Abstract
Summary: We present 3D-SURFER, a web-based tool designed to facilitate high-throughput comparison and characterization of proteins based on their surface shape. As each protein is effectively represented by a vector of 3D Zernike descriptors, comparison times for a query protein against the entire PDB take, on an average, only a couple of seconds. The web interface has been designed to be as interactive as possible with displays showing animated protein rotations, CATH codes and structural alignments using the CE program. In addition, geometrically interesting local features of the protein surface, such as pockets that often correspond to ligand binding sites as well as protrusions and flat regions can also be identified and visualized.
Availability: 3D-SURFER is a web application that can be freely accessed from: http://dragon.bio.purdue.edu/3d-surfer
Contact: dkihara@purdue.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Greater insight into the inner workings of the cellular machinery will become more critical as the current structural genomics initiatives for solving protein structures at high-throughput rates continue to rapidly progress. Given the huge number of experimentally solved but largely uncharacterized structures, the understanding of protein biochemical function assumes great importance. Although numerous representations of proteins have been used, surface-based approaches have been found to be quite useful both by way of analysis and visualization (Venkatraman et al., 2009, Fischer et al., 1993; Rosen et al., 1998).
Traditional protein structure comparison techniques make use of the pairwise alignment of protein Cα backbone or all atom structure representations. However, computing alignments has a high time complexity and is unsuitable for applications such as real-time structure database searches. To obviate this difficulty, methods such as 3D-BLAST encode the structure as a 1D sequence of alphabets (Yang and Tung, 2006; Supplementary Material). Light Field Descriptors, on the other hand, create 2D projections (combination of 2D Zernike and Fourier coefficients) rendered from uniformly distributed points around a sphere that surrounds the protein (Yeh et al., 2005). More recently, the development of 3D moment-based shape representations have shown promising performance for large-scale comparisons (Bustos et al., 2007). Among these, the 3D Zernike descriptors (3DZD) have been found to be suitable for the efficient comparison of protein surfaces (Sael et al., 2008). Unlike the previous two methods, which compare 1D or 2D representations, 3DZD are based on a 3D function expansion.
Here, we present 3D-SURFER, a web-based environment for high-throughput protein surface comparison and analysis. The server compares a single protein surface against all protein structures in PDB in just a couple of seconds (over 1 30 000 single chain structures, more than 55 000 total PDB entries, which are updated monthly). A performance comparison against other similar tools can be found in the previous work (Sael et al., 2008). In addition, local geometrical characteristics of a protein, which represent potential ligand binding sites, can be identified by the VisGrid algorithm (Li et al., 2008). Results shown include visual aids in the form of animated rotations of proteins along with the associated CATH codes (Orengo et al., 2002), and structure alignment calculations using the Combinatorial Extension (CE) algorithm (Shindyalov and Bourne 1998).
2 METHODS
2.1 3D Zernike descriptors
3DZD are utilized for the efficient comparison of protein surfaces across the entire PDB. The calculation of the invariants starts by voxelizing the protein molecular surface that is triangulated by MSROLL version 3.9.3 (Connolly, 1993). The mesh is then discretized to generate a cubic grid. 3DZD, a vector of 121 numbers, is then computed for the protein surface represented by the grid voxels. A single protein represented as a vector can be compared with other structures simply using the Euclidean distance. An example of the retrieval by 3DZD is shown in Table 1.
Table 1.
Top 10 results using the query 2mta-A
| Rank | PDB ID | CATH Code | Euclidean Distance | RMSDa (Å) |
|---|---|---|---|---|
| 1 | 1t5k-A | 2.60.40.420 | 1.575 | 0.48 |
| 2 | 2idq-A | 2.60.40.420 | 1.643 | 0.44 |
| 3 | 1t5k-B | 2.60.40. 420 | 1.777 | 0.77 |
| 4 | 1t5k-C | 2.60.40. 420 | 1.778 | 0.38 |
| 5 | 1aan-A | 2.60.40. 420 | 1.840 | 0.55 |
| 6 | 2idu-A | 2.60.40. 420 | 2.160 | 0.89 |
| 7 | 1id2-C | 2.60.40. 420 | 2.201 | 0.60 |
| 8 | 1aaj-A | 2.60.40. 420 | 2.215 | 0.53 |
| 9 | 1id2-A | 2.60.40. 420 | 2.320 | 0.56 |
| 10 | 2idt-A | 2.60.40. 420 | 2.322 | 1.00 |
aStructural alignments calculated using CE
We have shown in our previous work that structure retrieval by 3DZD agrees well with main-chain comparison by CE (Sael et al., 2008). Also it was found that surface comparison by 3DZD can identify functionally related proteins that cannot be discovered otherwise, due to distant evolutionary relationship (Sael et. al, 2008).
2.2 Analyzing local surface geometry
A protein can be interactively analyzed by VisGrid, which identifies geometric features of protein surfaces, i.e. pockets, protrusions, hollow spaces and flat regions (Li et al., 2008), which are often associated with binding sites. VisGrid uses a novel visibility criterion, which essentially indicates the fraction of open directions for a given point on the protein surface. The three largest pockets and protrusions are reported. The Qhull program (Barber et al., 1996) is used to calculate volumes and surface area of the pockets identified.
2.3 Input
3D-SURFER takes a PDB ID as an input structure to compare against the entire PDB. PDB IDs may be followed by a character representing the chain. For example, if the PDB structure 2MTA and chain A is of interest, the text entry should be 2MTA-A. Alternatively, a custom PDB structure may be uploaded and utilized as the query. In either case, a search against the entire structure database is executed on-the-fly. Additionally, the user can specify two types of filtering: CATH filtering that avoids displaying structures with similar CATH levels, and length filtering, in charge of displaying proteins whose lengths are similar to the query structure.
2.4 Output
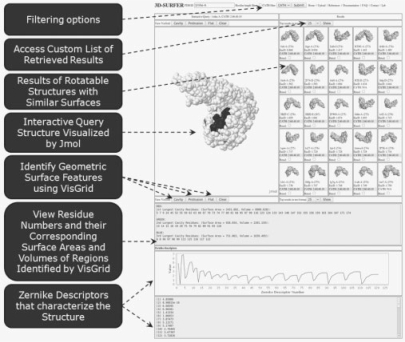
The right section of the results panel lists the structures identified as similar by 3DZD (Fig. 1). The distance of each retrieved protein to a query is shown after the label, EucD. CATH codes for each of the results are also displayed. Each reported result displays the corresponding PDB ID and is directly linked to the PDB web site. Root mean squared deviations (RMSD) values, calculated using CE, can be viewed by selecting the Rmsd checkbox and visualized by clicking on the Rmsd button. The protein surface analysis results can be viewed on the left panel (Jmol applet), which can be used to color the surface by clicking on the buttons called Cavity, Protrusion or Flat. The interface will render the surface in three different colors based on their rank in terms of geometric visibility: Red (1st), Green (2nd) and Blue (3rd). The volumes and surface areas of each region are also shown.
Fig. 1.
The 3D-SURFER user interface.
3 SUMMARY
3D-SURFER provides a platform to perform both global and local structure analysis in real time. Similarity in global structure infers evolutionary relationship in many cases, which can give a clue for the function of the protein. We plan to incorporate protein pocket database search into our platform in the future. In addition, protein surface properties such as electrostatic potentials, hydrophobicity and conservation will be integrated into 3D-SURFER for detailed analysis designed to assist investigating function of proteins.
4 SUPPORTED PLATFORMS
All latest web browsers are supported. The Java plug-in, and appropriate configuration, is required for visualization using Jmol.
Funding: National Institutes of Health (R01 GM075004); National Science Foundation (DMS 0604776, DMS0800568).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Barber CB, et al. The Quickhull algorithm for convex hulls. ACM Trans. Math. Softw. 1996;22:469–483. [Google Scholar]
- Bustos B, et al. Content-based 3D object retrieval. IEEE Comput. Graph. Appl. 2007;27:22–27. doi: 10.1109/mcg.2007.80. [DOI] [PubMed] [Google Scholar]
- Connolly ML. The molecular surface package. J. Mol. Graph. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- Fischer D, et al. Surface motifs by a computer vision technique: searches, detection, and implications for protein-ligand recognition. Proteins. 1993;16:278–292. doi: 10.1002/prot.340160306. [DOI] [PubMed] [Google Scholar]
- Li B, et al. Characterization of local geometry of protein surfaces with the visibility. Proteins. 2008;71:670–683. doi: 10.1002/prot.21732. [DOI] [PubMed] [Google Scholar]
- Orengo CA, et al. The CATH protein family database: a resource for structural and functional. Proteomics. 2002;2:11–21. [PubMed] [Google Scholar]
- Rosen M, et al. Molecular shape comparisons in searches for active sites and functional similarity. Protein Eng. 1998;11:263–277. doi: 10.1093/protein/11.4.263. [DOI] [PubMed] [Google Scholar]
- Sael L, et al. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins. 2008;72:1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Venkatraman V, et al. Potential for protein surface shape analysis using spherical harmonics and 3D Zernike descriptors. Cell Biochem. Biophys. 2009;54:23–32. doi: 10.1007/s12013-009-9051-x. [DOI] [PubMed] [Google Scholar]
- Yang JM, Tung CH. Protein structure database search and evolutionary classification. Nucleic Acids Res. 2006;34:3646–3659. doi: 10.1093/nar/gkl395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JS, et al. A web-based three-dimensional protein retrieval system by matching visual. Bioinformatics. 2005;21:3056–3057. doi: 10.1093/bioinformatics/bti458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.