method will become smaller as more and more microsatellite loci are included in the experiment. Our simulations show that with perfect detection, the magnitude of the bias will be only a few percent, provided one includes three or more loci (Figure 1). Substantial bias does not appear until the number of loci included in the analysis is less than 2 and is greatest (although still modest) for inbreeding factors greater than 0.2. Given the number of loci sampled in this study, the number of alleles detected in the population, and the results of these simulations, bias caused by shared alleles between family members is most likely low. Our missed detection rate (30–32%) has the effect of reducing the number of loci being measured per sample from four to three. From Figure 1, our computer simulations indicate that given our use of four microsatellite loci and our 30–32% missed detection rate, we can expect the proportion of multiple meals reported by the three-allele method to underestimate the true proportion of multiple meals by approximately 3–5%, depending on the amount of inbreeding in the population of humans represented by the blood meals. The magnitude of the bias is well below the 95% CIs of our measured rate and cannot explain the disparity between our rates and the previously published rates.
Because An. gambiae s.s. and An. funestus are not found in Macha, it is unclear whether the disparity can be explained by the variability of An. arabiensis foraging behavior, local ecology, or whether it is caused by differences in the human population, such as behavior, sleeping arrangements, or a greater ability to detect multiple blood meals caused by allelic diversity. It is possible that this disparity could be caused by a high number of newly emerged, nulliparous mosquitoes. Gillies showed that nearly all An. gambiae and An. funestus require more than one blood meal to lay their first egg batch, and that 65% and 26%, respectively, took their first blood meal prior to fertilization.28,29 We have yet to investigate the age structure or parity of the An. arabiensis population in Macha, and this possibility warrants further study. A second possibility is that An. arabiensis is comparatively larger than An. gambiae30 or An. funestus,20 and may therefore require a larger volume of blood for egg production.
There was no significant difference in multiple feedings between village areas, between seasons, or between collection methods. Therefore, we believe that it is reasonable to generalize this figure to An. arabiensis in the Macha area. Additionally, we found no evidence for a dependence of multiple feeding on P. falciparum infection status in An. arabiensis. This finding is in contrast to reported results in An. gambiae,13,14 and could be caused by the low number of P. falciparum-positive samples in our collections (n = 28), giving reduced statistical power.
It is possible that heterogeneity in biting affected our measure of the multiple biting rate. In an infinite host population, heterogeneity in biting increases R0, the basic reproductive number of the vector-borne pathogen.31 In small populations, such as Macha, heterogeneity in biting may actually lead to a decrease in R0 caused by most bites landing on a subset of the population that is re-infected.32 Multiple feeding, by increasing the human biting rate, would increase malaria transmission,13 but most likely within the subset of the population that is bitten more frequently. If this is the case, the probability of a mosquito taking multiple feeds from one person is higher, but these multiple blood meals would not be detectable by our methods, leading to an underestimate of the multiple feeding rate. Increases in heterogeneity, for instance by the introduction of insecticide-treated bed nets that would decrease the available host population, would lower the apparent multiple feeding rate.
Our time-course experiment demonstrated that human microsatellite DNA in blood meals was detectable as long as 36 hours post-blood feeding, and complete profiles were obtained as late as 30 hours. Microsatellite loci were successfully amplified at later time points than those reported by Mukabana and others (less than 50% after 15 hours),33 possibly caused by the use of different microsatellite loci or our greater ability to visualize products using fluorescent capillary electrophoresis, rather than electrophoresis and silver staining on polyacrylamide gels. Weather conditions in Macha during the rainy season are variable, with night time temperatures as low as 16°C. Therefore, blood meal digestion may be slowed, and human microsatellite DNA may be detectable for longer time periods in field samples. Regardless, this indicates that it is possible to detect microsatellite DNA from blood meals aquired more than a day before mosquito collection. Therefore, it is difficult to determine if multiple blood meals were taken on the same night or on consecutive nights.
Finally, we note that the 18.9% multiple blood meal rate reported here has a potentially large effect on the EIR, the number of infective bites that a person receives each year. The EIR is typically calculated as the human biting rate measured by human landing catch and multiplied by the P. falciparum sporozoite infection rate in mosquitoes. Raw human landing catch counts will underestimate the actual EIR if mosquitoes are biting more than one person per gonotrophic cycle. For example, the EIRs for the Lupata and Chidakwa village areas during the 2005–2006 season were 18.3 and 1.6 infective bites per season, respectively.8 If multiple biting is taken into account, this linearly increases the human biting rate, and thus the EIR. In this instance, a multiple feeding rate of 18.9% would increase these EIRs to 21.8 and 1.9 infective bites per season, respectively. Because EIR directly corresponds to malaria risk, neglecting multiple blood meals by malaria vectors could greatly underestimate potential for disease transmission.
Acknowledgments
We thank R. Kent, H. Hamapumbu, and S. Habbanti for assisting with sample collection and processing.
Footnotes
Financial support: This study was supported in part by funding from the Johns Hopkins Malaria Institute to Douglas E. Norris and Fernando J. Pineda, by grant T32 AI 007417 from the National Institutes of Health to Laura C. Norris, and by a Sommer Scholar award to Christen M. Fornadel.
Authors' addresses: Laura C. Norris, Christen M. Fornadel, Wei-Chien Hung, Fernando J. Pineda, and Douglas E. Norris, The W. Harry Feinstone Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, 615 North Wolfe Street, Baltimore, MD 21205, E-mails: lnorris@jhsph.edu, cfornade@jhsph.edu, wehung@jhsph.edu, fernando.pineda@jhu.edu, and dnorris@jhsph.edu.
References
- 1.Gillies MT, DeMeillon B. The Anophelinae South of the Sahara (Ethiopian Zoological Region) Johannesburg: South African Institute for Medical Research; 1968. [Google Scholar]
- 2.Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today. 2000;16:74–77. doi: 10.1016/s0169-4758(99)01563-x. [DOI] [PubMed] [Google Scholar]
- 3.Fontenille D, Lochouarn L, Diatta M, Sokhna C, Dia I, Diagne N, Lemasson JJ, Ba K, Tall A, Rogier C, Trape JF. Four years' entomological study of the transmission of seasonal malaria in Senegal and the bionomics of Anopheles gambiae and A. arabiensis. Trans R Soc Trop Med Hyg. 1997;91:647–652. doi: 10.1016/s0035-9203(97)90506-x. [DOI] [PubMed] [Google Scholar]
- 4.Shililu JI, Maier WA, Seitz HM, Orago AS. Seasonal density, sporozoite rates and entomological inoculation rates of Anopheles gambiae and Anopheles funestus in a high-altitude sugarcane growing zone in western Kenya. Trop Med Int Health. 1998;3:706–710. doi: 10.1046/j.1365-3156.1998.00282.x. [DOI] [PubMed] [Google Scholar]
- 5.Mendis C, Jacobsen JL, Gamage-Mendis A, Bule E, Dgedge M, Thompson R, Cuamba N, Barreto J, Begtrup K, Sinden RE, Hogh B. Anopheles arabiensis and An. funestus are equally important vectors of malaria in Matola coastal suburb of Maputo, southern Mozambique. Med Vet Entomol. 2000;14:171–180. doi: 10.1046/j.1365-2915.2000.00228.x. [DOI] [PubMed] [Google Scholar]
- 6.Ijumba JN, Mosha FW, Lindsay SW. Malaria transmission risk variations derived from different agricultural practices in an irrigated area of northern Tanzania. Med Vet Entomol. 2002;16:28–38. doi: 10.1046/j.0269-283x.2002.00337.x. [DOI] [PubMed] [Google Scholar]
- 7.Okello PE, Van Bortel W, Byaruhanga AM, Correwyn A, Roelants P, Talisuna A, D'Alessandro U, Coosemans M. Variation in malaria transmission intensity in seven sites throughout Uganda. Am J Trop Med Hyg. 2006;75:219–225. [PubMed] [Google Scholar]
- 8.Kent RJ, Thuma PE, Mharakurwa S, Norris DE. Seasonality, blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in southern Zambia. Am J Trop Med Hyg. 2007;76:267–274. [PMC free article] [PubMed] [Google Scholar]
- 9.White GB, Magayuka SA, Boreham PFL. Comparative studies on sibling species of the Anopheles gambiae Giles complex (Dipt., Culicidae): binomics and vectorial activity of species A and species B at Segera, Tanzania. Bull Entomol Res. 1972;62:295–317. [Google Scholar]
- 10.Ameneshewa B, Service MW. Resting habits of Anopheles arabiensis in the Awash River Valley of Ethiopia. Ann Trop Med Parasitol. 1996;90:515–521. doi: 10.1080/00034983.1996.11813077. [DOI] [PubMed] [Google Scholar]
- 11.Ralisoa Randrianasolo BO, Coluzzi M. Genetical investigations on zoophilic and exophilic Anopheles arabiensis from Antananarivo area (Madagascar) Parassitologia. 1987;29:93–97. [PubMed] [Google Scholar]
- 12.Larkin GL, Thuma PE. Congenital malaria in a hyperendemic area. Am J Trop Med Hyg. 1991;45:587–592. doi: 10.4269/ajtmh.1991.45.587. [DOI] [PubMed] [Google Scholar]
- 13.Scott TW, Githeko AK, Fleisher A, Harrington LC, Yan G. DNA profiling of human blood in Anophelines from lowland and highland sites in western Kenya. Am J Trop Med Hyg. 2006;75:231–237. [PubMed] [Google Scholar]
- 14.Koella JC, Sorensen FL, Anderson RA. The malaria parasite, Plasmodium falciparum, increases the frequency of multiple feeding of its mosquito vector, Anopheles gambiae. Proc Bio Sci. 1998;1398:763–768. doi: 10.1098/rspb.1998.0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soremekun S, Maxwell C, Zuwakuu M, Chen C, Michael E, Curtis C. Measuring the efficacy of insecticide treated bed nets: the use of DNA fingerprinting to increase the accuracy of personal protection estimates in Tanzania. Trop Med Int Health. 2004;9:663–672. doi: 10.1111/j.1365-3156.2004.01250.x. [DOI] [PubMed] [Google Scholar]
- 16.De Benedictis J, Chow-Schaffer E, Costero A, Clark GG, Edman JD, Scott TW. Identification of the people from whom engorged Aedes aegypti took blood meals in Florida, Puerto Rico using PCR-based DNA profiling. Am J Trop Med Hyg. 2003;68:447–452. [PubMed] [Google Scholar]
- 17.Service MW. Field Sampling Methods. London: Elsevier Applied Science; 1976. (Mosquito Ecology). [Google Scholar]
- 18.Beier JC. Vector incrimination and entomological inoculation rates. Methods Mol Med. 2002;72:3–11. doi: 10.1385/1-59259-271-6:01. [DOI] [PubMed] [Google Scholar]
- 19.Sudia WD, Chamberlain RW. Battery-operated light trap, an improved model. J Am Mosq Control Assoc. 1988;4:536–538. [PubMed] [Google Scholar]
- 20.Gillies MT, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara. Johannesburg: South African Institute for Medical Research; 1987. [Google Scholar]
- 21.Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am J Trop Med Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- 22.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 23.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 24.Fornadel CM, Norris DE. Increased endophily by the malaria vector Anopheles arabiensis in southern Zambia and identification of digested blood meals. Am J Trop Med Hyg. 2008;79:876–880. [PMC free article] [PubMed] [Google Scholar]
- 25.Chemical Science and Technology Laboratory National Institute of Standards and Technology. Overview of STR Fact Sheets. 2009. http://www.cstl.nist.gov/div831/strbase/str_fact.htm Available at. Accessed May 1, 2009.
- 26.Agresti A, Coull BA. Approximate is better than ‘exact’ for interval estimation of binomial proportions. Am Stat. 1998;52:119–126. [Google Scholar]
- 27.Chow-Shaffer E, Sina B, Hawley WA, De Benedictis J, Scott TW. Laboratory and field evaluation of polymerase chain reaction-based forensic DNA profiling for use in identification of human blood meal sources of Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2000;37:492–502. doi: 10.1603/0022-2585-37.4.492. [DOI] [PubMed] [Google Scholar]
- 28.Gillies MT. The recognition of age-groups within populations of Anopheles gambiae by the pre-gravid rate and the sporozoite rate. Ann Trop Med Parasitol. 1954;48:58–74. doi: 10.1080/00034983.1954.11685599. [DOI] [PubMed] [Google Scholar]
- 29.Gillies MT. The pre-gravid phase of ovarian development in Anopheles funestus. Ann Trop Med Parasitol. 1955;49:320–325. doi: 10.1080/00034983.1955.11685681. [DOI] [PubMed] [Google Scholar]
- 30.Petrarca V, Sabatinelli G, Touré YT, Di Deco MA. Morphometric multivariate analysis of field samples of Anopheles arabiensis and Anopheles gambiae s.s. (Diptera: Culicidae) J Med Entomol. 1998;35:16–25. doi: 10.1093/jmedent/35.1.16. [DOI] [PubMed] [Google Scholar]
- 31.Dye C, Hasibeder G. Population dynamics of mosquito-borne disease: effects of flies which bite some people more frequently than others. Trans R Soc Trop Med Hyg. 1986;80:69–77. doi: 10.1016/0035-9203(86)90199-9. [DOI] [PubMed] [Google Scholar]
- 32.Smith DL, McKenzie FE, Snow RW, Hay SI. Revisiting the basic reproductive number for malaria and its implications for malaria control. PLoS Biol. 2007;5:e42. doi: 10.1371/journal.pbio.0050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukabana WR, Takken W, Seda P, Killeen GF, Hawley WA, Knols BG. Extent of digestions affects the success of amplifying human DNA from blood meals of Anopheles gambiae (Diptera: Culicidae) Bull Entomol Res. 2002;92:233–239. doi: 10.1079/BER2002164. [DOI] [PubMed] [Google Scholar]


