ABSTRACT
Cryptococcus neoformans and Cryptococcus gattii are globally distributed human fungal pathogens and the leading causes of fungal meningitis. Recent studies reveal that myo-inositol is an important factor for fungal sexual reproduction. That C. neoformans can utilize myo-inositol as a sole carbon source and the existence of abundant inositol in the human central nervous system suggest that inositol is important for Cryptococcus development and virulence. In accord with this central importance of inositol, an expanded myo-inositol transporter (ITR) gene family has been identified in Cryptococcus. This gene family contains two phylogenetically distinct groups, with a total of 10 or more members in C. neoformans and at least six members in the sibling species C. gattii. These inositol transporter genes are differentially expressed under inositol-inducing conditions based on quantitative real-time PCR analyses. Expression of ITR genes in a Saccharomyces cerevisiae itr1 itr2 mutant lacking inositol transport can complement the slow-growth phenotype of this strain, confirming that ITR genes are bona fide inositol transporters. Gene mutagenesis studies reveal that the Itr1 and Itr1A transporters are important for myo-inositol stimulation of mating and that functional redundancies among the myo-inositol transporters likely exist. Deletion of the inositol 1-phosphate synthase gene INO1 in an itr1 or itr1a mutant background compromised virulence in a murine inhalation model, indicating the importance of inositol sensing and acquisition for fungal infectivity. Our study provides a platform for further understanding the roles of inositol in fungal physiology and virulence.
IMPORTANCE
Cryptococcus neoformans is an AIDS-associated human fungal pathogen that causes over 1 million cases of meningitis annually and is the leading cause of fungal meningitis in immunosuppressed patients. The initial cryptococcal infection is caused predominantly via inhalation of sexual spores or desiccated yeast cells from the environment. How this fungus completes its sexual cycle and produces infectious spores in nature and why it frequently infects the central nervous system to cause fatal meningitis are critical questions that remain to be understood. In this study, we demonstrate that inositol acquisition is important not only for fungal sexual reproduction but also for fungal virulence. We identified an expanded inositol transporter gene family that contains over 10 members, important for both fungal sexual reproduction and virulence. Our work contributes to our understanding of how fungi respond to the environmental inositol availability and its impact on sexual reproduction and virulence.
INTRODUCTION
Inositol is essential for all eukaryotes, including model and pathogenic fungi. myo-Inositol is the precursor of phosphatidylinositol (PI), which plays key roles in both cellular structure and intracellular signal regulation. The intracellular inositol level is precisely regulated by specific PI kinases, phosphatases, and phospholipases. Inositol polyphosphates derived from PI orchestrate myriad cell functions, including nuclear export, telomere length, chromatin remodeling, and transcription (1–5). Inositol-derived products have been reported to be important for fungal pathogenicity. Some enzymes involved in sphingolipid biosynthesis and degradation pathways have also been found to promote pathogenicity in Cryptococcus neoformans, such as inositol-phosphoryl ceramide synthase 1 (Ipc1) (6) and inositol phosphosphingolipid-phospholipase C1 (Isc1) (7). The diacyglycerol (DAG)-protein kinase C1 (Pkc1) branch signaling pathway is also critical for virulence factor production and pathogenicity in C. neoformans (8–11). Inositol also functions as the precursor for phospholipomannan, a glycophosphatidylinositol (GPI)-anchored glycolipid on the cell surface of Candida albicans that binds to human macrophages and is necessary for pathogenicity (12).
Inositol can serve as a carbon source for fungi. There are two main sources by which fungal cells can acquire inositol. One route involves the conversion of intracellular glucose into myo-inositol by a multiple-step inositol biosynthesis pathway (13). Inositol 1-phosphate synthase (Ino1) is the key enzyme for this synthetic route and converts glucose 6-phosphate to inositol 3-phosphate in the rate-determining step (14). Inositol can also be imported from the extracellular environment via inositol transporters. The myo-inositol transporter gene family is part of the sugar transporter superfamily and plays an important role in inositol acquisition in fungi, including Saccharomyces cerevisiae (15–20), C. albicans (13, 21, 22), and Schizosaccharomyces pombe (23).
There are two myo-inositol transporters (Itrs) in S. cerevisiae, which were first isolated by complementation of a yeast mutant defective in myo-inositol uptake. Itr1 is the major transporter, and its abundant mRNA is transcriptionally and posttranslationally repressed by inositol and choline. Itr2 is a minor transporter that is constitutively expressed at a low level (18, 19). Depletion of inositol from the growth medium stimulates ITR1 expression, while addition of inositol to the medium triggers repression of ITR1 expression and inhibits uptake activity (15). The C. albicans myo-inositol transporter CaItr1 exhibits high substrate specificity for inositol, and interactions between the C-2, C-3, and C-4 hydroxyl groups of myo-inositol and the transporter are critical for substrate recognition and binding (22). A recent study showed that, similar to S. cerevisiae, C. albicans can generate inositol de novo through Ino1 and also import it from the environment through CaItr1. C. albicans may utilize these two complementary mechanisms to obtain inositol during host infection (21). There is another phylogenetically distinct transporter (orf19.5447) that may represent a second ITR gene, but it is expressed at a low level and less well studied (21). The fission yeast S. pombe is a natural auxotroph for inositol due to the absence of Ino1 and therefore cannot grow in the absence of inositol. Two transporters, Itr1 and Itr2, are involved in inositol uptake in S. pombe. High concentrations of inositol in the culture medium stimulate mating and sporulation, while a low concentration supports only vegetative growth (23). It was reported that myo-inositol regulates the production of pheromone P and the response of cells to pheromones, but production of pheromone M is inositol independent. It is likely that inositol or one of its metabolites is involved in pheromone P secretion and pheromone signaling and thereby influences sexual reproduction (24).
Cryptococcus neoformans is a major AIDS-associated human fungal pathogen that often infects immunocompromised individuals to cause fatal meningoencephalitis. Inositol metabolism is important for the growth and development of C. neoformans and may be involved in its survival both in environmental niches, including plants and soil, and in humans and other hosts (25, 26). The development of cryptococcosis is thought to be initiated by inhalation of spores and/or desiccated yeasts from the environment, as no human-to-human transmission (other than iatrogenic) has been reported, and spores are small enough to lodge in the alveoli of the lung (27). Recent studies also demonstrate that spores are fully virulent (28, 29). Our recent study revealed that high concentrations of inositol on plants (~6.5 µg/cm2 on the Eucalyptus camaldulensis leaf surface) and in media stimulate Cryptococcus to complete the sexual cycle and produce spores, which could provide an explanation as to how spores are produced in nature (26). Understanding how inositol triggers fungal mating will provide insights into the interactions between C. neoformans and its environmental niches.
The high rate of cryptococcal meningitis may be related to the elevated inositol levels found in the mammalian brain and the preference of the fungus for proliferation in this tissue (30–32). Inositol concentrations in human cerebrospinal fluid (CSF) are ~25 mg/liter compared to an average concentration of 4.3 mg/liter in plasma (33). Serial analysis of gene expression (SAGE) has been applied to characterize gene expression profiles during experimental cryptococcal meningitis in a rabbit model (34). The INO1 gene and inositol monophosphatase were found to be abundant in a SAGE library generated from RNAs isolated during brain infection, suggesting that the myo-inositol internal synthesis pathway is functional and that inositol could be important for the development of meningitis (34). Inositol metabolism has also been linked to PKA signaling, which is critical for the virulence of C. neoformans (35). Another unique feature of C. neoformans is that it can utilize inositol as a sole carbon source, which was first described in 1976 (36, 37). We recently identified an undefined myo-inositol transporter gene family that contains seven members with high similarity based on the ITR sequences in S. cerevisiae (26). It is unusual to find this expanded family of inositol transporters in C. neoformans, considering that most other fungi have only two or three related inositol transporters; this finding also supports the importance of inositol for Cryptococcus physiology and virulence.
In this study, we describe the full suite of expanded ITR genes found in Cryptococcus. Our results with real-time PCR showed that these ITR genes are differentially expressed in response to myo-inositol induction. Phenotypic complementation of an S. cerevisiae itr1 itr2 double mutant strain by some ITR genes from C. neoformans supports the idea that these ITR homologs are bona fide inositol transporters. Phenotypic analyses of itr mutants revealed that inositol acquisition is important for fungal mating and that virulence and functional redundancy exist among ITR genes and between the ITR gene family and the inositol internal biosynthesis pathway. Our results further support the hypothesis that inositol is important for C. neoformans sexual reproduction and virulence.
RESULTS
myo-Inositol is a key compound for triggering mating and sporulation.
In previous studies, we discovered a defined medium, modified MS (Murashige and Skoog) medium, which supports the efficient mating of both C. neoformans and Cryptococcus gattii (26). By omission and readdition of components, myo-inositol was shown to be the active component stimulating mating. Standard MS medium contains 100 mg/liter myo-inositol. myo-Inositol was added at different concentrations to MS inositol dropout medium, and a concentration as low as 15 mg/liter myo-inositol was found to stimulate mating. A medium made from V8 juice is classically used for mating assays in C. neoformans. In related studies, the myo-inositol concentration in V8 juice has been measured (38). Based on this information, standard V8 mating medium contains 16.675 mg/liter inositol, and this level is sufficient to support mating in MS medium. The addition of more myo-inositol to V8 medium triggers increased mating filament and spore production (26). These observations suggest that myo-inositol is an active compound in V8 medium that stimulates mating. Compared with mating on V8 medium, mating on MS medium shows significantly increased sporulation efficiency.
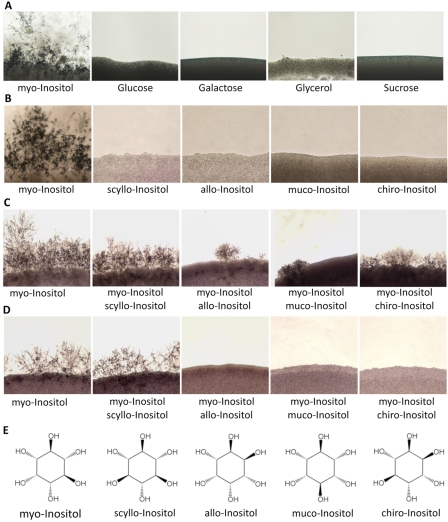
Because myo-inositol is the main carbon source in MS medium, we replaced myo-inositol with other carbon sources, including glucose, sucrose, galactose, and glycerol. None of those sugars stimulated mating at the same concentration, indicating that myo-inositol elicits mating in addition to its known ability to serve as a carbon source (Fig. 1A).
FIG 1 .
Mating stimulation is myo-inositol specific. (A) Mating assays between strains H99 and KN99a were performed in MS medium containing different carbon sources at a concentration of 100 mg/liter, including myo-inositol, glucose, galactose, glycerol, or sucrose. No other tested carbon source except myo-inositol stimulated fungal mating. (B) Mating assays were performed in MS medium containing different inositol isomers, including myo-, scyllo-, allo-, muco-, and chiro-inositol. No other isomers except myo-inositol stimulated mating at a concentration of 100 mg/liter. (C and D) Mating assays were performed in MS medium containing 50 mg/liter (C) or 20 mg/liter (D) myo-inositol and 100 mg/liter of each other form, such as scyllo-, allo-, muco-, or chiro-inositol. The presence of allo-, muco-, or chiro-inositol competed with myo-inositol and inhibited mating. No obvious inhibition by scyllo-inositol was observed. (E) Structures of the inositol isomers studied.
We also tested several other inositol isomers, including scyllo-inositol, allo-inositol, muco-inositol, and chiro-inositol. None stimulated mating, indicating that mating stimulation is specific to myo-inositol (Fig. 1B). We have also investigated whether any of these inositol isoforms can compete to inhibit myo-inositol-stimulated mating. By adding 100 mg/liter of each isomer to MS medium containing 50 mg/liter myo-inositol, respectively (yielding a 2:1 ratio), a clear inhibition of mating by allo-, muco-, or chiro-inositol was observed (Fig. 1C). Lowering the myo-inositol concentration to 20 mg/liter in these experiments to yield a 5:1 ratio (at a constant concentration of 100 mg/liter of the inositol isomers) completely blocked mating filament production (Fig. 1D). These results suggest that allo-, muco-, and chiro-inositol antagonize mating by competing with myo-inositol. However, no antagonistic effect was observed by adding scyllo-inositol to MS medium with 20 mg/liter or 50 mg/liter myo-inositol (Fig. 1C and D). Because scyllo-inositol is an inactive form of inositol, these results may reflect an inability of scyllo-inositol to compete with myo-inositol for binding.
Identification of an expanded ITR gene family.
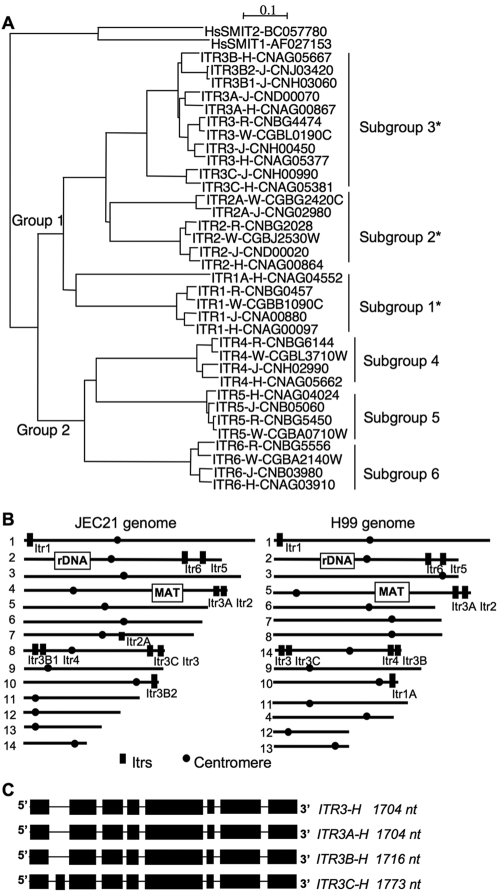
Fungal cells can acquire inositol from the following two main sources: internal inositol synthesis from glucose or transport from the environment via inositol transporters. We have identified a large myo-inositol transporter (ITR) gene family in Cryptococcus as encoding potential inositol permeases or sensors based on sequence identity shared with inositol transporter genes identified in S. cerevisiae and S. pombe (26). Because the ITR genes belong to the sugar transporter superfamily, here we identified all sugar transporter homologs from the genomes of the C. neoformans strain H99, S. cerevisiae, and C. albicans, and the phylogenetic relationships among them were compared (see Fig. S1 in the supplemental material). Homologs of Itrs in S. cerevisiae and C. albicans form a cluster in the phylogenetic tree generated. A total of 10 proteins in H99 were considered ITR candidates and included in this cluster. We then compared all of the Itr homologs identified from the following four complete Cryptococcus genome sequences: C. neoformans var. grubii H99 (serotype A, VNI), C. neoformans var. neoformans JEC21 (serotype D, VNIV), C. gattii WM276 (serotype B, VGI), and C. gattii R265 (serotype B, VGII) (Table 1). The Itr homologs can be divided into two distinct groups (Fig. 2A). The group 1 genes are homologs of Itr genes identified from S. cerevisiae and S. pombe and of the Itr1 gene from C. albicans. The group 2 genes are homologs of the HGT19 (orf19.5447) gene from C. albicans, which also exhibits myo-inositol transporter activity based on computational analysis presented in the Candida Genome Database (http://www.candidagenome.org/) but has not been extensively studied (21). The first group may have undergone recent duplication because the numbers of Itr paralogs differ among Cryptococcus strains, while the second group is well conserved among all of the strains that were compared. Based on the similarity among these two groups of Itrs in Cryptococcus, they can be further divided into six different subgroups (Itr1 to -6). Among these, group 1 Itrs can be further divided into subgroups 1, 2, and 3, while group 2 Itrs can be divided into subgroups 4, 5, and 6. Subgroups 4, 5, and 6 are well conserved, and each subgroup has one Itr (Itr4, Itr5, and Itr6) from each Cryptococcus strain. Interestingly, subgroup 3 has expanded in C. neoformans to include four homologs in H99 (ITR3, ITR3A, ITR3B, and ITR3C) and five homologs in JEC21 (ITR3, ITR3A, ITR3B1, ITR3B2, and ITR3C) but only one each for R265 and WM276 (Fig. 2A; Table 1).
TABLE 1 .
Potential myo-inositol transporters in Cryptococcus species
| Strain | Gene name | Locus tag | Chromosome |
|---|---|---|---|
| H99 | ITR1-H | CNAG_00097 | 1 |
| H99 | ITR1A-H | CNAG_04552 | 10 |
| H99 | ITR2-H | CNAG_00864 | 5 |
| H99 | ITR3-H | CNAG_05377 | 14 |
| H99 | ITR3A-H | CNAG_00867 | 5 |
| H99 | ITR3B-H | CNAG_05667 | 14 |
| H99 | ITR3C-H | CNAG_05381 | 14 |
| H99 | ITR4-H | CNAG_05662 | 14 |
| H99 | ITR5-H | CNAG_04024 | 2 |
| H99 | ITR6-H | CNAG_03910 | 2 |
| JEC21 | ITR1-J | CNA00880 | 1 |
| JEC21 | ITR2-J | CND00020 | 4 |
| JEC21 | ITR2A-J | CNG02980 | 7 |
| JEC21 | ITR3-J | CNH00450 | 8 |
| JEC21 | ITR3A-J | CND00070 | 4 |
| JEC21 | ITR3B1-J | CNH03060 | 8 |
| JEC21 | ITR3B2-J | CNJ03420 | 10 |
| JEC21 | ITR3C-J | CNH00990 | 8 |
| JEC21 | ITR4-J | CNH02990 | 8 |
| JEC21 | ITR5-J | CNB05060 | 2 |
| JEC21 | ITR6-J | CNB03980 | 2 |
| WM276 | ITR1-W | CGB_B1090C | 2 |
| WM276 | ITR2-W | CGB_J2530W | 10 |
| WM276 | ITR2A-W | CGB_G2420C | 7 |
| WM276 | ITR3-W | CGB_L0190C | 12 |
| WM276 | ITR4-W | CGB_L3710W | 12 |
| WM276 | ITR5-W | CGB_A0710W | 1 |
| WM276 | ITR6-W | CGB_A2140W | 1 |
| R265 | ITR1-R | CNBG_0457 | Supercontig 1 |
| R265 | ITR2-R | CNBG_2028 | Supercontig 4 |
| R265 | ITR3-R | CNBG_4474 | Supercontig 11 |
| R265 | ITR4-R | CNBG_6144 | Supercontig 23 |
| R265 | ITR5-R | CNBG_5450 | Supercontig 16 |
| R265 | ITR6-R | CNBG_5556 | Supercontig 16 |
FIG 2 .
Phylogenetic analysis of myo-inositol transporter (Itr) proteins in C. neoformans and C. gattii. (A) Potential myo-inositol transporters from H99, JEC21, WM276, and R265 were identified using BLASTn and BLASTp analyses. The phylogram was generated using ClustalX 2.0.1 and viewed with the TreeView program. Two human sodium-dependent myo-inositol transporters (HsSMIT1 and HsSMIT2) served as outgroup controls. *, genes with closely related duplicated paralogs. (B) ITR gene locations in the genomes of JEC21 and H99. All ITR homologs in JEC21 and H99 were labeled at the appropriate locations on the 14 chromosomes of the genomes. (C) Subgroup 3 ITR genes of H99 have similar intron locations. Transcripts of ITR3, ITR3A, ITR3B, and ITR3C are shown in black, and lines indicate introns in each gene. nt, nucleotides.
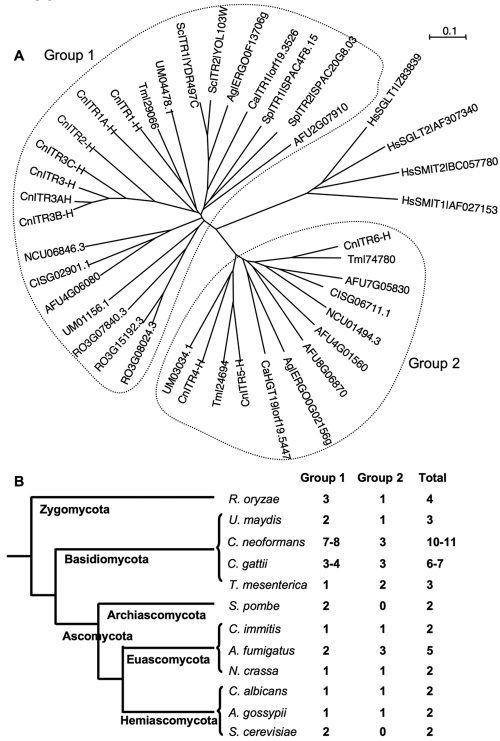
When the group 1 and group 2 Itrs from Cryptococcus were compared with related Itr homologs from other fungi, group 1 was found to share a high level of identity with the Itrs in other yeasts, such as S. cerevisiae and S. pombe. There are two Itrs in S. cerevisiae and S. pombe, and both cluster within group 1. In most other ascomycetes with completed genome sequences, only one ITR gene was found in this group, including in hemiascomycetes (C. albicans and Ashbya gossypii) and euascomycetes (Coccidioides immitis and Neurospora crassa) (Fig. 3). Two basidiomycete genomes were used for comparison. The closely related species Tremella mesenterica has only one ITR homolog, while the corn smut fungus Ustilago maydis has two (Fig. 3). One exception in this group is the zygomycete Rhizopus oryzae, which has three Itr paralogs in group 1.
FIG 3 .
Phylogenetic analysis of myo-inositol transporter (Itr) proteins among different fungi. (A) myo-Inositol transporter homologs from the Hemiascomycota (S. cerevisiae, A. gossypii, and C. albicans), Euascomycota (N. crassa, Aspergillus fumigatus, and C. immitis), Archiascomycota (S. pombe), Basidiomycota (U. maydis and C. neoformans var. grubii), and Zygomycota (R. oryzae) were identified based on BLAST searches. The human sodium-dependent inositol transporters served as outgroup controls. (B) Numbers of Itr homologs in different fungi.
There are three Itrs in the four Cryptococcus strains that belong to group 2. Most fungi analyzed have only one Itr that belongs to group 2, including C. albicans, A. gossypii, C. immitis, N. crassa, U. maydis, and R. oryzae. T. mesenterica has two Itrs that belong to group 2, and interestingly, A. fumigatus has three group 2 representatives.
Based on the comparative genomic analysis, it is clear that the ITR gene family in Cryptococcus has expanded (Fig. 3). Because C. neoformans is an environmental microorganism that associates with a variety of niches, the expansion of the ITR gene family may reflect adaptation to nutrient-limited niches, such as survival on plants or in soil containing plant debris, where free inositol or phytic acid (IP6) is abundant. C. neoformans can utilize inositol as a sole carbon source, which may require this fungus to have more efficient inositol transport systems both to survive in natural habitats and to successfully infect the host and cause infection.
Most of the ITR genes in C. neoformans are located in telomeric regions of the chromosomes, which may have contributed to the expansion of this gene family (Fig. 2B) via recombination-mediated exchange of telomeric and subtelomeric regions between different chromosomes. Interestingly, in JEC21 chromosome 8 and its corresponding chromosome, 14, in H99, four ITR genes are located in telomeric regions, and three of these belong to subgroup 3, the least conserved transporter cluster. In addition, all four genes in subgroup 3 of H99 have very similar intron locations in the genes except ITR3C, which contains an extra exon (Fig. 2C). Comparison of 20-kb flanking sequence regions of the ITR genes in JEC21 and H99 revealed synteny between ITR3A (chromosome 4) and ITR3B2 (chromosome 10) in JEC21, an indication that this genomic region underwent duplication (see Fig. S2 in the supplemental material). These observations suggest that the subgroup 3 ITR genes on these chromosomes may have undergone recent duplications. In this study, we focused further effort on functional analyses of the seven group 1 Itr homologs in H99 to understand their roles in mating and virulence of C. neoformans.
Inositol transporters are differentially expressed under different culture conditions.
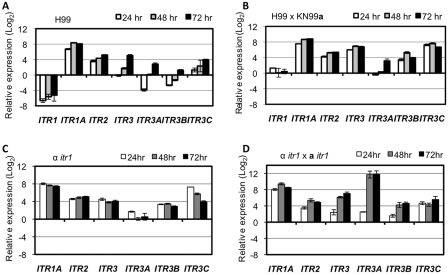
To understand how the Itrs are involved in inositol sensing and transport, quantitative real-time PCR (qRT-PCR) was performed to detect the expression of ITR genes during growth under high-inositol conditions and during mating on MS medium containing inositol. In this study, we designed specific primers for seven ITR genes of H99 that belong to group 1 and that are closely related to well-studied ITR genes in S. cerevisiae and S. pombe. Cells of strain H99 and mating coculture of H99 and KN99a were inoculated on MS medium containing 100 mg/liter inositol. Cells were recovered from the medium surface at 24 h, 48 h, and 72 h postinoculation. Total RNAs were purified, and first-strand cDNAs were synthesized. Each cDNA sample was used as a template to amplify all seven ITR genes using gene-specific primers in a qRT-PCR. Our qRT-PCR results showed that these ITR genes are differentially expressed both during mating and during growth as a single strain in the presence of inositol (Fig. 4). Within the first 24 h of incubation, the mating mixture undergoes cell fusion, and all seven ITR genes were found to be expressed. Increased expression was observed when incubation was continued for 48 h or 72 h. The expression of ITR1 at all time points tested was significantly repressed in the single-strain culture but was not repressed during mating. Meanwhile, ITR1A and ITR2 were constitutively highly induced under both culture conditions, especially ITR1A, which represented the highest expression at all conditions; this suggests that these two may play important roles in inositol uptake. ITR3, ITR3B, and ITR3C are highly induced only during mating but not in the single-strain culture, possibly also consistent with a role in mating. ITR3A was expressed at a low level under both culture conditions, and thus, it may not play a significant role in inositol transport.
FIG 4 .
Comparison of the expression of seven ITR genes in C. neoformans at both mating and growth stages in the presence of inositol. H99 cultures (A), H99 × KN99a mating mixtures (B), the α itr1 cultures (C), and itr1 × itr1 mating cultures (D) were spotted on agar plates and collected at 24, 48, and 72 h after plating. Gene expression was measured by qRT-PCR in triplicate, and the comparative CT method was used for the relative quantification. Values are expressed as relative expression (log2) (means ± SD) of ITR genes, normalized to the GAPDH gene endogenous reference and relative to the 0-h time point (H99 overnight liquid culture was considered the 0-h time point). Error bars show standard deviations of three repeats.
Overall, our qRT-PCR results suggest that there is a mating effect on the expression of ITR genes, since several ITR genes are expressed at a much higher level in mating culture than in single-strain culture. Our expression profiles of the inositol transporter gene family also provide an important indication that this gene family may play redundant but distinguishable roles in response to inositol, during both mating coculture and growth in monoculture. It will be of interest to investigate the expression profiles of these ITR genes at different sexual development stages or in other strain backgrounds, such as in ino1 and itr mutant strains.
Inositol transporters (Itrs) from C. neoformans complement the growth defect of S. cerevisiae itr1 itr2 double mutants.
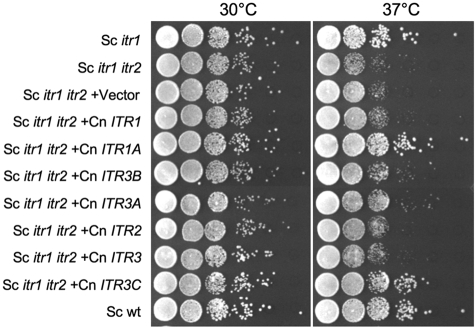
To understand the potential role of the C. neoformans ITR gene family in inositol uptake, we generated S. cerevisiae itr1 itr2 double mutants lacking both inositol transporters. Such double mutants showed a growth defect in yeast extract-peptone-dextrose (YPD) at 37°C. Full-length cDNAs for all seven ITR genes from the C. neoformans H99 strain were cloned into the yeast expression vector pTH19 under the control of the ADH1 promoter and expressed in an S. cerevisiae itr1 itr2 double mutant strain. The heterologous expression of all seven ITR genes of H99 was confirmed by RT-PCR analyses (data not shown). Growth assays were performed for these S. cerevisiae strains expressing C. neoformans ITR genes, and the potential complementation of the S. cerevisiae itr1 itr2 growth defect phenotype was investigated. Expression of ITR1A and ITR3C fully complemented the growth defect of the S. cerevisiae itr1 itr2 mutant strain, while ITR3A and ITR3B partially rescued the slow growth on YPD medium (Fig. 5). Yeast strains expressing ITR1, ITR2, or ITR3, on the other hand, showed growth rates similar to that of the original S. cerevisiae itr1 itr2 mutant strain, suggesting no obvious complementation. These results suggest that Itr1A and Itr3C are important for inositol uptake, which is consistent with our qRT-PCR results suggesting that both ITR1A and ITR3C are highly expressed in response to the availability of environmental myo-inositol. The ITR genes that did not complement the growth defect of S. cerevisiae itr1 itr2 might be caused by either insufficient inositol uptake or expressed proteins that are unstable or not functional.
FIG 5 .
ITR genes from C. neoformans complement the growth defect of an S. cerevisiae itr1 itr2 mutant strain. S. cerevisiae itr1, itr1 itr2, and itr1 itr2 expressing vector pTH19 or one of the seven C. neoformans ITR genes were cultured in YPD or SD-Ura medium. Concentrations of overnight cultures were determined by measuring the optical density at 600 nm (OD600) and adjusted to the same cell density with YPD. Serial 10-fold dilutions were prepared, and 5 µl of each dilution was spotted on YPD plates and incubated at 30°C or 37°C for 48 h before photography. This assay was repeated multiple times, with similar results. Sc, S. cerevisiae; Cn, C. neoformans; wt, wild type.
Itr1 and Itr1A are important for fungal mating.
Gene deletion mutations were generated by biolistic transformation and homologous recombination for all seven ITR genes in C. neoformans var. grubii in both the α and a mating-type backgrounds. Each individual mutation was not sufficient to block sexual reproduction based on mating assays with both MS and V8 mating media. Normal mating hyphae and spores were observed in all bilateral mutants by mutant mating assays. The exception was that bilateral mating between itr1 mutants exhibited reduced mating hypha production (Fig. 6A). Cell fusion assays showed that itr1 mutants have cell fusion efficiency similar to that of wild-type strains, suggesting that the mating defect could be caused by post-cell fusion events, such as insufficient dikaryotic hypha production or a meiosis defect (Fig. 6C). Because expression of ITR1 in an S. cerevisiae itr1 itr2 mutant strain failed to rescue the growth defect of that yeast strain, Itr1 may not be important for inositol uptake but may play other roles, such as functioning as an inositol sensor. The fact that ITR1 expression is repressed by high concentrations of inositol also suggests that Itr1 may be functional as an inositol sensor. To further understand the potential role of Itr1 in the regulation of other Itrs, qRT-PCR analysis was performed to test the expression of the other six ITR genes in the itr1 mutant background, both as single cultures and during mating, using the same approach as that used for wild-type strains (Fig. 4C and D). Our results revealed that ITR3, ITR3B, and ITR3C are expressed at a much higher level in itr1 single cultures than in the H99 wild-type strain. During mating, most ITR genes were highly expressed, with similar expression patterns in both itr1 mating cultures and wild-type strains, except that ITR3A was highly expressed only in itr1 mating cultures (Fig. 4). These results indicate that the ITR3 subgroup genes may be regulated by Itr1, which further suggests that Itr1 could be an inositol sensor that regulates other Itrs.
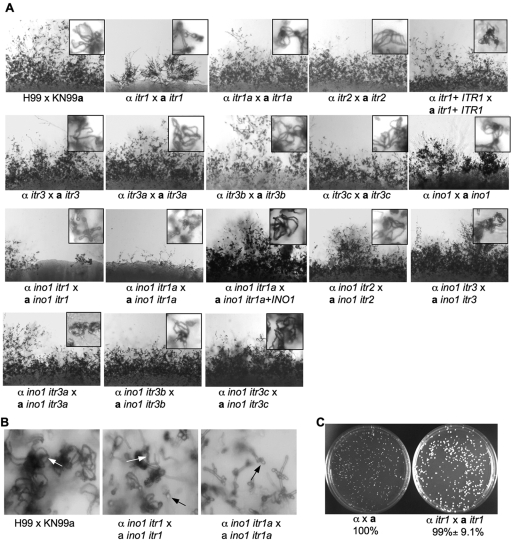
FIG 6 .
Itr1 and Itr1A promote mating in response to inositol. (A) Bilateral mating assays were performed in MS medium with all seven inositol transporter mutants and with double mutants lacking ino1 and each itr gene. Mating cultures were incubated at room temperature in the dark for 7 days before being examined by microscopy and photographed. Both mating filaments (40× magnification) and spores (insets, 400× magnification) were observed for all mating cultures. Mating of itr1, ino1 itr1, and ino1 itr1a mutants showed a defect in mating filament production and sporulation, suggesting an important role in inositol sensing. (B) Bilateral mating cultures of wild-type strains, ino1 itr1 mutants, and ino1 itr1a mutants were incubated under the same conditions as described in the legend to panel A for 14 days, and sporulation results were photographed under 200× magnification. White arrows indicate basidiospore chains; black arrows indicate basidia. (C) Cell fusion assays were performed for wild-type strains (YSB119 and YSB121) and itr1 mutants (CDX175 and CUX50). Cell fusion results were quantified from repeat assays, and representative images are shown.
We have also tested the development of virulence factors and found that all mutants produced normal amounts of capsule and melanin and had normal growth at 37°C (data not shown). The absence of notable phenotypes may indicate that these transporters could be functionally redundant. Another possibility is that although one Itr could play a major role in inositol uptake, the inositol internal biosynthesis pathway might produce sufficient inositol to compensate for reduced inositol uptake from the environment in itr mutants.
The key enzyme for the myo-inositol biosynthetic pathway, inositol 1-phosphate synthase (Ino1), has been identified, and ino1 mutants have been generated, thereby blocking the inositol internal biosynthesis pathway. Importantly, ino1 mutants failed to grow on medium without myo-inositol (data not shown). No obvious mating defect was observed in an ino1 × ino1 bilateral mating assay using MS medium (Fig. 6A). To study the function of Itrs without the potential interference of the inositol biosynthesis pathway, ino1 with itr1, itr1a, itr2, itr3, itr3a, itr3b, or itr3c double mutants were generated in both of the mating-type backgrounds by genetic crossing. While none of the three major virulence factors were significantly altered in these double mutants, mating filament production and sporulation were clearly reduced in bilateral matings of ino1 itr1 × ino1 itr1 and ino1 itr1a × ino1 itr1a double mutants, suggesting that Itr1 and Itr1A are important for inositol transport or sensing (Fig. 6A). Compared with wild-type strains, the ino1 itr1 double mutant produced sporadic mating hyphae and reduced spore production, while the ino1 itr1a double mutant produced evenly distributed mating filaments but at much lower hyphal density and produced very few spores in MS medium. The sporulation defect in the ino1 itr1a double mutant is much more severe than in the ino1 itr1 double mutant (Fig. 6B). Bilateral mating of other double mutants produced normal mating structures, including dikaryotic hyphae and basidiospores. The identification of the ITR gene family and generation of itr and ino1 single and double mutants provide a valuable system to further study how C. neoformans senses and acquires inositol.
Virulence study of inositol transporters in a murine systemic infection model.
Although none of the itr mutants generated showed significant effects on several key virulence factors, such as capsule and melanin production and growth at 37°C (data not shown), their potential involvement in fungal virulence was assessed because virulence is a complex trait. All seven itr mutants (itr1, itr1a, itr2, itr3, itr3a, itr3b, and itr3c) were examined in a murine inhalation model of systemic C. neoformans infection. Groups of 10 female A/JCr mice were intranasally inoculated with 105 yeast cells from each strain, and animals were monitored daily. As previously demonstrated, all mice infected with the wild-type strain H99α survived between 16 and 22 days (39, 40). Mice infected with the mutants exhibited mortality rates similar to those of mice infected with the wild-type H99 strain, indicating that none of these seven itr mutants are attenuated for virulence of C. neoformans under these conditions (Fig. 7). It is possible that the functional redundancy of this gene family or the existence of the inositol biosynthesis pathway is sufficient for the virulence of each single mutant.
FIG 7 .
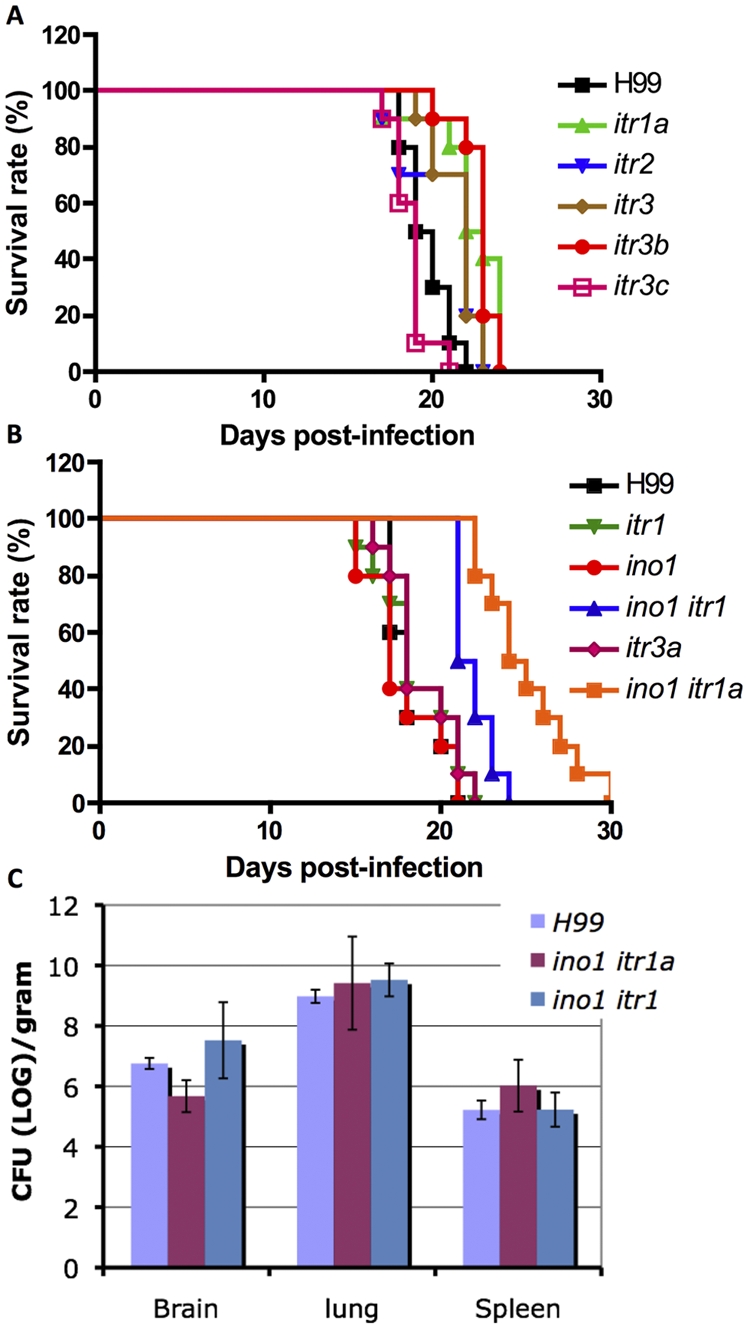
itr mutant strains are still virulent in mice. (A and B) Female A/JCr mice were inoculated intranasally with the following strains: H99, itr1 mutant (CUX7), itr1a mutant (CDX99), itr2 mutant (CDX103), itr3 mutant (CDX105), itr3a mutant (CDX102), itr3b mutant (CDX101), itr3c mutant (CUX42), ino1 mutant (UBCINO1), ino1 itr1 double mutant (CUX17), and ino1 itr1a double mutant (CUX23). Two experiments were performed to finish the virulence test for all above-mentioned strains. Groups of 10 mice were infected with each strain. Animals were monitored for clinical signs of cryptococcal infection and sacrificed at predetermined clinical end points that predict imminent mortality. (C) Fungal burdens in organs infected by ino1 itr1 and ino1 itr1a mutants were compared with those in organs infected by H99 at the end point of the infection. The numbers of colony-forming units (CFU) per gram of organ tissue were measured in brain, lung, and spleen homogenates. Each error bar indicates the standard error of the mean for values obtained from testing three animals.
Virulence studies were also conducted in the murine model for an ino1 mutant, an ino1 itr1 double mutant, and an ino1 itr1a double mutant. While the ino1 mutant was fully virulent, the ino1 itr1 double mutant was moderately attenuated for virulence, with statistical significance (P = 0.001) (Fig. 7). The median survival time was delayed from 18 days in H99 to 21.5 days in the ino1 itr1 double mutant. Interestingly, the ino1 itr1a double mutant exhibited an even more severe virulence attenuation (P < 0.0001), and the median survival time was prolonged to 24.5 days (Fig. 7). Evaluation of fungal burdens at the end point of the infection revealed that brains infected by the ino1 itr1a mutant contained 10-fold fewer yeast cells than those with H99 infection, which may indicate that the ino1 itr1a mutant either has a defect in central nervous system (CNS) entry or proliferates at a lower rate in the brain (Fig. 7C). These results indicate that both Itr1 and Itr1A are important for inositol acquisition in C. neoformans and that inositol acquisition is important for fungal virulence.
Genes involved in inositol metabolic pathways are highly upregulated by inositol.
To investigate how inositol affects Cryptococcus mating, the effect of inositol on the global transcriptional profile was evaluated by conducting whole-genome microarray experiments. Global gene expression profiles during mating under inositol induction conditions were evaluated in both wild-type strains (H99 and KN99a) and ino1 mutants (α ino1 and a ino1). Mating assays were performed in MS medium without inositol and in MS medium with 100 mg/liter inositol for 48 h before cells were collected and RNA was purified. C-3- and C-5-labeled cDNAs were hybridized to JEC21 70-mer whole-genome arrays. When the transcriptomes of wild-type mating mixtures in MS medium were compared with that in MS medium without inositol, only a small set of genes (less than 50 genes) were upregulated over 2-fold by inositol. A large proportion of these genes encode putative proteins (see Fig. S3 in the supplemental material). Among these, two inositol oxygenase genes (180.m00186 and 177.m03138) were upregulated over 10-fold. An α-glucoside transporter gene was also significantly upregulated (Table 2). When the transcriptomes of ino1 mutant mating mixtures in MS medium with and without inositol were compared, over 300 genes were found to be upregulated over 2-fold by inositol. Inositol oxygenase and glucoside transporter genes were also among the genes with the highest induction levels. The pheromone receptor Ste3 was found to be upregulated in both of the experiments. In the ino1 mutant results, more genes involved in inositol sensing and metabolism were found to be upregulated, including a phosphatidylinositol 3-kinase gene and several inositol transporter genes, including ITR1, ITR2, ITR3A, and ITR3B2, indicating that ino1 mutant cells are more responsive to environmental inositol than wild-type cells. Results obtained from such systemic analyses provide rich information for further understanding of inositol acquisition and cellular function in C. neoformans, especially with respect to sexual reproduction and fungal virulence.
TABLE 2 .
Genes upregulated by inositol during mating
| Gene and function | Product/description | Average fold change in gene upregulation |
|
|---|---|---|---|
| α ino1 × a ino1 | H99 × KN99a | ||
| Metabolism | |||
| 180.m00186 | myo-Inositol oxygenase | 38.05 | 11.81 |
| 177.m03138 | Inositol oxygenase | 14.82 | 17.79 |
| 184.m04662 | Isocitrate lyase | 4.93 | 1.78 |
| 184. m04739 | Phosphatidylinositol 3-kinase | 3.41 | 0.86 |
| 162.m02888 | Phosphoenolpyruvate carboxylase | 3.14 | 4.59 |
| 181.m07830 | Fructose-1,6-bisphosphatase | 2.85 | 2.23 |
| 162. m02888 | Phosphoenolpyruvate carboxylase | 3.14 | 4.59 |
| 185.m02442 | Pyruvate decarboxylase-related protein | 2.32 | 2.74 |
| 179.m00355 | Cytochrome c oxidase | 1.04 | 3.05 |
| 181.m08517 | Acetate-coenzyme A ligase | 1.55 | 2.88 |
| 177.m03189 | Malate dehydrogenase | 1.12 | 2.44 |
| 180.m00160 | Formate dehydrogenase | 1.57 | 2.75 |
| Transporter/receptor | |||
| 180.m00244 | α-Glucoside transporter | 11.51 | 5.16 |
| 180.m00262 | α-Glucoside transporter | 3.01 | 0.97 |
| 162.m02883 | α-Glucoside transporter | 3.44 | 1.01 |
| 177.m03407 | Itr2 | 3.02 | 0.70 |
| 185.m02698 | Itr3B2 | 2.78 | 1.17 |
| 181.m07866 | Itr1 | 2.34 | 1.4 |
| 163.m03771 | Itr3A | 1.99 | 1.75 |
| 179.m00178 | Putative sugar transporter | 3.38 | 0.71 |
| 186.m03862 | Ste3 | 2.05 | 2.07 |
| 164.m01642 | Amino acid transporter, putative | 2.53 | 0.76 |
| 177.m03009 | GDP-mannose transporter | 2.59 | 1.03 |
| 183.m01606 | Hxt1, putative | 3.45 | 0.85 |
| 185.m02415 | Uracil permease | 2.59 | 1.31 |
DISCUSSION
Inositol metabolism and catabolism have been found to play important roles in both cellular structure and intracellular signaling and, thus, are critical for cell development. Our previous study identified myo-inositol as an important elicitor for Cryptococcus mating, both in media and on living plants (26). The importance of myo-inositol for fungal mating and the facts that Cryptococcus can utilize myo-inositol as a sole carbon source and that the human brain contains a high concentration of myo-inositol all indicate that myo-inositol is important for this fungus. In accord, we found that Cryptococcus has an unusually large, expanded inositol transporter gene family. These findings raise the following questions. How do fungal cells sense and transport inositol, and how does inositol affect fungal mating? What is the role of each inositol transporter? Does inositol sensing/transport play a role in fungal virulence? Is there a correlation or causative role between inositol sensing and the high rate of cryptococcal meningitis?
It is likely that the presence of a large ITR gene family in Cryptococcus is the result of coevolution between this yeast and its environmental niches and host conditions. As one major environmental niche, soil often contains abundant decayed plant materials that are rich in phytic acid (IP6) and other inositol-derived compounds. Although the addition of phytic acid to media did not yield a clear effect on fungal development or mating (data not shown), this may be because IP6 is charged and cannot enter fungal cells directly; IP6 may be converted into other inositol forms that can be utilized by Cryptococcus. Cryptococcus species, especially C. gattii, are commonly isolated from living plants, including Eucalyptus species, as environmental niches. We found that free myo-inositol exists on several living plant surfaces, and the concentration on Eucalyptus is much higher than on Arabidopsis, which could have significant implications for how this human pathogen completes its sexual cycle in nature, since Eucalyptus is the major environmental niche for C. gattii (26).
Sexual reproduction in Cryptococcus is important for both genetic manipulation and the production of the suspected primary infectious particles (basidiospores) and has been extensively studied (28, 29, 41–44). In Cryptococcus species that infect humans and animals, C. neoformans strains are mostly fertile. C. neoformans var. neoformans (serotype D) is even more fertile than C. neoformans var. grubii (serotype A), while most isolates of the sibling species C. gattii strains (serotype B and serotype C) have lower levels of mating efficiency or are even sterile. We found that the serotype D strain JEC21 has 11 potential ITR genes, more ITR homologs than any other Cryptococcus strain, while the serotype A strain H99 contains 10 ITR genes, the serotype B VGI strain WM276 has 7, and the serotype B VGII strain R265 has only 6 ITR homologs. Thus, there is an interesting potential correlation between the number of ITR genes in each serotype strain and fertility. Further functional studies are required to verify whether all of the candidate ITR genes indeed encode functional inositol transporters. The comparison of ITR genes among different Cryptococcus strains clearly indicates that subgroup 3 has undergone a major expansion in C. neoformans compared to those in C. gattii. The facts that most of the ITR genes in this subgroup are localized in the telomeric regions of a single chromosome and that all of them share similar intron locations provide further insight into this expansion. Besides environmental niches, human and animal brains have been widely reported to have high concentrations of myo-inositol (30–32, 45, 46). Because the major lethal infection caused by Cryptococcus is in the CNS and results in cryptococcal meningoencephalitis, there may be a potential correlation between inositol sensing and acquisition and cryptococcal pathogenesis. It is possible that the organism adapted to inositol-rich environments by expanding the ITR gene family to promote survival under harsh host conditions.
The subtelomeric location of the ITR gene family is interesting and may explain the rapid expansion of ITR genes in C. neoformans, since genes in such a chromosomal location are commonly rapidly evolving. Our functional genomic analysis also revealed gene duplication events within the genomes of H99 and JEC21 (see Fig. S2 in the supplemental material). Other large gene families have also been observed to have telomeric or subtelomeric locations. The EPA (epithelial adhesin) gene family in Candida glabrata is important for cell adhesin and contains at least 17 members in the CBS138 strain and 23 in the BG2 strain, and most of them are located in telomeric or subtelomeric regions (47). Interestingly, many are subject to chromatin-based transcriptional silencing that involves telomere-associated proteins, including the Rap1, Hdf1, and Sir proteins (Sir2, Sir3, and Sir4) (47–49). In contrast, we did not observe such transcriptional silencing for the ITR genes we studied, because all seven ITR genes in H99 were expressed based on our RT-PCR results (Fig. 4).
We identified two groups of ITR homologs based on the known inositol transporter sequences in other yeasts; functional studies are important to verify whether they are indeed involved in inositol transport or sensing. In this study, we focused on the seven ITR homologs in group 1 of H99. Our quantitative RT-PCR results showed that all seven ITR genes are expressed with different expression patterns in response to inositol induction, an indication that this gene family might have both functional redundancy and specificity. Overall, most ITR genes were expressed at higher levels at the later time points, suggesting that at later incubation stages, fungal cells require more inositol from the environment as internal inositol and other stored carbon sources are depleted, triggering the induction of ITR expression. Also, most ITR genes were induced at a higher level in strains during mating than when individual strains were grown alone, suggesting that mating may require more inositol, either as an energy source, a signaling compound, or both. ITR1A and ITR3C were highly induced by inositol, and both of them complemented the growth defect of the S. cerevisiae itr1 itr2 double mutant in YPD at 37°C, suggesting that these two are bona fide ITR genes and play important roles in inositol uptake in H99. Inositol uptake assays in this yeast heterologous expression system will be important to understand the inositol affinity and specificity for each Itr from C. neoformans.
Interestingly, single-gene deletion mutants of the first seven ITR genes in group 1 still undergo cell fusion, mating filament production, and sporulation, indicating that none of these Itrs is essential for fungal mating. However, the mating dikaryotic hyphal production in the bilateral itr1 × itr1 mutant mating was reduced, especially at early time points, indicating that Itr1 may be important for fungal inositol acquisition. In our qRT-PCR study, ITR1 is the only ITR gene whose expression was consistently repressed when grown in MS medium as a single strain. Because the expression of the major inositol transporter in S. cerevisiae, ITR1, is also repressed when additional inositol is available (18), Itr1 could also be important for inositol transport or sensing in C. neoformans. The fact that expression of ITR1 in the S. cerevisiae itr1 ir2 double mutant strain failed to rescue the YPD growth mutant phenotype suggests that Itr1 may be a sensor rather than a transporter. In S. cerevisiae, a large glucose transporter gene family has been identified to differentially control glucose uptake, and two permease homologs, Snf3 and Rgt2, function as sensors rather than transporters; these two proteins play important roles in regulating the expression of the other hexose transporters (50, 51). A similar paradigm may exist in Cryptococcus to sense inositol using one or more members of the expanded ITR gene family. The significant changes in other expression patterns of ITR genes in the itr1 mutant background provide further evidence that Itr1 could be an inositol sensor that regulates the function of other Itrs. It is also possible that the expression of ITR1 from C. neoformans in S. cerevisiae may not be stable or functional. On the other hand, Itr1A is clearly an important inositol transporter in C. neoformans because ITR1A is constitutively highly expressed under inositol induction conditions and it also complemented the growth defect of the S. cerevisiae itr1 itr2 double mutant.
There are two possible explanations as to why none of the itr mutants was completely blocked in mating or significantly altered for virulence factor production. First, potential functional redundancy of these ITR genes could compensate for a defect conferred by any ITR single mutation. Second, it is also possible that since the internal inositol biosynthesis pathway is functional in C. neoformans and converts glucose-6-phosphate into myo-inositol, this may compensate for any defect in inositol uptake caused by an itr mutation. Although none of the seven itr single mutants tested showed an obvious effect on fungal virulence in a murine systemic infection model, the mating and virulence defects observed in the ino1 itr1 and ino1 itr1a double mutants provide evidence for a coordinated inositol acquisition mechanism that includes inositol internal biosynthesis and environmental inositol sensing/transport pathways. The virulence attenuation in these two double mutants supports our hypothesis that inositol acquisition is important not only for fungal sexual reproduction but also for fungal disease development. Interesting, although Itr3C could complement the growth defect of the S. cerevisiae itr1 itr2 mutant strain, ino1 itr3c double mutants did not show any obvious phenotype in either mating or virulence factor development. It will still be of interest to test this double mutant in the murine model in the future to determine whether Itr3c is also involved in virulence. It will also be important and necessary in future studies to generate multiple itr mutations in the ino1 mutant strain background (such as ino1 itr1 itr1a) to understand the function of each Itr in C. neoformans.
We found that ino1 mutants, similar to itr single mutants, C. neoformans are still fully virulent and have normal in vitro growth, which is consistent with what has been reported in C. albicans (21) but different from what has been reported in the bacterial pathogen Mycobacterium tuberculosis (52) and the parasite Trypanosoma brucei (53). Similar to C. neoformans, both M. tuberculosis and T. brucei maintain functional inositol biosynthetic pathways and active inositol transporters, but the imported inositol is not sufficient for full cell activity, and the Ino1 protein is required for cell growth and full virulence. However, despite the fact that the C. albicans itr1 and ino1 homozygous single mutants have normal growth and full virulence, similar to what we found in C. neoformans, inositol is essential for the viability of C. albicans, and the itr1/itr1 ino1/ino1 double mutant is inviable. The lethality of such double mutations suggests that C. albicans also utilizes both the internal biosynthetic pathway and environmental uptake mechanism to acquire inositol, and these two pathways can complement each other (13, 21). Such inositol acquisition machinery could be conserved in other pathogens as well. Because itr1 ino1 double mutants of C. albicans are inviable, it is a challenge to score an effect on virulence. The conditional double mutant (ino1/ino1 itr1/PMET3::ITR1) was found to be avirulent, suggesting that inositol is also important for the development of candidiasis (21). In C. neoformans, ino1 itr1 and ino1 itr1a double mutants have normal in vitro growth and show virulence attenuation, which indicates that Cryptococcus has a more well-developed system for acquiring inositol than Candida albicans or other pathogens and thus provides a valuable system to evaluate the effect of inositol sensing/transporting on fungal disease development.
It is still unclear how these mutations affect virulence. Although ino1 itr1 and ino1 itr1a double mutants show virulence attenuation, they still produce normal virulence factors in vitro and cause lethal infection. One possibility is that the high concentrations of inositol in the human central nervous system (CNS) play a role in the development of cryptococcal meningitis. C. neoformans cells may sense inositol via a mechanism similar to chemotaxis in other microorganisms. Such a chemotactic response could attract yeast cells to cross the blood-brain barrier (BBB) and to reach the CNS, where large amounts of inositol are present. Once inside the brain, yeast cells could utilize inositol in the brain, either as a carbon source, signaling molecule, or both; thus, inositol may facilitate fungal proliferation and accelerate the development of meningitis. Itr1 and Itr1A may be required for yeast cells to cross the BBB and/or proliferate in the CNS. In fact, our preliminary animal study results have shown that the ino1 itr1a double mutant produced ~10 times fewer CFU in the brain (average, 1.7 × 105 cells/g organ) than the wild-type strain (average, 7.1 × 106 cells/g organ) at the end point of the infection, while in the lung and spleen, both the mutant and wild-type strains reached equivalent levels of fungal burden (Fig. 7C). This result suggests that the ino1 itr1a mutant strain may be slower to disseminate and cross the BBB and/or proliferate in the CNS, and thus, mice infected with the ino1 itr1a mutant survived longer in our experiments. It would be very interesting to further investigate this hypothesis, which may ultimately contribute to our understanding of why C. neoformans so frequently causes lethal cryptococcal meningitis.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The C. neoformans strains used in this study are listed in Table 3. Strains were grown at 30°C in yeast extract-peptone-dextrose (YPD) agar medium and synthetic defined (SD) medium. V8 medium (pH 5.0) was used for mating assays. Modified MS medium was used for mating and sporulation assays and prepared as previously described, with modification (26). Niger seed medium was used to test for melanin production. Dulbecco modified Eagle’s (DME) medium for assessing capsule production was prepared as previously described (54). All other media were prepared as described previously (55–57).
TABLE 3 .
Strains used in this study
| C. neoformans var. grubii strain | Description | Reference |
|---|---|---|
| H99 | MATα wild type | (69) |
| KN99a | MAT a wild type | (70) |
| CDX175 | MATα itr1::NAT | This study |
| CUX7 | MAT a itr1::NAT | This study |
| CUX50 | MAT a itr1::NEO | This study |
| CUX53 | MATα itr1::nat ITR1-NEO | This study |
| CUX54 | MAT a itr1::nat ITR1-NEO | This study |
| CDX99 | MATα itr1a::NAT | This study |
| CDX100 | MAT a itr1a::NEO | This study |
| CDX103 | MATα itr2::NAT | This study |
| CDX104 | MAT a itr2::NEO | This study |
| CDX105 | MATα itr3::NAT | This study |
| CDX106 | MAT a itr3::NEO | This study |
| CDX196 | MATα itr3a::NEO | This study |
| CDX197 | MAT a itr3a::NEO | This study |
| CDX101 | MATα itr3b::NAT | This study |
| CDX102 | MAT a itr3b::NEO | This study |
| CDX166 | MATα itr3c::NAT | This study |
| CDX167 | MAT a itr3c::NAT | This study |
| UBCINO1 | MATα ino1::NEO | This study |
| CUX8 | MAT a ino1::NEO | This study |
| UBCINO11 | MATα ino1::NEO INO1 | This study |
| CUX17 | MATα ino1::NEO itr1::NAT | This study |
| CUX18 | MAT a ino1::NEO itr1::NAT | This study |
| CUX23 | MATα ino1::NEO itr1a::NAT | This study |
| CUX24 | MAT a ino1::NEO itr1a::NAT | This study |
| CUX29 | MATα ino1::NEO itr2::NAT | This study |
| CUX30 | MAT a ino1::NEO itr2::NAT | This study |
| CUX15 | MATα ino1::NEO itr3::NAT | This study |
| CUX16 | MAT a ino1::NEO itr3::NAT | This study |
| CUX27 | MATα ino1::NEO itr3a::NAT | This study |
| CUX28 | MAT a ino1::NEO itr3a::NAT | This study |
| CUX25 | MATα ino1::NEO itr3b::NAT | This study |
| CUX26 | MAT a ino1::NEO itr3b::NAT | This study |
| CUX33 | MATα ino1::NEO itr3c::NAT | This study |
| CUX34 | MAT a ino1::NEO itr3c::NAT | This study |
| YSB119 | MATα aca1::NAT ura5 ACA1-URA5 | (54) |
| YSB121 | MAT a aca1::NEO ura5 ACA1-URA5 | (54) |
Database and sequence information.
All fungal ITR gene sequences and other sequences were obtained from a variety of databases. All sequences for C. neoformans var. grubii and C. gattii were obtained from the Broad Institute (http://www.broadinstitute.org/), and sequences for C. neoformans var. neoformans were obtained from the TIGR database (http://www.tigr.org/). Related ITR sequences in S. cerevisiae and C. albicans were obtained from the Saccharomyces genome database and Candida genome database. The well-defined ITR genes in S. cerevisiae, S. pombe, and C. albicans were used as queries to identify the initial ITR homologs in the H99 genome via the BLAST algorithm with the default setting, and the output was sorted with top hits ranked by BLASTp scores (58). Phylogeny trees were generated using ClustalX 2.0 (59) and viewed via TreeView X (60).
To identify ITR gene homologs/paralogs in different Cryptococcus species, the identified ITR gene homologs in H99 were used as queries to conduct BLASTn and BLASTx searches in the genome database for C. neoformans strains H99 and JEC21 and C. gattii strains R265 and WM276. BLAST results were parsed using an in-house Perl script. Alignment of the deduced amino acid sequences of all ITR genes was performed with ClustalW (61). The aligned amino acid sequences were imported to PhyML to construct their phylogenetic organization using the maximum likelihood method (62). The generated phylogenetic tree was viewed and edited with FigTree software (http://tree.bio.ed.ac.uk/software/figtree/).
Detection of ITR gene expression using quantitative RT-PCR.
To test how the ITR genes respond to the presence of environmental myo-inositol, we measured the mRNA levels for all seven ITR genes under inositol induction conditions via quantitative real-time PCR (qRT-PCR). Mating assays were performed by mixing H99 and KN99a and coculturing on MS medium containing 100 mg/liter myo-inositol. A single-strain H99 culture was also grown in MS medium. Plates were incubated at room temperature in the dark. Mating mixtures and H99 cultures were collected from agar surfaces at 24 h, 48 h, or 72 h after plating using cell scrapers. Collected cells were washed with distilled water (dH2O), and pellets were used for total RNA extraction. Total RNAs were extracted using Trizol reagents (Invitrogen) and purified with the Qiagen RNeasy cleanup kit (Qiagen) by following the manufacturer’s instructions. Purified RNAs were quantified using a NanoDrop instrument (Thermo Scientific). The same approach was used to prepare RNAs from itr1 mutant strains.
First-strand cDNAs were synthesized using a SuperScript III cDNA synthesis kit (Invitrogen) by following the instructions provided by the manufacturer. Expression of the ITR genes and the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was analyzed using Brilliant SYBR green quantitative PCR reagents (Stratagene). The gene expression level was normalized using the endogenous control GAPDH gene, and the relative levels were determined using the comparative threshold cycle (CT) method. Real-time PCRs were performed using a Mx4000 quantitative PCR system (Stratagene), with an initial denaturation step at 95°C for 10 min, followed by 40 PCR cycles in which each cycle consisted of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. SYBR green fluorescence emissions were monitored after each cycle. The primers for each gene are listed in Table S1 in the supplemental material. Amplification of specific transcripts was confirmed by melting curve profiles (cooling the sample to 60°C and heating it slowly to 95°C, with measurement of fluorescence) at the end of each PCR. The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis and sequencing.
Generation of S. cerevisiae itr1 itr2 double mutants and heterologous expression of C. neoformans ITR genes.
S. cerevisiae itr1 and itr2 single mutants were obtained from Open Biosystems. A genetic cross between S. cerevisiae α itr1 and S. cerevisiae a itr2 was conducted in YPD medium, and the mating mixture was colony purified in yeast nitrogen base (YNB) supplemented with histidine, leucine, and uracil and incubated at 30°C for 2 days. Single colonies were inoculated in sporulation medium and incubated for 5 days, and meiotic products were then isolated by tetrad dissection. Five tetrads were analyzed, and two double mutants (S. cerevisiae itr1 itr2) were confirmed by PCR using gene-specific primers (see Table S1 in the supplemental material) and selected for further study. The growth rate of these double mutants was compared with those of wild-type yeast strain BY4742 and S. cerevisiae itr1 and itr2 single mutants at both 30°C and 37°C.
Seven ITR genes from C. neoformans were cloned into the yeast expression vector pTH19 (63), under the control of the ADH1 promoter. Each of the seven ITR expression plasmids was introduced into an S. cerevisiae itr1 itr2 double mutant background, and colonies were selected on SD-Ura medium. The expression of all of these ITR genes in this yeast heterologous system was verified with RT-PCR using gene-specific primers (see Table S1 in the supplemental material). Yeast strains were tested for growth in YPD at 30°C and 37°C.
Generation of inositol transporter mutants and ino1 mutants.
Seven inositol transporters have been identified in the genome of H99 that share significant identity with the Itrs of S. cerevisiae (Itr1 and Itr2), S. pombe (Itr1 and Itr2), and C. albicans (Itr1). itr mutants were generated in the congenic C. neoformans serotype A α (H99) and a (KN99a) strains by overlap PCR as previously described (64). The 5′ and 3′ regions of each ITR gene were amplified with primers F1 and R1 and primers F2 and R2, respectively (see Table S1 in the supplemental material for primer sequences), from H99 or KN99a genomic DNA. The dominant selectable markers (Natr or Neor) were amplified with the M13 primers (M13F/M13R) from plasmid pNATSTM#122 or pJAF1 (65), respectively. Each target gene replacement cassette was generated by overlap PCR with primers F1 and R2 (see Table S1 in the supplemental material). Purified overlap PCR products were precipitated onto 600-µg gold microcarrier beads (0.8 µm [Bioworld Inc.] or 0.6 µm [Bio-Rad]), and strains H99 and KN99a were biolistically transformed as described previously (66). Stable transformants were selected in YPD medium containing nourseothricin (100 mg/liter) or G418 (200 mg/liter). To screen for mutants of each ITR gene, diagnostic PCR was performed by analyzing the 5′ junction of the disrupted mutant alleles with primers F4 and JH8994 (see Table S1 in the supplemental material). Positive transformants identified by PCR screening were further confirmed by Southern blot analysis. The ino1 deletion mutants in the H99 background were generated using the same strategy used to generate itr mutants, as described above. Because the ino1 mutant is myo-inositol autotrophic, all mutants were tested for growth in YNB medium without myo-inositol to verify the deletion of the INO1 gene.
To generate complemented strains of itr1 mutants, a genomic DNA fragment that contains a 1.5-kb upstream region of the ITR1 open reading frame (ORF) and its 500 bp downstream region was amplified with a PCR using primers JH19630 and JH19631. This PCR fragment was fused with the Neor selective marker gene at its C-terminal end with an overlap PCR using primers JH19630 and M13R. The overlap PCR product was biolistically transformed in both α itr1 and a itr1 mutant strains. Mating assays were performed to identify the transformants that complemented the itr1 phenotype.
Generation of ino1 itr double mutants via genetic crosses.
In a mating assay, C. neoformans cells of opposite mating types were mixed and cocultured in V8 or MS agar medium at 25°C in the dark for 10 days, and filamentation was examined by light microscopy. Spore production was also visualized by microscopy and photographed. Basidiospores were also dissected from mating performed in MS medium.
To generate ino1 itr1 double mutants, a mating assay between α ino1::NEO and a itr1::NAT mutants was conducted, and spores were visualized and isolated with an MSM system (Singer Instruments, England). All progeny that grew in YPD with both nourseothricin and G418 were tested in YNB without inositol, and those that failed to grow were subjected to genomic DNA extraction. PCR was used to screen for ITR gene deletion with primers F4/JH8994 and F3/R3 (see Table S1 in the supplemental material). INO1 gene mutations for all potential double mutants were also tested with primers JH18517/JH18518 and confirmed by Southern blotting. The same approach was used to generate double mutants between ino1 and other itr mutants.
Microarray analyses for genes regulated by inositol.
To understand the effect of inositol on gene expression profiles during cell development, microarray experiments were performed to monitor the genes regulated during mating by inositol. Overnight cultures of H99 and KN99a were mixed in equal cell amounts and washed with dH2O once, and cell mixtures were inoculated on MS medium with or without inositol. Cells were collected from MS plates 48 h postinoculation and washed with dH2O, and total RNA was purified. Total RNAs were extracted using Trizol reagents (Invitrogen) and purified using the Qiagen RNeasy cleanup kit (Qiagen). Cy3- and Cy5-labeled cDNA were generated by incorporating amino-allyl-dUTP during reverse transcription of 5 µg of total RNA as described previously (67) and competitively hybridized to a JEC21 whole-genome array generated previously at Washington University in Saint Louis, MO. After hybridization, arrays were scanned with a GenePix 4000B scanner (Axon Instruments) and analyzed by using GenePix Pro version 4.0 and BRB array tools (developed by Richard Simon and Amy Peng Lam at the National Cancer Institute [http://linus.nci.nih.gov/BRB-ArrayTools.html]) as described previously (68).
Assays for melanin and capsule production.
Melanin production was assayed by inoculating C. neoformans strains into 2 ml of YPD liquid medium, incubating the culture overnight at 30°C, and spotting 5 µl of each culture on Niger seed agar medium. The agar plates were incubated at 30°C or 37°C for 2 days, and pigmentation of fungal colonies was assessed and photographed. To examine capsule production, 5 µl of overnight cultures was inoculated on DME agar medium and incubated at 30°C for 3 days. The capsule was visualized with India Ink negative staining and observed with a 100× Olympus CX41 equipped with an Infinity digital camera (Olympus).
Virulence study.
Yeast strains were grown at 30°C overnight, and cultures were washed twice with 1× phosphate-buffered saline (PBS) and resuspended at a final concentration of 2 × 106 CFU/ml. Groups of 10 female A/JCr mice (NCI-Frederick, MD) were intranasally infected with 105 yeast cells of each strain as previously described (39). Animals that appeared to be moribund or in pain were sacrificed by CO2 inhalation. Survival data obtained from the murine experiments were statistically analyzed between paired groups using the log rank test and PRISM program 4.0 (GraphPad Software) (P values of <0.01 were considered significant). At the end point of the infection, infected lungs, brains, and spleens were also isolated and homogenized using a homogenizer in 1× PBS buffer. Resuspensions were diluted, 50 µl of each dilution was spread on YPD medium with antibiotics, and colonies were counted after 3 days of incubation at 30°C.
Supplemental material
ACKNOWLEDGMENTS
We thank Alex Idnurm for constructing the Agrobacterium-mediated mutagenesis library of C. neoformans. We thank Anna Averette for assistance with microarray experiments, Qing Chen for valuable technique assistance on qRT-PCR experiments, Lukasz Kozubowski for assistance with yeast tetrad dissection, John Perfect and Jason Stajich for valuable suggestions and discussions on bioinformatics analyses. We also acknowledge use of the C. neoformans genome sequences at Duke University (Fred Dietrich), the Broad Institute, and the TIGR database and use of the Tremella mesenterica genome sequence at the DOE-JGI.
This work was supported by National Institute of Health R01 grant AI39115 to J.H. and R21 grant AI070230 to J.H. and C.X. This work was also supported by the new PI institutional start-up fund from UMDNJ to C.X.
Footnotes
Citation Xue, C., T. Liu, L. Chen, W. Li, I. Liu, et al. 2010. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 1(1):e00084-10. doi:10.1128/mBio.00084-10.
REFERENCES
- 1. Bhandari R., Saiardi A., Ahmadibeni Y., Snowman A. M., Resnick A. C., Kristiansen T. Z., Molina H., Pandey A., Werner J. K., Jr., Juluri K. R., Xu Y., Prestwich G. D., Parang K., Snyder S. H. 2007. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U. S. A. 104:15305–15310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubois E., Scherens B., Vierendeels F., Ho M. M., Messenguy F., Shears S. B. 2002. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 277:23755–23763 [DOI] [PubMed] [Google Scholar]
- 3. Seeds A. M., York J. D. 2007. Inositol polyphosphate kinases: regulators of nuclear function. Biochem. Soc. Symp. 74:183–197 [DOI] [PubMed] [Google Scholar]
- 4. Steger D. J., Haswell E. S., Miller A. L., Wente S. R., O'Shea E. K. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299:114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. York S. J., Armbruster B. N., Greenwell P., Petes T. D., York J. D. 2005. Inositol diphosphate signaling regulates telomere length. J. Biol. Chem. 280:4264–4269 [DOI] [PubMed] [Google Scholar]
- 6. Luberto C., Toffaletti D. L., Wills E. A., Tucker S. C., Casadevall A., Perfect J. R., Hannun Y. A., Del Poeta M. 2001. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 15:201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shea J. M., Kechichian T. B., Luberto C., Del Poeta M. 2006. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74:5977–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerik K. J., Bhimireddy S. R., Ryerse J. S., Specht C. A., Lodge J. K. 2008. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 7:1685–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerik K. J., Donlin M. J., Soto C. E., Banks A. M., Banks I. R., Maligie M. A., Selitrennikoff C. P., Lodge J. K. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393–408 [DOI] [PubMed] [Google Scholar]
- 10. Heung L. J., Kaiser A. E., Luberto C., Del Poeta M. 2005. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J. Biol. Chem. 280:28547–28555 [DOI] [PubMed] [Google Scholar]
- 11. Heung L. J., Luberto C., Plowden A., Hannun Y. A., Del Poeta M. 2004. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. J. Biol. Chem. 279:21144–21153 [DOI] [PubMed] [Google Scholar]
- 12. Trinel P. A., Plancke Y., Gerold P., Jouault T., Delplace F., Schwarz R. T., Strecker G., Poulain D. 1999. The Candida albicans phospholipomannan is a family of glycolipids presenting phosphoinositolmannosides with long linear chains of beta-1,2-linked mannose residues. J. Biol. Chem. 274:30520–30526 [DOI] [PubMed] [Google Scholar]
- 13. Reynolds T. B. 2009. Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: making it and taking it. Microbiology 155:1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donahue T. F., Henry S. A. 1981. Inositol mutants of Saccharomyces cerevisiae: mapping the ino1 locus and characterizing alleles of the ino1, ino2 and ino4 loci. Genetics 98:491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai K., Bolognese C. P., Swift S., McGraw P. 1995. Regulation of inositol transport in Saccharomyces cerevisiae involves inositol-induced changes in permease stability and endocytic degradation in the vacuole. J. Biol. Chem. 270:2525–2534 [DOI] [PubMed] [Google Scholar]
- 16. Lai K., McGraw P. 1994. Dual control of inositol transport in Saccharomyces cerevisiae by irreversible inactivation of permease and regulation of permease synthesis by INO2, INO4, and OPI1. J. Biol. Chem. 269:2245–2251 [PubMed] [Google Scholar]
- 17. Nikawa J., Hosaka K. 1995. Isolation and characterization of genes that promote the expression of inositol transporter gene ITR1 in Saccharomyces cerevisiae. Mol. Microbiol. 16:301–308 [DOI] [PubMed] [Google Scholar]
- 18. Nikawa J., Hosaka K., Yamashita S. 1993. Differential regulation of two myo-inositol transporter genes of Saccharomyces cerevisiae. Mol. Microbiol. 10:955–961 [DOI] [PubMed] [Google Scholar]
- 19. Nikawa J., Tsukagoshi Y., Yamashita S. 1991. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266:11184–11191 [PubMed] [Google Scholar]
- 20. Robinson K. S., Lai K., Cannon T. A., McGraw P. 1996. Inositol transport in Saccharomyces cerevisiae is regulated by transcriptional and degradative endocytic mechanisms during the growth cycle that are distinct from inositol-induced regulation. Mol. Biol. Cell 7:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y. L., Kauffman S., Reynolds T. B. 2008. Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect. Immun. 76:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin J. H., Seyfang A. 2003. High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology 149:3371–3381 [DOI] [PubMed] [Google Scholar]
- 23. Niederberger C., Graub R., Schweingruber A. M., Fankhauser H., Rusu M., Poitelea M., Edenharter L., Schweingruber M. E. 1998. Exogenous inositol and genes responsible for inositol transport are required for mating and sporulation in Schizosaccharomyces pombe. Curr. Genet. 33:255–261 [DOI] [PubMed] [Google Scholar]
- 24. Voicu P. M., Poitelea M., Schweingruber E., Rusu M. 2002. Inositol is specifically involved in the sexual program of the fission yeast Schizosaccharomyces pombe. Arch. Microbiol. 177:251–258 [DOI] [PubMed] [Google Scholar]
- 25. Franzot S. P., Doering T. L. 1999. Inositol acylation of glycosylphosphatidylinositols in the pathogenic fungus Cryptococcus neoformans and the model yeast Saccharomyces cerevisiae. Biochem. J. 340(Pt. 1):25–32 [PMC free article] [PubMed] [Google Scholar]
- 26. Xue C., Tada Y., Dong X., Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273 [DOI] [PubMed] [Google Scholar]
- 27. Sukroongreung S., Kitiniyom K., Nilakul C., Tantimavanich S. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36:419–424 [PubMed] [Google Scholar]
- 28. Giles S. S., Dagenais T. R., Botts M. R., Keller N. P., Hull C. M. 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 77:3491–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Velagapudi R., Hsueh Y. P., Geunes-Boyer S., Wright J. R., Heitman J. 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 77:4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shetty H. U., Holloway H. W., Schapiro M. B. 1996. Cerebrospinal fluid and plasma distribution of myo-inositol and other polyols in Alzheimer disease. Clin. Chem. 42:298–302 [PubMed] [Google Scholar]
- 31. Spector R., Lorenzo A. V. 1975. Myo-inositol transport in the central nervous system. Am. J. Physiol. 228:1510–1518 [DOI] [PubMed] [Google Scholar]
- 32. Spector R., Lorenzo A. V. 1975. The origin of myo-inositol in brain, cerebrospinal fluid and choroid plexus. J. Neurochem. 25:353–354 [DOI] [PubMed] [Google Scholar]
- 33. Vincent V. L., Klig L. S. 1995. Unusual effect of myo-inositol on phospholipid biosynthesis in Cryptococcus neoformans. Microbiology 141(Pt. 8):1829–1837 [DOI] [PubMed] [Google Scholar]
- 34. Steen B. R., Zuyderduyn S., Toffaletti D. L., Marra M., Jones S. J., Perfect J. R., Kronstad J. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu G., Steen B. R., Lian T., Sham A. P., Tam N., Tangen K. L., Kronstad J. W. 2007. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 3(3):e42 doi:10.1371/journal.ppat.0030042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barnett J. A. 1976. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 32:125–234 [DOI] [PubMed] [Google Scholar]
- 37. Healy M. E., Dillavou C. L., Taylor G. E. 1977. Diagnostic medium containing inositol, urea, and caffeic acid for selective growth of Cryptococcus neoformans. J. Clin. Microbiol. 6:387–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clements R. S., Darnell B. 1980. Myo-inositol content of common foods: development of a high-myo-inositol diet. Am. J. Clin. Nutr. 33:1954–1967 [DOI] [PubMed] [Google Scholar]
- 39. Cox G. M., Mukherjee J., Cole G. T., Casadevall A., Perfect J. R. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xue C., Bahn Y. S., Cox G. M., Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lengeler K. B., Davidson R. C., D'Souza C., Harashima T., Shen W. C., Wang P., Pan X., Waugh M., Heitman J. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McClelland C. M., Chang Y. C., Varma A., Kwon-Chung K. J. 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 12:208–212 [DOI] [PubMed] [Google Scholar]
- 43. Wang P., Heitman J. 1999. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2:358–362 [DOI] [PubMed] [Google Scholar]
- 44. Wickes B. L. 2002. The role of mating type and morphology in Cryptococcus neoformans pathogenesis. Int. J. Med. Microbiol. 292:313–329 [DOI] [PubMed] [Google Scholar]
- 45. Berry G. T., Wu S., Buccafusca R., Ren J., Gonzales L. W., Ballard P. L., Golden J. A., Stevens M. J., Greer J. J. 2003. Loss of murine Na+/myo-inositol cotransporter leads to brain myo-inositol depletion and central apnea. J. Biol. Chem. 278:18297–18302 [DOI] [PubMed] [Google Scholar]
- 46. Spector R. 1988. Myo-inositol transport through the blood-brain barrier. Neurochem. Res. 13:785–787 [DOI] [PubMed] [Google Scholar]
- 47. Kaur R., Domergue R., Zupancic M. L., Cormack B. P. 2005. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 8:378–384 [DOI] [PubMed] [Google Scholar]
- 48. Castano I., Pan S. J., Zupancic M., Hennequin C., Dujon B., Cormack B. P. 2005. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 55:1246–1258 [DOI] [PubMed] [Google Scholar]
- 49. Penas A., Pan S. J., Castano I., Alder J., Cregg R., Cormack B. P. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ozcan S., Johnston M. 1999. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63:554–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Santangelo G. M. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Movahedzadeh F., Smith D. A., Norman R. A., Dinadayala P., Murray-Rust J., Russell D. G., Kendall S. L., Rison S. C., McAlister M. S., Bancroft G. J., McDonald N. Q., Daffe M., Av-Gay Y., Stoker N. G. 2004. The Mycobacterium tuberculosis ino1 gene is essential for growth and virulence. Mol. Microbiol. 51:1003–1014 [DOI] [PubMed] [Google Scholar]
- 53. Martin K. L., Smith T. K. 2005. The myo-inositol-1-phosphate synthase gene is essential in Trypanosoma brucei. Biochem. Soc. Trans. 33:983–985 [DOI] [PubMed] [Google Scholar]
- 54. Bahn Y. S., Hicks J. K., Giles S. S., Cox G. M., Heitman J. 2004. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3:1476–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alspaugh J. A., Perfect J. R., Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bahn Y. S., Kojima K., Cox G. M., Heitman J. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Granger D. L., Perfect J. R., Durack D. T. 1985. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 76:508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 60. Page R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 61. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guindon S., Delsuc F., Dufayard J. F., Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537:113–137 [DOI] [PubMed] [Google Scholar]
- 63. Harashima T., Heitman J. 2005. Galpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol. Biol. Cell 16:4557–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., D'Souza C., Wang P., Heitman J. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607–2615 [DOI] [PubMed] [Google Scholar]
- 65. Fraser J. A., Subaran R. L., Nichols C. B., Heitman J. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Davidson R. C., Cruz M. C., Sia R. A., Allen B., Alspaugh J. A., Heitman J. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38–48 [DOI] [PubMed] [Google Scholar]
- 67. Kraus P. R., Boily M. J., Giles S. S., Stajich J. E., Allen A., Cox G. M., Dietrich F. S., Perfect J. R., Heitman J. 2004. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot. Cell 3:1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ko Y. J., Yu Y. M., Kim G. B., Lee G. W., Maeng P. J., Kim S., Floyd A., Heitman J., Bahn Y. S. 2009. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot. Cell 8:1197–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Perfect J. R., Ketabchi N., Cox G. M., Ingram C. W., Beiser C. L. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 31:3305–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nielsen K., Cox G. M., Wang P., Toffaletti D. L., Perfect J. R., Heitman J. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.