Abstract
Universal childhood hepatitis B vaccination was introduced in Indonesia in 1997; by 2008, coverage was estimated to be 78%. This study aimed to investigate the serologic status and virologic characteristics of hepatitis B virus (HBV) among the children in East Java. A total of 229 healthy children born during 1994–1999 were enrolled in this study. Overall, 3.1% were positive for hepatitis B surface antigen (HBsAg) and 23.6% were positive for antibody to HBsAg (anti-HBs). HBV DNA was detected in 5 of 222 HBsAg-negative carriers, which were suggested to be cases of occult HBV infection. A single amino substitution (T126I) in the S region was frequently found. HBV infection remains endemic, and the prevalence of anti-HBs remains insufficient among children in East Java, Indonesia.
Introduction
A safe and effective vaccine against hepatitis B has been available since 1982. The introduction of a childhood immunization program in many countries has dramatically reduced the carrier rate of hepatitis B virus (HBV) and significantly decreased the incidence of hepatocellular carcinoma (HCC).1–3 However, HBV infection, which can lead to acute or fulminant hepatitis, chronic hepatitis, cirrhosis, and HCC,4 remains endemic in many parts of the world.5
The potential significance of surface gene mutants for vaccination failure has been studied in several endemic countries.6–8 Hepatitis B surface antigen (HBsAg) is a major component of the hepatitis virion envelope. The a determinant is a key region for HBV antigenicity, and it has been suggested that amino acid changes in this region affect immune responses.9 Surface gene variants have been detected in vaccinated children, liver-transplant recipients treated with monoclonal and polyclonal antibodies to HBsAg (anti-HBs), and chronic carriers with occult HBV infection.10,11 Moreover, previous studies have suggested that variations in the core region may be responsible for the lack of HBsAg.12,13
The status of HBV is of particular concern in Indonesia, an area with intermediate-to-high endemicity for HBV infection and a carrier rate of 5–20% in the general population.5 Most chronic HBV carriers were infected in early infancy. Indonesia launched a nationwide hepatitis B universal childhood vaccination program in 1997, and the coverage (defined as the percentage of children receiving at least three doses of hepatitis B vaccine) in Indonesia in 2007 was estimated by World Health Organization/United Nations Children's Fund to be 78%.14 However, the current serologic status of HBV in older children has not been fully investigated.
The aim of this study was to investigate the status of HBV infection and to elucidate the prevalence and significance of HBV variants in children in Lamongan, an area in East Java, Indonesia, with intermediate-to-high endemicity for HBV.
Materials and Methods
Study subjects.
Four of 18 elementary schools in the Lamongan District, a rural area of East Java, were randomly selected. All children in grades 4–6 (n = 229, 113 boys and 116 girls 8–13 years of age) were enrolled in this study. Serum samples were obtained during November–December 2007 and stored at –20°C until use. The coverage rate among 8–10-year-old children (n = 118) was estimated to be 90% on the basis of local records for the childhood hepatitis B vaccination program during 1997–1999. Written informed consent was obtained from parents of all children. No individual hepatitis B vaccination records remained. The study protocol was reviewed and approved by the Ethics Committees of Kobe University in Japan and Airlangga University in Indonesia.
Serologic markers of HBV infection.
All refrigerated samples were tested for HBsAg by Reversed Passive Hemagglutination (R-PHA) (Mycell II HBsAg; Institute of Immunology, Tokyo, Japan) and for anti-HBs by Passive Hemagglutination (PHA) (Mycell II anti-HBs; Institute of Immunology). To differentiate vaccine-induced antibody from naturally acquired antibody and to identify occult HBV infections, the prevalence of antibody to hepatitis B core antigen (anti-HBc) was assessed by PHA (Mycell anti-rHBc; Institute of Immunology).
Detection of HBV DNA and nucleotide sequence analysis.
After being assayed for HBV serologic status, HBsAg-positive serum samples (n = 7) and serum samples negative for HBsAg but positive for either anti-HBs or anti-HBc (n = 89) were subjected to HBV genetic analysis to confirm infection and identify surface antigen and pre-core/core variants. After serologic testing, serum samples were stored at –80°C until genetic testing. DNA was extracted from 100 μL of serum samples by using a DNA extractor kit (QIAamp DNA Blood Mini Kit; QIAGEN, Tokyo, Japan). The presence of HBV DNA was assayed by two nested polymerase chain reaction (PCR) assays with primer pairs from the surface and pre-core/core promoter genes of the viral genome, as described.15,16 Amplified fragments were directly sequenced by using the Big Dye Deoxy Terminator cycle sequencing kit with an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA). Viral load was assessed by real-time PCR using an ABI Prism 7300 Real Time PCR System (Applied Biosystems). HBV was amplified by using a primer and probe set, as described previously.17 The HBV nucleotide sequences of the S gene were translated to amino acids and aligned with reference sequences.
Confirmation of HBsAg status in HBV DNA-positive children.
To minimize the possibility of false-negative results for HBsAg and confirm occult HBV infection, an immunochromatography method based on the principle of an enzyme immunoassay (EIA) (Espline HBsAg; Fuji Rebio, Tokyo, Japan) was used for samples that were HBsAg negative by R-PHA but HBV DNA positive by PCR.
HBV genotyping.
The HBV genotypes were determined by using the phylogenetic tree of the S region. Reference sequences were retrieved from the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank database. Alignments were performed by using CLUSTAL X software (www.clustal.org), phylogenetic trees were constructed by using the neighbor-joining method, and bootstrap resampling was performed 1,000 times. Analyses were conducted by using the Molecular Evolutionary Genetics Analysis (MEGA) software program.18
Statistical analysis.
Statistical analysis was performed by using the chi-square test or Fisher's exact test for categorical variables. A P value < 0.05 was considered statistically significant.
Results
A total of 229 children were screened for serologic markers of HBV infection. Because of limited sample volumes, 208 of 229 samples were tested for anti-HBc. Overall, positivity rates for HBsAg and anti-HBs were 3.1% (7 of 229) and 23.6% (54 of 229), respectively. Among 8–10-year-old children (n = 118) born after introduction of the universal vaccination program, anti-HBs was found in 28 (23.7%). Of these children, anti-HBc testing was performed on 21 children; 16 children were negative. Of 54 anti-HBs-positive children, 28 were negative for anti-HBc. All 7 HBsAg-positive children (L17, L23, L69, L70, L74, L103, and L216) were negative for anti-HBs and had a high HBV viral load (4.9–7.4 logcopies/mL), as shown in Table 1.
Table 1.
Demographic, serologic, and virologic characteristics of 12 HBV DNA-positive children, East Java*
| ID | Age, years | Sex | Genotype (subgenotype) | HBsAg | Anti-HBs | Anti-HBc | Viral load (log copies/mL) |
|---|---|---|---|---|---|---|---|
| L17 | 11 | M | B3 | + | – | + | 7.2 |
| L23 | 11 | F | B3 | + | – | NT | 6.9 |
| L69 | 9 | M | B3 | + | – | – | 4.9 |
| L70 | 9 | M | B3 | + | – | NT | NT |
| L74 | 9 | M | B3 | + | – | – | UD |
| L103 | 11 | M | B3 | + | – | NT | 7.4 |
| L216 | 11 | M | UD | + | – | + | UD |
| L29 | 10 | F | B3 | – | – | + | 4.8 |
| L33 | 10 | F | B3 | – | – | + | 4.8 |
| L44 | 9 | F | C6 | – | + | – | UD |
| L62 | 9 | M | B3 | – | + | + | 4.5 |
| L119 | 11 | F | B3 | – | – | + | 6.1 |
HBV = hepatitis B virus; ID = identification; HBsAg = hepatitis B surface antigen; anti-HBs = antibody to HBsAg; anti-HBc = antibody to hepatitis B core antigen; NT = not tested; UD = undetermined.
Five cases of occult HBV infection were found in this study (Table 2). Nucleotide sequence analysis of the core promoter gene showed that the A1762T/G1764A mutation was observed in only one child (L44, Table 2). Children L29, L62, and L119 had two amino acid substitutions, T126I and T143S, in the a determinant region (Figure 1 and Table 2). The T143S substitution is invariably associated with T126I. Moreover, these 3 strains had other substitutions (S113T, Y161F, V177A, M213I, and T227I), which were specific to three strains that showed changes outside the a determinant region but within the S gene (Figure 1). Child L33 also had the substitutions K24R, Q56P, I57T, and C64S, as did children L29 and L119. No substitution was found in the a determinant region of child L44. Two (L44 and L62) of five children with occult HBV infections were positive for anti-HBs (Tables 1 and 2). The HBV viral load was detected in four of the five children and ranged from 4.5 to 6.1 log copies/mL (Table 1).
Table 2.
Genomic variability of HBV in HBsAg-negative carriers, East Java, Indonesia*
| ID | Anti-HBs/ Anti-HBc | Amino acid mutation in the Pre-S/S region | Nucleotide mutation in the Pre-C/CP region | ||
|---|---|---|---|---|---|
| Pre-S region | S region | A1762T/ G1764A | G1896T | ||
| L29 | –/+ | K24R,Q56P,I57T,C64S, S113T | T126I, T143S | – | – |
| L33 | –/+ | K24R,Q56P,I57T,C64S | Wild | UD | UD |
| L44 | +/− | K24R,Q56P,I57T,C64S | Wild | + | – |
| L62 | +/+ | S113T | T126I, T143S | – | – |
| L119 | –/+ | K24R,Q56P,I57T,C64S, S113T | T126I, T143S | – | – |
HBV = hepatitis B virus; HBsAg = hepatitis B surface antigen; ID = identification; anti-HBs = antibody to HBsAg; anti-HBc = antibody to hepatitis B core antigen; S = surface; C = core; CP = core promoter; UD = undetermined.
Figure 1.
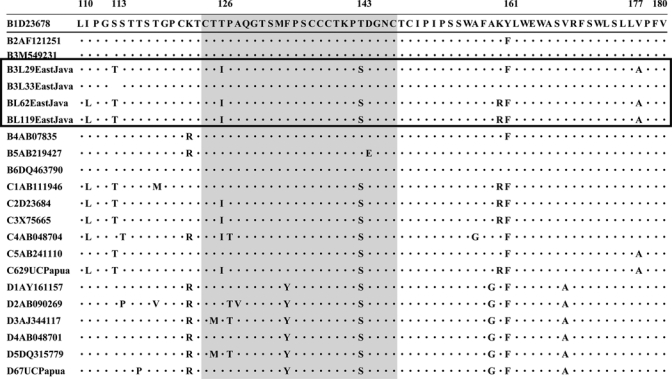
Surface antigen a determinant amino acid sequence alignment for hepatitis B virus (HBV). The first line sequence is the consensus sequence corresponding to an HBV subgenotype (B1; accession no. D23678) reference strain retrieved from the DNA Data Bank of Japan/GenBank database. Dots in the alignment indicate positions with amino acids identical to the HBV/B1 consensus sequence. The a determinant region in the S gene is indicated with a different color.
We found 12 HBV DNA-positive children in this study (Table 1). The HBV genotypes were successfully determined for 11 (91.7%) of the 12 HBV DNA-positive children (Figure 2). The HBV genotype B (HBV/B) was identified in 10 (83.3%) children, HBV/C was identified in 1 (8.3%) child, and was not identified in 1 child.
Figure 2.
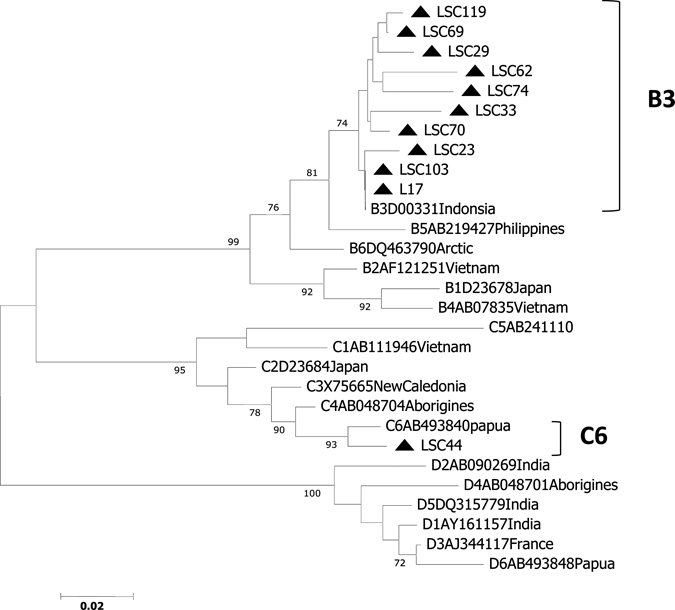
Phylogenetic trees of the surface gene in hepatitis B virus (HBV) strains isolated from 11 children in East Java, Indonesia, and 18 reference strains. Numbers in the tree indicate bootstrap reliability. Lengths of horizontal bars indicate number of nucleotide substitutions per site. Isolates from the database are indicated by their accession number, and relevant country names have been added to each HBV/B, HBV/C, and HBV/D strain.
Discussion
A universal childhood vaccination program was launched in Indonesia in 1997 and its coverage was estimated to be high (approximately 90%) on the basis of the local records for Lamongan in East Java, Indonesia. The Indonesian government provided hepatitis B vaccines produced in Indonesia to health centers called puskesmas. Most infants, especially in rural areas, were taken to puskesmas to be vaccinated. Plasma-derived HBV vaccine was produced in Indonesia until 1997, when it was replaced by a recombinant hepatitis B vaccine. In this study, some children had received the plasma-derived vaccine and others had received the recombinant vaccine during infancy. Although a higher loss rate of anti-HBs was detected when the recombinant vaccine was used, it may provide better protection than the plasma-derived vaccine.19 In the initial vaccination program in Indonesia, three doses of recombinant vaccine were administered, with the first dose being given within 12 hours of birth to prevent perinatal HBV transmission, and the second and third doses being given at one and six months of age, respectively, in accordance with national guidelines. However, the government now recommends the first dose of HBV vaccine to given within seven days of birth.
Presently in Indonesia, an HBV-diphtheria-pertusis-tetanus combination vaccine is given at birth, followed by a monovalent HBV vaccine. In Indonesia, recombinant hepatitis B vaccines that include pre-S1 and pre-S2 regions currently unavailable. However, this new triple-antigen vaccine is more efficient than the single-antigen vaccine,20,21 and prevents infection from vaccine-escape mutants.22 The triple-antigen vaccine should be considered in our study area where the prevalence of anti-HBs remains insufficient among children.
This study was unable to assess the actual coverage rate because no individual vaccination records remained. For this reason, efficacy of vaccination was not evaluated in this study. However, this study did show that acquiring protective antibody against HBV infection was insufficient among children born after introduction of the universal vaccination program. There are two possible explanations for this finding. First, the first dose of vaccination may have been delayed until after birth. To prevent perinatal transmission, the first dose should be given as soon as possible after birth.23 In reality, the first dose of HBV vaccine was often given several months after birth because the vaccinators and parents were fearful that the infant might have been killed by being immunized so soon after birth.5 Moreover, because giving birth at home is still common, such children have less opportunity to receive the vaccine than those born in a hospital.4 Failure to administer the vaccine in a timely manner will reduce the impact of vaccination in countries with a high prevalence of HBV infection,5 such as Indonesia. Second, loss of protective antibody against HBV infection may have occurred in some children. In previous studies in Taiwan, protective anti-HBs titers gradually decreased by age 12; a higher loss rate of anti-HBs was demonstrated in children immunized with the recombinant vaccine.24–26
This study identified seven HBsAg-positive children, and we suggest that these children were naturally infected during infancy. The prevalence of HBsAg (3.1%) in this study was similar to that among the general population in Indonesia.5
Causes of the failure to detect HBsAg in serum may include mutations in the S regions of the virus genome, which is also known as occult HBV infection.27 In this study, the R-PHA assay was used to screen for HBsAg positivity. This assay was useful but less sensitive (around 93%) than an EIA.28 In addition, accuracy of the EIA assay was higher than that of the R-PHA.29,30 Thus, we confirmed the presence of occult HBV infection by using the EIA.
We identified five HBsAg-negative children who were positive for HBV DNA and considered them to be children with occult HBV infections. The status of the five children with undetectable HBsAg was as follows: child L44 had recovered from wild-type HBV infection because of production of anti-HBs and a low viral load; child L62 had a vaccine-escape mutant; and children L29, L33, and L109 were naturally infected with vaccine-escape mutants and did not show production of anti-HBs after vaccination.31 These results suggest that potential vaccine-escape mutants exist within normally infected HBV carriers and that such carriers do not produce detectable HBsAg.
Mutations in the a determinant region (amino acids 121–149) affect the structure of the S region and are known to cause vaccine-escape mutations during immunization. The G145R mutation is the most common vaccine-escape mutant after immunoprophylaxis and in nature.32 Another amino acid substitution, T126I, which is unique to genotype C, has also been reported to affect the antigenicity of HBsAg.33–35 T143S was invariably associated with T126I in this study, suggesting that amino acid substitutions (not T126I itself, but in combination with T143S) affect the antigenicity of HBsAg. In this study, T126I appeared in only HBV/B and was not specific to genotype C, in contrast to findings of a previous study. Because the T126I substitution involves the largest change in chemical properties, it is most likely to cause structural changes in HBsAg.33–35 In addition, A1762T/G1764A was observed in only one child, who had recovered from wild-type HBV infection. Although previous studies have reported A1762T/G1764A mutations in occult HBV infection,11–13 the correlation between these mutations and occult HBV infection remains unclear. No other mutations were found in the core region in this study.
All 10 HBV/B strains were classified into subgenotype B3 (HBV/B3), which is prevalent in Indonesia. One strain of HBV/C had high homology with HBV/C6, which was reported as a novel strain from Papua (accession no. AB493840).36,37
HBV infection in children is still endemic in East Java. We detected several variants in the a determinant region in children who were HBsAg negative, and T126I might be one of the viral mechanisms that help the virus to escape from current hepatitis B vaccines. Emergence of viruses capable of escaping neutralization by vaccine-induced antibodies poses a serious threat to our ability to control hepatitis B.31 This pilot study indicates the presence of unique HBV among children in East Java. Because this study was small, an analysis of a larger population is necessary to confirm our findings. Empirical data are essential for future policy changes on the maintenance of the vaccination program.
Acknowledgments
We thank Dr. Shinzo Izumi and Professor Indropo Agusni (Institute of Tropical Disease, Airlangga University, Surabaya, Indonesia) for advice concerning sample collection.
Footnotes
Financial support: This study was supported by a grant-in-aid from the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Authors' addresses: Takako Utsumi, Indonesia-Japan Collaborative Research Center for Emerging and Re-emerging Infectious Diseases, Institute of Tropical Disease, Airlangga University, Campus C, Jl. Mulyoreijo, Surabaya 60115, Indonesia, and Center for Infectious Diseases, Kobe University Graduate School of Medicine, Kusunoki-cho 7-5-1, Chuo-ku, Kobe, 650-0017, Japan, E-mail: tutsumi@people.kobe-u.ac.jp. Maria Inge Lusida, Mochamad Amin, and Soetjipto, Indonesia-Japan Collaborative Research Center for Emerging and Re-emerging Infectious Diseases, Institute of Tropical Disease, Airlangga University, Campus C, Jl. Mulyoreijo, Surabaya 60115, Indonesia, E-mails: ingelusida@yahoo.com, amin_tdc@yahoo.co.id, and soetjiptobr@sby.centrin.net.id. Yoshihiko Yano and Hak Hotta, Center for Infectious Diseases, Kobe University Graduate School of Medicine, Kusunoki-cho 7-5-1, Chuo-ku, Kobe, 650-0017, Japan, E-mails: yanoyo@med.kobe-u.ac.jp and hotta@kobe-u.ac.jp. Yoshitake Hayashi, Center for Infectious Diseases, Kobe University Graduate School of Medicine, Kusunoki-cho 7-5-1, Chuo-ku, Kobe, 650-0017, Japan and Kobe University Graduate School of Health Sciences, Tomogaoka 7-10-2, Suma-ku, Kobe, 654-0412, Japan, E-mail: hayashiy@med.kobe-u.ac.jp.
References
- 1.Ni YH, Chang MH, Huang LM, Chen HL, Hsu HY, Chiu TY, Tsai KS, Chen DS. Hepatitis B virus infection in children and adolescents in a hyperendemic area: 15 years after mass hepatitis B vaccination. Ann Intern Med. 2001;135:796–800. doi: 10.7326/0003-4819-135-9-200111060-00009. [DOI] [PubMed] [Google Scholar]
- 2.Hsu HY, Chang MH, Ni YH, Chen HL. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut. 2004;53:1499–1503. doi: 10.1136/gut.2003.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou J, Liu Z, Gu F. Epidemiology and prevention of hepatitis B virus infection. Int J Med Sci. 2005;2:50–57. doi: 10.7150/ijms.2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 5.Creati M, Saleh A, Ruff TA, Stewart T, Otto B, Sutanto A, Clements CJ. Implementing the birth dose of hepatitis B vaccine in rural Indonesia. Vaccine. 2007;25:5985–5993. doi: 10.1016/j.vaccine.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 6.Carman WF, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345:1406–1407. doi: 10.1016/s0140-6736(95)92599-6. [DOI] [PubMed] [Google Scholar]
- 7.Lee PI, Chang LY, Lee CY, Huang LM, Chang MH. Detection of hepatitis B surface gene mutation in carrier children with or without immunoprophylaxis at birth. J Infect Dis. 1997;176:427–430. doi: 10.1086/514060. [DOI] [PubMed] [Google Scholar]
- 8.Tsebe KV, Burnett RJ, Hlungwani NP, Sibara MM, Venter PA, Mphahlele MJ. The first five years of universal hepatitis B vaccination in South Africa: evidence for elimination of HBsAg carriage in under 5-year-olds. Vaccine. 2001;19:3919–3926. doi: 10.1016/s0264-410x(01)00120-7. [DOI] [PubMed] [Google Scholar]
- 9.Howard CR. The structure of hepatitis B envelope and molecular variants of hepatitis B virus. J Viral Hepat. 1995;2:165–170. doi: 10.1111/j.1365-2893.1995.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 10.Allain JP. Occult hepatitis B virus infection. Transfus Clin Biol. 2004;11:18–25. doi: 10.1016/j.tracli.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Mu SC, Lin YM, Jow GM, Chen BF. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J Hepatol. 2009;50:264–272. doi: 10.1016/j.jhep.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Kao JH, Chen PJ, Lai MY, Chen DS. Sequence analysis of pre-S/surface and pre-core/core promoter genes of hepatitis B virus in chronic hepatitis C patients with occult HBV infection. J Med Virol. 2002;68:216–220. doi: 10.1002/jmv.10188. [DOI] [PubMed] [Google Scholar]
- 13.Pinarbasi B, Onel D, Cosan F, Akyuz F, Dirlik N, Cakaloglu Y, Badur S, Besisik F, Demir K, Okten A, Kaymakoglu S. Prevalence and virological features of occult hepatitis B virus infection in female sex workers who work uncontrolled in Turkey. Liver Int. 2009;29:227–230. doi: 10.1111/j.1478-3231.2008.01809.x. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization Review of National Immunization Coverage 1980–2008. 2009. http://www.who.int/immunization_monitoring/data/idn.pdf Available at. Accessed August 6, 2009.
- 15.Sugauchi F, Mizokami M, Orito E, Ohno T, Kato H, Suzuki S, Kimura Y, Ueda R, Butterworth LA, Cooksley WGE. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: complete genome sequence and phylogenetic relatedness. J Gen Virol. 2001;82:883–892. doi: 10.1099/0022-1317-82-4-883. [DOI] [PubMed] [Google Scholar]
- 16.Yamamura T, Tanaka E, Matsumoto A, Rokuhara A, Orii K, Yoshizawa K, Miyakawa Y, Kiyosawa K. A case-control study for early prediction of hepatitis B e antigen seroconversion by hepatitis B virus DNA levels and mutations in the precore region and core promoter. J Med Virol. 2003;70:545–552. doi: 10.1002/jmv.10429. [DOI] [PubMed] [Google Scholar]
- 17.Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M, Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899–2903. doi: 10.1128/jcm.37.9.2899-2903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 19.Huang ML, Liao WL, Ho MS. HBV serological markers of vaccinated children in remote areas of Taiwan: emphasis on factors contributing to vaccine failure. Vaccine. 2007;25:6326–6333. doi: 10.1016/j.vaccine.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Young MD, Schneider DL, Zuckerman AJ, Du W, Dickson B, Maddrey WC, US Hepacare Study Group Adult hepatitis B vaccination using a novel triple antigen recombinant vaccine. Hepatology. 2001;34:372–376. doi: 10.1053/jhep.2001.26167. [DOI] [PubMed] [Google Scholar]
- 21.Zuckerman JN, Zuckerman AJ, Symington I, Du W, Williams A, Dickson B, Young MD. Evaluation of a new hepatitis B triple-antigen vaccine in inadequate responders to current vaccines. Hepatology. 2001;34:798–802. doi: 10.1053/jhep.2001.27564. UK Hepacare Study Group. [DOI] [PubMed] [Google Scholar]
- 22.He C, Nomura F, Itoga S, Isobe K, Nakai T. Prevalence of vaccine-induced escape mutants of hepatitis B virus in the adult population in China: a prospective study in 176 restaurant employees. J Gastroenterol Hepatol. 2001;16:1373–1377. doi: 10.1046/j.1440-1746.2001.02654.x. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization Introduction of Hepatitis B Vaccine into Childhood Immunization Services. 2001. http://www.who.int/vaccines-documents/DocsPDF01/ Available at. Accessed August 6, 2009.
- 24.Chongsrisawat V, Yoocharoen P, Theamboonlers A, Tharmaphornpilas P, Warinsathien P, Sinlaparatsamee S, Paupunwatana S, Chaiear K, Khwanjaipanich S, Poovorawan Y. Hepatitis B seroprevalence in Thailand: 12 years after hepatitis B vaccine integration into the national expanded programme on immunization. Trop Med Int Health. 2006;11:1496–1502. doi: 10.1111/j.1365-3156.2006.01709.x. [DOI] [PubMed] [Google Scholar]
- 25.Mu SC, Lin YM, Jow GM, Chen BF. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J Hepatol. 2009;50:264–272. doi: 10.1016/j.jhep.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Kao JT, Wang JH, Hung CH, Yen YH, Hung SF, Hu TH, Lee CM, Lu SN. Long-term efficacy of plasma-derived and recombinant hepatitis B vaccines in a rural township of Central Taiwan. Vaccine. 2009;27:1858–1862. doi: 10.1016/j.vaccine.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 27.Chemin I, Trépo C. Clinical impact of occult HBV infections. J Clin Virol. 2005;34:S15–S21. doi: 10.1016/s1386-6532(05)80005-8. [DOI] [PubMed] [Google Scholar]
- 28.Thammanichanond D, Kunakorn M, Settaudom C, Khupulsup K. Reversed passive hemagglutination test fails to detect HBsAg in a number of HBeAg positive sera. Asian Pac J Allergy Immunol. 2002;20:135–137. [PubMed] [Google Scholar]
- 29.Lau DT, Ma H, Lemon SM, Doo E, Ghany MG, Miskovsky E, Woods GL, Park Y, Hoofnagle JH. A rapid immunochromatographic assay for hepatitis B virus screening. J Viral Hepat. 2003;10:331–334. doi: 10.1046/j.1365-2893.2003.00418.x. [DOI] [PubMed] [Google Scholar]
- 30.Sato K, Ichiyama S, Iinuma Y, Nada T, Shimokata K, Nakashima N. Evaluation of immunochromatographic assay systems for rapid detection of hepatitis B surface antigen and antibody, Dainascreen HBsAg and Dainascreen Ausab. J Clin Microbiol. 1996;34:1420–1422. doi: 10.1128/jcm.34.6.1420-1422.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto K, Horikita M, Tsuda F, Itoh K, Akahane Y, Yotsumoto S, Okamoto H, Miyakawa Y, Mayumi M. Naturally occurring escape mutants of hepatitis B virus with various mutations in the S gene in carriers seropositive for antibody to hepatitis B surface antigen. J Virol. 1994;68:2671–2676. doi: 10.1128/jvi.68.4.2671-2676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KM, Kim YS, Ko YY, Yoo BM, Lee KJ, Kim JH, Hahm KB, Cho SW. Emergence of vaccine-induced escape mutant of hepatitis B virus with multiple surface gene mutations in a Korean child. J Korean Med Sci. 2001;16:359–362. doi: 10.3346/jkms.2001.16.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren F, Tsubota A, Hirokawa T, Kumada H, Yang Z, Tanaka H. A unique amino acid substitution, T126I, in human genotype C of hepatitis B virus S gene and its possible influence on antigenic structural change. Gene. 2006;383:43–51. doi: 10.1016/j.gene.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Katsoulidou A, Paraskevis D, Magiorkinis E, Moschidis Z, Haida C, Hatzitheodorou E, Varaklioti A, Karafoulidou A, Hatzitaki M, Kavallierou L, Mouzaki A, Andrioti E, Veneti C, Kaperoni A, Zervou E, Politis C, Hatzakis A. Molecular characterization of occult hepatitis B cases in Greek blood donors. J Med Virol. 2009;81:815–825. doi: 10.1002/jmv.21499. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Tachiquín ME, Valdez-Salazar HA, Juárez-Barreto V, Dehesa-Violante M, Torres J, Muñoz-Hernández O, Alvarez-Muñoz MT. Molecular analysis of hepatitis B virus “a” determinant in asymptomatic and symptomatic Mexican carriers. Virol J. 2007;4:6. doi: 10.1186/1743-422X-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lusida MI, Nugrahaputra VE, Soetjipto Handajani R, Nagano-Fujii M, Sasayama M, Utsumi T, Hotta H. Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia. J Clin Microbiol. 2008;46:2160–2166. doi: 10.1128/JCM.01681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Utsumi T, Lusida MI, Yano Y, Nugrahaputra VE, Amin M, Juniastuti Soetjipto, Hayashi Y, Hotta H. Complete genome sequence and phylogenetic relatedness of hepatitis B virus isolates in Papua, Indonesia. J Clin Microbiol. 2009;47:1842–1847. doi: 10.1128/JCM.02328-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


