Abstract
Background
Matrix metalloproteinases (MMPs) are enzymes that degrade all the components of extra cellular matrix and collagen. Various types of MMPs are known to be expressed and activated in patients with oral submucous fibrosis (OSMF) as well as head and neck squamous cell carcinoma (HNSCC). The purpose of this study was to asses the association of the single nucleotide polymorphism (SNP) adenosine insertion/deletion polymorphism (-1171 5A->6A) in the MMP-3 promoter region in these lesions.
Methods
MMP-3 SNP was genotyped by polymerase chain reaction-restriction fragment polymorphism (PCR-RFLP) analysis in a case control study consisting of 362 participants; 101 cases of OSMF, 135 of HNSCC and 126 controls, compared for age, sex and habits. ROC distribution was plotted to assess the contributions of genetic variation in MMP-3 genotypes with relation to age.
Results
Analysis of MMP 3 (-1171 5A->6A) polymorphism revealed the frequency of 5A allele in OSMF, HNSCC and controls to be 0.15, 0.13 and 0.07, respectively. A significant difference was found in 5A genotype frequency between OSMF (5A genotype frequency = 0.15, p = 0.01, OR = 2.26, 95% CI = 1.22-4.20) and in controls (5A genotype frequency 0.07) as well as HNSCC (5A genotype frequency 0.13, p = 0.03,95%CI = 1.06-3.51) and controls (5A genotype frequency = 0.07) In this study, 5A genotype had greater than two fold risk for developing OSMF (OR = 2.26) and nearly the same in case of HNSCC (OR = 1.94) as compared to controls. In patients with OSMF as well as HNSCC, the ROC analysis between the MMP-3 genotype and age, 6A/6A allele was found to be significant in patients both over and under 45 years of age; while the 5A/5A carrier alleles showed an association only in patients less than 45 years of age.
Conclusions
This study concluded that the expression of MMP-3 genotype associated with the 5A alleles, it may have an important role in the susceptibility of the patients to develop OSMF and HNSCC.
Background
Matrix metalloproteinases (MMP) are a group of structurally related zinc-dependent endopeptidases and have the capability to degrade all components of extra cellular matrix (ECM) [1]. MMPs are synthesized mainly by fibroblasts and to a lesser extent by activated macrophages and keratinocytes adjacent to the sites of injury [2]. MMP-3, also known as stromelysin-1, it is responsible for degradation of collagen type IV [3]. Brinckerhoff et al reported that MMP-3 also plays an important role in activation of proMMP-1 into the active form of MMP-1 in malignant tissues [4]. MMP-3 expression is low in normal tissues but it is altered during tumour formation, where remodeling of the extra cellular matrix is required [5]. MMP-3 has wide substrate specificity for various ECM components and is involved in many biological functions, including extracellular matrix degradation and remodelling, cell proliferation, angiogenesis as well as induction of synthesis of other metalloproteinases such as MMP-1 and MMP-9 [6]. The author's group recently reviewed the molecular functions and single nucleotide polymorphic association of different MMPs such as MMP-1 (-1607 1G/2G), MMP-2 (-1306 C/T), MMP-3 (-1171 5A/6A), MMP-9 (-1562 C/T) and TIMP-2 (-418 G/C or C/C) and it's possible therapeutic aspects of these proteases and they concluded that MMPs may play an association with potentially malignant (OSMF) and malignant head and neck lesions (HNSCC). Further research is required for the development of their potential diagnostic and therapeutic possibilities [7].
The expression of MMP-3 in carcinogenesis is regulated primarily at the transcriptional level, but there is also evidence of modulation of mRNA stability in response to growth factors and cytokines secreted by tumour-infiltrating inflammatory cells as well as by tumour and stromal cells [3,5,8]. MMP-3 transcription is higher in head and neck squamous cell carcinoma (HNSCC) and several other types of malignancies including lung and breast carcinomas [9-12]. MMP-3 degrades types II, V, IX and X collagens, proteoglycans, gelatine, fibronectin, laminin and elastin [13]. MMP-3 can also activate other MMPs, including collagenase, matrilysin and gelatinase B.
The MMP-3 gene has been mapped to the long arm of chromosome 11q22.3 and the level of expression of this gene can be influenced by single nucleotide polymorphisms (SNPs) in the promoter region of their respective gene [14]. The promoter region of MMP3 is characterized by a 5A/6A promoter polymorphism at position -1171 in which one allele has six adenosine (6A) and the second has five adenosine (5A). A single adenosine insertion/deletion polymorphism (5A/6A) at position -1171 of the MMP-3 promoter region causes different transcription of MMP-3.
In-vitro assays of promoter activity showed that the 5A allele had a two-fold higher promoter activity than the 6A allele [15]. The 5A allele is more prevalent in the European population (frequency range 40-50%) as compared to East Asian population (frequency range 7-20%) [11,14,16]. At position -1171 of the promoter region of the MMP-3 gene, a polymorphism of 5 or 6 adenosines (5A/6A) affects its low transcription level. The 5A allele results in higher gene expression in fibroblasts and vascular smooth muscle cells compared to the 6A allele and the 6A allele has a lower promoter reactivity than the 5A allele in vitro [14].
The aim of this study was to find out the association of the -1171 5A->6A polymorphism in the MMP-3 gene in patients suffering from OSMF, HNSCC and healthy controls.
Methods
Patients with OSMF and HNSCC as well as controls from the Departments of Otorhinolaryngology and Pathology, Moti Lal Nehru Medical College, Allahabad, India, were included in the study from June 2007 to October 2009; this study was approved by the institutional ethical committee. This case-control study group included 362 participants. Two hundred and thirty six subjects (average age 43.4 ± 14.8 years) were included in this study, of which 101 patients suffered from OSMF (78 males and 23 females) and 135 patients from HNSCC (114 males and 21 females) and 126 healthy subjects who did not have a history of pre-malignant or malignant lesions. These (average age 37.01 ± 12.89 years, 105 males 21 females) healthy controls were compared for age, sex and habits. Detailed clinical and personal information of each patient was noted in a standardized proforma. Information regarding the patients name, age, sex, occupation, personal habits and present complaints was gathered. Emphasis was given to addictions like areca nut, tobacco and alcohol. None of the HNSCC patients gave a history of having OSMF prior to developing malignancy. Individuals with haematological diseases, previous malignancies, skin diseases and autoimmune disorders were excluded from the study. All cases were histopathologically confirmed. The blood samples were taken after obtaining the patients informed consent to participate in the study. 5 ml. blood was drawn from each subject into vacutainer tubes containing ethylene-di-amine-tetra-acetic acid (EDTA) and stored at 4°C till the samples were processed and isolation of genomic DNA was done.
Isolation of Genomic DNA
Genomic DNA was extracted from the blood samples by using the Qiagen QIAamp DNA Blood Mini Kit (Qiagen Inc. USA) The extracted genomic DNA was quantified and checked for purity by spectrophotometer (Spectro UV-Vis Double Beam PC, UVD Model 2950 LABOMED, Inc. CA, USA). Ethidium bromide (EtBr) stained 0.8% agarose gel electrophoresis was used to confirm the presence of genomic DNA in patient and controls samples.
Genotyping of the MMP-3 promoter polymorphism
The MMP-3 genotype was determined by the PCR-RFLP assay. The PCR primers used for amplifying the MMP-3 polymorphism were: forward primer (FP) 5'-GGTTCTCCATTCCTTTGATGGGGGGAAAGA-3' and reverse primer (RP) 5'-CTTCCTGGAATTCACATCACTGCCACCACT-3' [17]. An A to G mutation at the second nucleotide close to the 3' end of the FP was made to create a Tth111I (TakaRa Biotechnology Co. Ltd, Dalian, China) recognition site in the case of a 5A allele. PCR (MJ Research PTC 100, GMI, Inc, Minnesota, USA) was performed in a 25 μl volume containing 50 ng of genomic DNA template, 2.5 μl of 10× PCR buffer, 2.5 mmol of MgCl2, 1 U of Taq DNA polymerase (Fermentas Inc. Glen Burnie MD), 200 nmol of dNTPs and 200 nmol of forward and reverse primer. The PCR cycling conditions were 5 min at 94°C followed by 35 cycles of 45 s at 94°C, 45 s at 66°C and 45 s at 72°C and with a final step at 72°C for 15 min to allow for the complete extension of all PCR fragments. For a negative control, each PCR reaction used distilled water instead of DNA in the reaction mixture.
Restriction enzyme (Tth 111I) digestion of MMP-3 gene
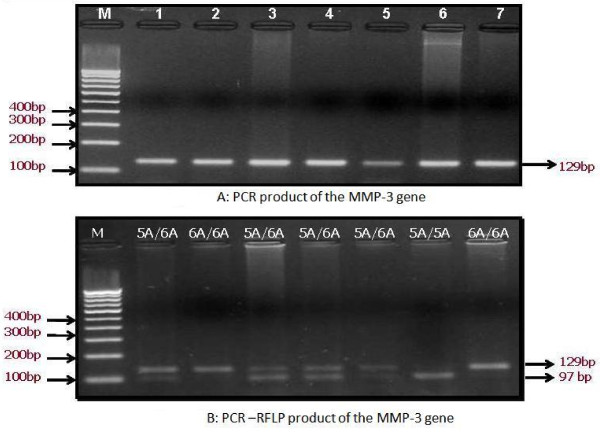
A 10 μl aliquot of PCR product was digested at 65°C for 5 hrs in a 15 μl reaction containing 10 U of Tth111 I and 1× reaction buffer. After digestion, the products were separated on 3.5% agarose gel stained with EtBr. On electrophoresis, the 5A alleles were represented by DNA bands of 97 and 32 bp, the 6A alleles were represented by a DNA band of 129 bp, whereas the heterozygote displayed a combination of both alleles (129, 97 and 32 bp) Fig. 1[A] and 1[B].
Figure 1.
A The expected PCR product size of MMP-3 gene by 100 bp marker [M], Lane 1-7 shows PCR product followed by separation on 2.0% agarose gel was confirmed; B: Genotyping of MMP-3 Promoter (-1171 5A->6A) Polymorphism by PCR-RFLP analysis followed by separation on 3. 5% agarose gel, Lane M = 100 bp marker; lane 1, 3, 4, 5 = 5A/6A; lane 2 = 5A/6A; lane 6 = 5A/5A.
Statistical analysis
The Chi-square test (χ2 test) was used to find out the difference in genotypic distribution of MMP-3 promoters between the OSMF, HNSCC and control groups. Fisher's exact test has also been applied in the statistical analysis. A p-value of < 0.05 was considered as statistically significant. For each parameter, patients with OSMF histopathological grade I, II, III, IV and HNSCC, risk was analyzed by odds ratios (OR) and 95% Confidence Intervals (95% CI). Distributions of MMP-3 genotype promoter polymorphisms in OSMF, HNSCC and control groups were analyzed by the Hardy-Weinberg equilibrium. The statistical analysis was performed using the SPSS 15.0 software package (SPSS Japan Inc., Tokyo, Japan).
Results
Patients of OSMF were diagnosed on the basis of clinical signs including trismus and presence of fibrotic bands in the oral cavity and malignancies. Figure 2[A, B, C] and 2[D]. MMP-3 promoter genotypes in patients with OSMF, HNSCC and controls were analyzed with respect to gender, age, habits like tobacco consumption with or without areca nut chewing, alcohol intake, as well as histopathological grade of OSMF, TNM staging of HNSCC and the location of the lesion.
Figure 2.
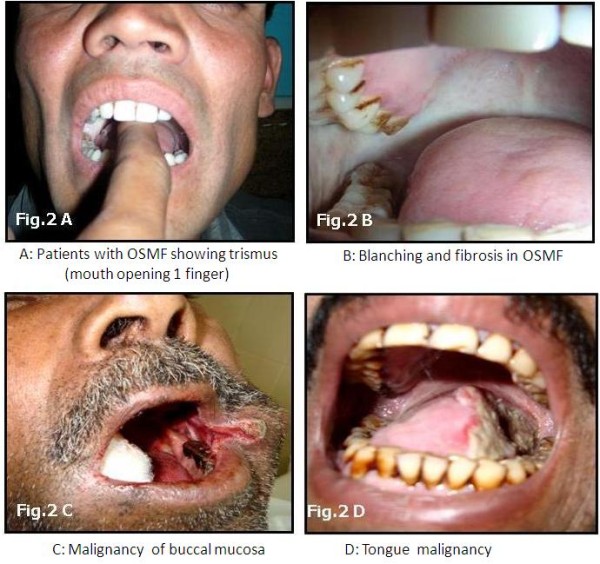
Clinical picture of patients with OSMF and HNSCC.
Out of 101 patients with OSMF, 74 (73.27%) had 6A/6A genotype, 24 (23.76%) 5A/6A and 3 (2.97%) 5A/5A, while in 135 patients with HNSCC, 106 (78.5%) had 6A/6A genotype, 23 (17.03%) 5A/6A and 6 (4.44%) 5A/5A. Of the male patients with OSMF, 62 had 6A/6A, 14 (5A/6A) and 2 (5A/5A) genotype and of the female patients, 12 had 6A/6A, 10 (5A/6A) and 1 (5A/5A) genotype. In male patients with HNSCC, 94 had 6A/6A, 15 (5A/6A) and 5 (5A/5A) genotype and of the females, 12 had 6A/6A, 8 (5A/6A) and 1 (5A/5A) genotype. Analyzing the different genotypes (6A/6A, 5A/6A and 5A/5A), it was found that the polymorphic association of these genotypes was statistically significant in both male and females suffering from OSMF (p = 0.001) and HNSCC (p = 0.01), according to the Fisher's exact test. This conclusion was drawn on analysis of the dataset illustrated in Table no 1.
Association of 5A/5A, 6A/6A and 5A/6A genotypes and tobacco chewing with areca nut chewers showed a significant association in OSMF and HNSCC cases respectively (p = 0.02,0.003) as did the OSMF (p = 0.001) and HNSCC histopathological grading (p = 0.001) as well as location, both in OSMF and HNSCC (p = 0.81 and p = 0.59 respectively) [Table 1].
Table 1.
Relation of MMP-3 genotype to clinical parameters and environmental factors among the OSMF, HNSCC and Control.
| Characteristics | OSMF (N = 101) |
HNSCC (N = 135) |
Total Control | Control (N = 126) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6A/6A genotype | 5A/6A genotype | 5A/5A genotype | p-value | 6A/6A genotype | 5A/6A genotype | 5A/5A genotype | p-value | 5A/5A genotype | 6A/6A genotype | 5A/6A genotype | (p-value) | |||
| Total | N = 236 | 74 (73.27%) |
24 (23.76%) |
3 (2.97%) |
106 (78.5%) |
23 (17.03%) |
6 (4.44%) |
126 | 2 (1.6%) |
110 (87.3%) |
14 (11.1%) |
|||
| Gender | ||||||||||||||
| Male | 192 (81.35) | 62(83.8) | 14(58.3) | 2(66.6) | 0.001 | 94 (88.67) | 15(65.21) | 5 (83.3) | 0.01 | 105(83.3) | 2 (100) | 101 (91.8) | 2(14.3) | 0.01 |
| Female | 44 (18.64) | 12(16.2) | 10 (41.6) | 1(33.3) | 12 (11.32) | 8(34.78) | 1 (16.6) | 21(16.7) | 0 | 9(8.2) | 12(85.7) | |||
| Age (Yrs) | ||||||||||||||
| Mean ± SD | 43.4 ± 14.8 | 38.8 ± 14.5 | 37.6 ± 14.2 | 36 ± 8.5 | 0.671 | 42 ± 16.5 | 47 ± 10.8 | 41 ± 9.86 | 0.42 | 37.0 ± 12.8 | 17.5 ± 0.7 | 36.7 ± 12.4 | 38.8 ± 16 | 0.41 |
| Tobacco with Areca nut | ||||||||||||||
| Chewer | 215(91.1) | 72(97.3) | 23 (95.8) | 2 (66.7) | 0.02 | 98(92.45) | 16(69.56) | 4 (66.6) | 0.003 | 85(67.5) | 2(100) | 71(64.5) | 12(85.7) | 0.12 |
| Non Chewer | 21(8.9) | 2(2.7) | 1 (4.2) | 1 (33.3) | 8(7.54) | 7(30.43) | 2 (33.3) | 41(32.5) | 0 | 39(35.5) | 2(14.3) | |||
| Smoking status | ||||||||||||||
| Smoker | 170(72.03) | 42(56.7) | 19(79.2) | 3 (100) | 0.077 | 88(83.01) | 15(65.21) | 4 (66.6) | 0.037 | 51(40.5) | 0 | 47(42.7) | 4(28.6) | 0.11 |
| Non-smoker | 66 (27.9) | 32(43.3) | 5 (20.8) | 0 | 18(16.98) | 8(34.78) | 2 (33.3) | 75(59.5) | 2(100) | 63(57.3) | 10(71.4) | |||
| Alcohol Intake | ||||||||||||||
| Drinker | 117(49.57) | 22(29.7) | 3 (12.5) | 2 (66.7) | 1.16 | 72(67.92) | 14(60.86) | 3 (50) | 0..81 | 26(20.6) | 0 | 25(22.7) | 1(7.1) | 0.24 |
| Non Drinker | 119(50.42) | 52(70.3) | 21 (87.5) | 1 (33.3) | 34(32.07) | 9(39.1) | 3 (50) | 100(79.4) | 2(100) | 85(77.3) | 13(92.9) | |||
| Location | ||||||||||||||
| Lip (lower &upper) | 51 (21.61) | 20(27) | 7 (29.2) | 1 (33.3) | 19(17.92) | 3(13.04) | 1(16.67) | |||||||
| Tongue | 38 (16.1) | 10(13.5) | 4 (16.7) | 0 | 21(19.81) | 2(8.69) | 1 (16.1) | |||||||
| Right & left Cheek | 32(13.56) | 11(15) | 3 (12.5) | 1 (33.3) | 0.81* | 14(13.20) | 3(13.04) | 1 (16.1) | 0.59* | - | - | - | - | - |
| Buccal mucosa | 34 (14.41) | 10(13.5) | 4 (16.6) | 0 | 15(14.15) | 3(13.04) | 1 (16.1) | |||||||
| Larynx/pharynx | 81 (34.32) | 23(31) | 6(25) | 1 (33.3) | 37(34.91) | 12(52.17) | 2 (33.3) | |||||||
| OSMF Grade | ||||||||||||||
| (N = 101) | ||||||||||||||
| I-II | 73 (72.28) | 55 (74.3) | 16 (66.6) | 2 (66.6) | 0.001 | - | - | - | - | - | - | - | - | |
| III-IV | 28 (27.72) | 19 (25.7) | 8 (33.3) | 1 (33.3) | ||||||||||
| HNSCC Grade | ||||||||||||||
| (N = 135) | ||||||||||||||
| T category | ||||||||||||||
| T1-3 | 37 (27.41) | 21 (19.81) | 14 (60.86) | 2 (33.3) | ||||||||||
| T4 | 98 (72.59) | - | - | - | - | 85 (80.18) | 9 (39.13) | 4 (66.6) | 0.001 | |||||
| N category | ||||||||||||||
| N0 | ||||||||||||||
| N1-3 | 42 (31.11) 93 (68.89) |
- | - | - | - | 30 (28.31) 76 (71.69) |
6 (26.1) 17 (73.9) |
1 (16.7) 5 (83.3) |
0.814 |
|||||
T & N categories [UICC TNM classification], Fisher's exact test & * Pearson's χ2 test were applied for statistical analysis.
The MMP-3 genotype distinguished homozygous 6A genotype (6A/6A), homozygous 5A genotype (5A/5A) and heterozygous genotype (5A/6A). The 5A allele frequencies in OSMF, HNSCC and controls were 0.15, 0.13 and 0.07 respectively (According to Hardy-Weinberg equilibrium). A significant difference was found in 5A allele frequency between OSMF and controls (p = 0.01), as well as HNSCC and controls (p = 0.03). 5A genotype had a more than two fold risk for OSMF (OR = 2.26) and little less in HNSCC (OR = 1.94) in relation to the control group. Frequency of 5A/6A or 5A/5A promoter genotypes having 5A alleles was associated with MMP-3 single nucleotide polymorphism (SNP) in OSMF (5A allele frequency = 0.15, p = 0.01, OR = 2.26, 95% CI = 1.22-4.20). Similar difference was also found in MMP-3 genotype distribution in HNSCC (5A allele frequency = 0.13, p = 0.03, OR = 1.94, 95% CI 1.06-3.51) as compared to controls (5A allele frequency = 0.07) [Table 2].
Table 2.
The genotype and allele frequency of MMP-3 in OSMF, HNSCC patients and controls.
| Groups | Total (N) |
5A/5A Genotype |
5A/6A Genotype |
6A/6A Genotype |
5A allele frequency | OR | 95%CI | P-value |
|---|---|---|---|---|---|---|---|---|
| Control | 126 | 2 (1.58%) | 14 (11.1%) | 110 (87.3%) | 0.07 | 1 | - | - |
| OSMF | 101 | 3(2.9%) | 24 (23.7%) | 74 (73.3%) | 0.15 | 2.26 | 1.22-4.20 | 0.01 |
| HNSCC | 135 | 6 (4.44%) | 23 (17.03%) | 106 (78.52%) | 0.13 | 1.94 | 1.06-3.51 | 0.03 |
[OR = Odd Ratio; CI = confidence interval; MMP = matrix metalloproteinase; OSMF = oral submucous fibrosis; HNSCC = Head and neck squamous cell carcinoma]
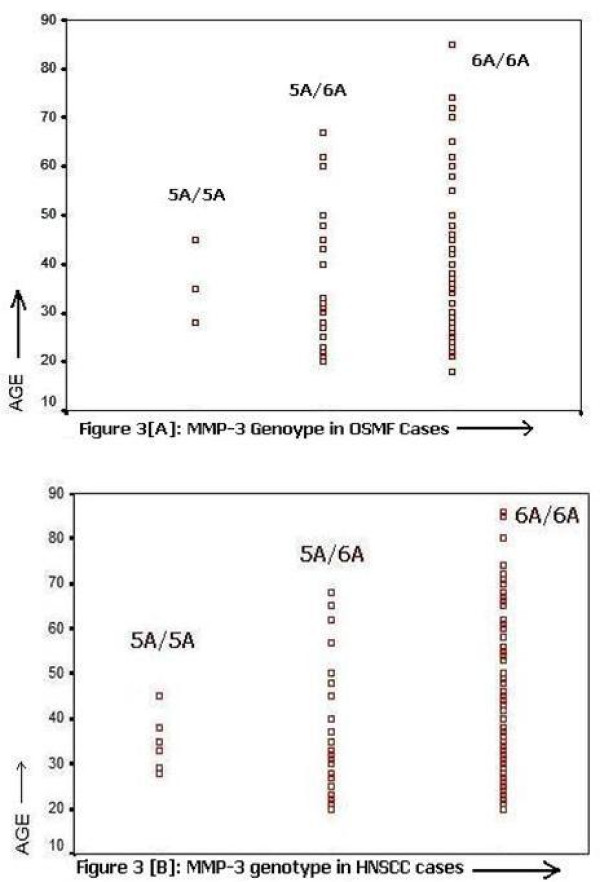
Receiver operating characteristics (ROC) showed a significant association between age and MMP-3 genotypes in OSMF and HNSCC. Figure 3[A] and 3[B] shows that 6A/6A allele was also found to be significant in subjects both over and under 45 years of age. While, 5A/5A carrier alleles showed an association only in patients less than 45 years of age. Fisher's exact test also revealed that patients with 5A alleles had a significant risk to develop OSMF (p = 0.01, OR = 2.51, 95%CI = 1.27-4.94). [Table 3 and 4]. On the other hand, significant difference was found in MMP-3 genotype distribution (6A/6A, 5A/5A and 5A/6A) between the areca nut with tobacco chewers and the non chewers both in OSMF (p = 0.02) and HNSCC (p = 0.003) patients vis-à-vis control group (p = 0.12) [Table 5 and 6]. However it should be noted that there are differences in age distribution, smoking and areca nut chewing between controls and patients.
Figure 3.
ROC 6A/6A allele was also found to be significant in subjects both over and under 45 years of age, while 5A/5A carrier alleles showed an association only in patients less than 45 years of age in case of [A] OSMF and [B] HNSCC.
Table 3.
Univariate analysis of predictive factors in case of OSMF.
| OSMF | CONTROL | p-value | OR | 95%CI | |
|---|---|---|---|---|---|
| Age | |||||
| <45 | 76 | 64 | 0.001 | 2.94 | 1.66-5.21 |
| ≥45 | 25 | 62 | |||
| Gender | |||||
| Male | 77 | 105 | 0.24 | 0.64 | 0.33-1.23 |
| Female | 24 | 21 | |||
| 6A* | |||||
| -ve | 3 | 2 | 0.79 | 1.89 | 0.05-6.93 |
| +ve | 98 | 124 | |||
| 5A** | |||||
| -ve | 74 | 110 | 0.01 | 2.508 | 1.27-4.94 |
| +ve | 27 | 16 | |||
*6A-denotes 5A/5A genotypes and 6A+denotes 6A/6A or 5A/6A. **: 5A-denotes 6A/6A genotype and 5A+ denotes 5A/5A or 5A/6A. Data were analyzed by Fisher's exact test.
Table 4.
Univariate analysis of predictive factors in case of HNSCC.
| HNSCC | CONTROL | p-value | OR | 95%CI | |
|---|---|---|---|---|---|
| Age | |||||
| <45 | 16 | 78 | 0.001 | 2.74 | 0.13-0.54 |
| ≥45 | 36 | 48 | |||
| Gender | |||||
| Male | 114 | 105 | 0.93 | 1.08 | 0.56-2.10 |
| Female | 21 | 21 | |||
| 6A* | |||||
| -ve | 6 | 2 | 0.03 | 2.93 | 0.58.79 |
| +ve | 129 | 126 | |||
| 5A** | |||||
| -ve | 106 | 110 | 0.02 | 1.8 | 0.96-3.66 |
| +ve | 29 | 16 | |||
* 6A-denotes 5A/5A genotypes and 6A+denotes 6A/6A or 5A/6A. **: 5A-denotes 6A/6A genotype and 5A+ denotes 5A/5A or 5A/6A. Data were analyzed by Fisher's exact test.
Table 5.
Age and habits distribution in relation to alleles in OSMF and Controls.
| Characteristics | OSMF (N = 101) |
Control (N = 126) |
||||||
|---|---|---|---|---|---|---|---|---|
| 6A/6A | 5A/6A | 5A/5A | p-value | 5A/5A | 6A/6A | 5A/6A | (p-value) | |
| Age (Yrs) | ||||||||
| Mean ± SD | 38.8 ± 14.5 | 37.6 ± 14.2 | 36 ± 8.5 | 0.671 | 17.5 ± 0.7 | 36.7 ± 12.4 | 38.8 ± 16 | 0.41 |
| Tobacco with Areca nut | ||||||||
| Chewer | 72(97.3%) | 23 (95.8%) | 2 (66.7%) | 0.02 | 2(100%) | 71(64.5%) | 12(85.7%) | 0.12 |
| Non Chewer | 2(2.7%) | 1 (4.2%) | 1 (33.3%) | 0 | 39(35.5%) | 2(14.3%) | ||
| Smoking status | ||||||||
| Smoker | 42(56.7%) | 19(79.2%) | 3 (100%) | 0.077 | 0 | 47(42.7%) | 4(28.6%) | 0.11 |
| Non-smoker | 32(43.3%) | 5 (20.8%) | 0 | 2(100%) | 63(57.3%) | 10(71.4%) | ||
| Alcohol Intake | ||||||||
| Drinker | 22(29.7%) | 3 (12.5%) | 2 (66.7%) | 1.16 | 0 | 25(22.7%) | 1(7.1%) | 0.24 |
| Non Drinker | 52(70.3%) | 21 (87.5%) | 1 (33.3%) | 2(100%) | 85(77.3%) | 13(92.9%) | ||
[Data was expressed as mean ± SD and %. Fischer exact test is used for habits in cases and control groups]
Table 6.
Age and habits distribution in relation to alleles in HNSCC and Controls
| Characteristics | HNSCC (N = 135) |
Control (N = 126) |
||||||
|---|---|---|---|---|---|---|---|---|
| 6A/6A | 5A/6A | 5A/5A | p-value | 5A/5A | 6A/6A | 5A/6A | (p-value) | |
| Age (Yrs) | ||||||||
| Mean ± SD | 42 ± 16.5 | 47 ± 10.8 | 41 ± 9.86 | 0.42 | 17.5 ± 0.7 | 36.7 ± 12.4 | 38.8 ± 16 | 0.41 |
| Tobacco with Areca nut | ||||||||
| Chewer | 98(92.45%) | 16(69.56%) | 4 (66.6%) | 2(100%) | 71(64.5%) | 12(85.7%) | 0.12 | |
| Non Chewer | 8(7.54%) | 7(30.43%) | 2 (33.3%) | 0.003 | 0 | 39(35.5%) | 2(14.3%) | |
| Smoking status | ||||||||
| Smoker | 88(83.01%) | 15(65.21%) | 4 (66.6%) | 0 | 47(42.7%) | 4(28.6%) | 0.11 | |
| Non-smoker | 18(16.98%) | 8(34.78%) | 2 (33.3%) | 0.037 | 2(100%) | 63(57.3%) | 10(71.4%) | |
| Alcohol Intake | ||||||||
| Drinker | 72(67.92%) | 14(60.86%) | 3 (50%) | 0..81 | 0 | 25(22.7%) | 1(7.1%) | 0.24 |
| Non Drinker | 34(32.07%) | 9(39.1%) | 3 (50%) | 2(100%) | 85(77.3%) | 13(92.9%) | ||
[Data was expressed as mean ± SD and %. Fischer exact test is used for habits in HNSCC cases and control groups]
Discussion
Carcinogenesis is a multistage process, which involves various molecular mechanisms associated to the fundamental alterations in cell physiology such as growth signals, escape from apoptosis, uncontrolled replication and growth-inhibitory signals [18]. MMPs are involved in the degradation of extra cellular matrix (ECM) components that take part in several molecular mechanisms such as invasion, angiogenesis, metastasis and progression of malignancies. Nagata et al suggested that expression levels of that molecules which are involved in tissue remodeling and cell-ECM adhesion, especially MMP-1 and integrin-3, may provide an accurate biomarker diagnosis for predicting the risk of cervical lymph node metastasis in oral squamous cell carcinoma (OSCC) [19]. Lin et al reported that functional promoter polymorphisms of MMP-1 and MMP-2 gene might be associated with the risk for development of OSCC but not in OSMF [20,21].
The MMP-3 promoter polymorphism is associated with a higher MMP-3 transcription level, Hashimoto et al reported that the MMP-3 SNP was not associated with the progression of OSCC as reflected by TNM and advances in stage [9]. In this study, a higher frequency of 5A allele was found in OSMF and HNSCC as compared to controls. This study supports the hypothesis that adenine insertion/deletion polymorphism in the MMP-3 promoter region was associated with the increased chance of development of OSMF. Tu et al also earlier reported the similar findings in OSMF patients from Taiwan [22].
MMPs play an important role in fibrosis of the oral cavity. OSMF is a premalignant disorder, which can be induced by areca nut chewing [23]. Due to excess chewing of areca nut; arecoline may increase the production of collagen fibers in the sub epidermal layer of buccal cavity and causes fibrosis. Chiu et al reported that areca chewing as well as polymorphism of genes associated with collagen homeostasis was correlated with risk of OSMF [24]. Shin et al suggested that polymorphism of transforming growth factor (TGF-α) related to an innate immune response was also found to associated with the risk of OSMF [25]. In this study, a significant difference was found in MMP-3 genotype polymorphism between controls, OSMF and HNSCC patients.
Areca nut chewers having a 5A allele had a higher risk for OSMF and HNSCC (OR = 2.26 and 1.94 respectively) in relation to the control group. Tu et al reported a significant difference in MMP-3 genotypic polymorphism between patients with OSMF vis-à-vis controls in male areca chewers, however not in patients with OSCC from Taiwan [22]. On the other hand, Vairaktaris et al from Greece also reported a significant difference in the 5A allele in oral cancer patients who smoked [26].
The presence of the 5A allele has been reported to be associated with susceptibility for oral cancer, lung carcinoma, esophageal squamous cell carcinoma, head and neck carcinoma, ovarian cancer as well as breast carcinomas [22,27-31]. On the other hand, a patient with lower expression of 6A allele has been associated with increased risk for colorectal cancer [32]. Nishijima et al reported that the anti-MMP-3 antibody is a serological marker that reflects the severity of systemic sclerosis and also suggested that it may contribute to the development of fibrosis by inhibiting MMP-3 activity and reducing the ECM [33]. Murawaki et al 1999 reported that measurement of serum MMP-3 is of little use for assessing fibrolysis in chronically diseased livers [34].
Tissue inhibitors of metalloproteinases (TIMPs) are the main endogenous inhibitors of the MMPs [35]. Strup-Perrot et al reported that expression of collagens; MMPs and TIMPs were increased simultaneously due to effect of collagen deposition in abdominal and pelvic cancers in radiation enteritis [36]. Chang et al found that arecoline acted not only as an inhibitor of gelatinolytic activity of MMP-2, but also a stimulator for TIMP-1 activity in OSMF [37]. Their study concluded that MMPs transcription activity might be associated with genesis of OSMF in younger areca nut chewers.
In this study, 6A/6A alleles was found to be significant both in OSMF and HNSCC patients less than and more than 45 years of age, while 5A/5A carrier alleles showed an association in OSMF and HNSCC patients less than 45 years of age. Nishizawa et al reported that apparent reduction of the MMP-1 1G/1G and 1G/2G genotypes distribution among the early onset OSCC cases under the ages of 45 years [38]. Since there were differences in age distribution, smoking and areca chewing between control and patients, the result need to be cautiously interpreted.
McCawley et al reported that high MMP-3 expression levels in OSCC may play an important role in oncogenesis and especially in invasion [39] Vairaktaris et al reported that the genotype containing 5A allele had a two-fold risk of oral cancer development in smokers [26]. In this study, a significant difference was found in MMP-3 genotypic polymorphism between controls and HNSCC (OR = 1.94), which is in concordance with results of Varkataris et al [26].
Conclusions
We conclude that adenosine insertion/deletion polymorphism (-1171 5A->6A) in the MMP-3 promoter region may play an important role in the development, initiation and progression of these lesions. To the best of our knowledge, this is the first study dealing with MMP-3 polymorphism in OSMF and HNSCC patients of Indian origin. Studies with a larger number of subjects are recommended to elucidate the genetic and polymorphic contribution of MMPs in these lesions, as this may have important role in designing future therapeutic strategies.
Abbreviations
(MMP): Matrix metalloproteinase; (ECM): Extra cellular Matrix; (SNP): single nucleotide polymorphism; (PCR): Polymerase chain reaction; (RFLP): Restricion fragment length polymorphism; (OSMF): Oral submucous fibrosis; (HNSCC): head and neck squamous cell carcinoma; (OR): odds ratio; (95% CI): 95% Confidence Intervals; (ROC): receiver operating characteristic curve.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AKC carried out the experimental work, analysis and drafted the manuscript. RM and Mamta Singh conceived of the study, participated in its design and coordination as well as helped to draft the manuscript. MS, ACB and SS participated in coordination of the study and helped to draft the manuscript. AKS participated in the statistical analysis. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Ajay K Chaudhary, Email: ajaygenome@gmail.com.
Mamta Singh, Email: mrspath25@gmail.com.
Alok C Bharti, Email: bhartiac@icmr.org.in.
Mangal Singh, Email: mangalsingh@yahoo.co.in.
Shirish Shukla, Email: shirishshukla31@gmail.com.
Atul K Singh, Email: atulksingh@sancharnet.in.
Ravi Mehrotra, Email: rm8509@gmail.com.
Acknowledgements
The authors thank the University Grant Commission (UGC), New Delhi, for providing financial support (Grant No.32-188/2006-SR) to AKC (D. Phil Scholar) for this study. The authors wish to express their gratitude to Shruti Pandya, SRF & D. Phil Scholar, in the Department of Pathology, MLN Medical College, University of Allahabad, for collection of the samples. Written consent was obtained for publication from the all patients or their relatives.
References
- Vihinen P, Kähäri VM. Matrix metalloproteinases in cancer: prognostic markers and therapeutic targets. Int J Cancer. 2002;99(2):157–66. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- Matrisian LM. Cancer biology. Extracellular proteinases in malignancy. Curr Biol. 1999;9:776–778. doi: 10.1016/S0960-9822(00)80011-1. [DOI] [PubMed] [Google Scholar]
- Impola U, Uitto VJ, Hietanen J, Hakkinen L, Zhang L, Larjava H, Isaka K, Saarialho-Kere U. Differential expression of matrilysin-1 (MMP-7), 92 kD gelatinase (MMP-9), and metalloelastase (MMP-12) in oral verrucous and squamous cell cancer. J Pathol. 2004;202(1):14–22. doi: 10.1002/path.1479. [DOI] [PubMed] [Google Scholar]
- Brinckerhoff CE, Rutter JL, Benbow U. Interstitial collagenases as markers of tumor progression. Clin Cancer Res. 2000;6(12):4823–4830. [PubMed] [Google Scholar]
- Johanssona N, Ahonenb M, Kähäri VM. Matrix metalloproteinases in tumor invasion. Cell Mol Life Sci. 2000;57:5–15. doi: 10.1007/s000180050495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage EH, Reed M, Funk SE, Truong T, Steadele M, Puolakkainen P, Maurice DH, Bassuk JA. Cleavage of the matricellular protein SPARC by matrix metalloproteinase 3 produces polypeptides that influence angiogenesis. J Biol Chem. 2003;278:37849–37857. doi: 10.1074/jbc.M302946200. [DOI] [PubMed] [Google Scholar]
- Chaudhary AK, Singh M, Bharti AC, Asotra K, Sundaram S, Mehrotra R. Genetic polymorphisms of matrix metalloproteinases and their inhibitors in potentially malignant and malignant lesions of the head and neck. J Biomed Sci. 2010;17:10. doi: 10.1186/1423-0127-17-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahari VM, Saarialho-Kere U. Matrix metalloproteinases and their inhibitors in tumour growth and invasion. Ann Med. 1999;31:34–45. doi: 10.3109/07853899909019260. [DOI] [PubMed] [Google Scholar]
- Zinzindohouém F, Blons H, Hans S, Loriot MA, Houllier AM, Brasnu D, Laccourreye O, Tregouet DA, Stucker I, Laurent-Puig P. Single nucleotide polymorphisms in MMP1 and MMP3 gene promoters as risk factor in head and neck squamous cell carcinoma. Anticancer Res. 2004;24(3b):2021–6. [PubMed] [Google Scholar]
- Hashimoto T, Uchida K, Okayama N, Imate Y, Suehiro Y, Hamanaka Y, Ueyama Y, Shinozaki F, Yamashita H, Hinoda Y. Association of matrix metalloproteinase (MMP)-1 promoter polymorphism with head and neck squamous cell carcinoma. Cancer Lett. 2004;211(1):19–24. doi: 10.1016/j.canlet.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Fang S, Jin X, Wang R, Li Y, Guo W, Wang N, Wang Y, Wen D, Wei L, Zhang J. Polymorphisms in the MMP1 and MMP3 promoter and non-small cell lung carcinoma in North China. Carcinogenesis. 2005;26:481–486. doi: 10.1093/carcin/bgh327. [DOI] [PubMed] [Google Scholar]
- Ghilardi G, Biondi ML, Caputo M, Leviti S, DeMonti M, Guagnellini E, Scorza R. A single nucleotide polymorphism in the matrix metalloproteinase-3 promoter enhances breast cancer susceptibility. Clin Cancer Res. 2002;8(12):3820–3823. [PubMed] [Google Scholar]
- Ashworth JL, Murphy G, Rock MJ. Fibrillin degradation by matrix metalo protienase: Implication for connective tissue remodelling. Biochem J. 1999;340(Pt 1):171–81. doi: 10.1042/0264-6021:3400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Eriksson P, Hamsten A, Kurkinen M, Humphries SE, Henney AM. Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression. J Biol Chem. 1996;271(22):13055–13060. doi: 10.1074/jbc.271.22.13244. [DOI] [PubMed] [Google Scholar]
- Ye S, Watts GF, Mandalia S, Humphries SE, Henney AM. Preliminary report; genetic variation in the human stromelysin promoter is associated with progression of coronary atherosclerosis. Br Heart J. 1995;73:209–15. doi: 10.1136/hrt.73.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Zhou W, Asomaning K, Lin X, Wain JC, Lynch TJ, Liu G, Christiani DC. Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis. 2006;27:1024–1029. doi: 10.1093/carcin/bgi283. [DOI] [PubMed] [Google Scholar]
- Gnasso A, Motti C, Irace C, Carallo C, Liberatoscioli L, Bernardini S, Massoud R, Mattioli PL, Federici G. Genetic variation in human stromelysin gene promoter and common carotid geometry in healthy male subjects. Arterioscler Thromb Vasc Biol. 2000;20:1600–1605. doi: 10.1161/01.atv.20.6.1600. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2003;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y, Ohnishi M, Ohyama T, Shingaki S, Kaji M. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer. 2003;106(5):683–689. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- Lin SC, Lo SS, Liu CJ, Chung MY, Huang JW, Chang KW. Functional genotype in matrix metalloproteinases-2 promoter is a risk factor for oral carcinogenesis. J Oral Pathol Med. 2004;33:405–9. doi: 10.1111/j.1600-0714.2004.00231.x. [DOI] [PubMed] [Google Scholar]
- Lin SC, Chung MY, Huang JW, Shieh TM, Liu CJ, Chang KW. Correlation between functional genotypes in the matrix metalloproteinases-1 promoter and risk of oral squamous cell carcinomas. J Oral Pathol Med. 2004;33:323–6. doi: 10.1111/j.1600-0714.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- Tu HF, Liu CJ, Chang CS, Lui MT, Kao SY, Chang CP, Liu TY. The functional -1171 (5A/6A) polymorphisms of matrix metalloproteinase 3 gene as a risk factor for oral submucous fibrosis among male areca users. Oral Pathol Med. 2006;3(5):99–103. doi: 10.1111/j.1600-0714.2006.00370.x. [DOI] [PubMed] [Google Scholar]
- Pandya S, Chaudhary AK, Sngh M, Mehrotra R. Correlation of histopathological diagnosis with habits and clinical findings in oral submucous fibrosis. Head & neck Oncology. 2009;1(1):11. doi: 10.1186/1758-3284-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CJ, Chiang CP, Chang ML. Association between genetic polymorphism of tumour necrosis factor-alpha and risk of oral submucous fibrosis, a pre cancerous condition of oral cancer. J Dent Res. 2001;80(12):2055–2059. doi: 10.1177/00220345010800120601. [DOI] [PubMed] [Google Scholar]
- Shin YN, Lin CJ, Chang KW, Lee YJ, Liu HF. Association of CTLA-4 gene polymorphism with oral submucous fibrosis in Taiwan. J Oral Pathol Med. 2004;33:200–3. doi: 10.1111/j.0904-2512.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Vairaktaris E, Yapijakis C, Vasiliou S, Derka S, Nkenke E, Serefoglou Z, Vorris E. Association of -1171 promoter polymorphism of matrix metalloproteinase-3 with increase risk for oral cancer. Anticancer Res. 2007;27(6B):4095–100. [PubMed] [Google Scholar]
- Fang S, Jin X, Wang R, Li Y, Guo W, Wang N, Wang Y, Wen D, Wei L, Zhang J. Polymorphisms in the MMP1 and MMP3 promoter and non-small cell lung carcinoma in North China. Carcinogenesis. 2005;26:481–486. doi: 10.1093/carcin/bgh327. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jin X, Fang S, Li Y, Wang R, Guo W, Wang N, Wang Y, Wen D, Wei L, Kuang G, Dong Z. The functional SNP in the matrix metalloproteinase-3 promoter modifies susceptibility and lymphatic metastasis in esophageal squamous cell carcinoma but not in gastric cardiac adenocarcinoma. Carcinogenesis. 2004;25:2519–2524. doi: 10.1093/carcin/bgh269. [DOI] [PubMed] [Google Scholar]
- Blons H, Gad S, Zinzindohoué F, Manière I, Beauregard J, Tregouet D, Brasnu D, Beaune P, Laccourreye O, Laurent-Puig P. Matrix metalloproteinase 3 polymorphism: a predictive factor of response to neoadjuvant chemotherapy in head and neck squamous cell carcinoma. Clin Cancer Res. 2004;10(8):2594–9. doi: 10.1158/1078-0432.CCR-1116-03. [DOI] [PubMed] [Google Scholar]
- Smolarz B, Szyłło K, Romanowicz-Makowska H, Niewiadomski M, Kozłowska E, Kulig A. PCR analysis of matrix metalloproteinase 3 (MMP-3) gene promoter polymorphism in ovarian cancer. Pol J Pathol. 2003;54(4):233–8. [PubMed] [Google Scholar]
- Yoon S, Tromb G, Vongpunsawad S, Ronkainen A, Juvonen T, Kuivaniemi H. Genetic analysis of MMP3 and MMP9 and PAI-1 in Finnish patients with abdominal aortic or intracranial aneurysms. Biochem Biophys Res Communic. 1999;265:563–568. doi: 10.1006/bbrc.1999.1721. [DOI] [PubMed] [Google Scholar]
- Hinoda Y, Okayama N, Takano N, Fujimura K, Suehiro Y, Hamanaka Y, Hazama S, Kitamura Y, Kamatani N, Oka M. Association of functional polymorphisms of matrix metalloproteinase-1 (MMP-1) and MMP-3 genes with colorectal cancer. Int J Cancer. 2002;102:526–529. doi: 10.1002/ijc.10750. [DOI] [PubMed] [Google Scholar]
- Nishijima C, Hayakawa I, Matsushita T, Komura K, Hasegawa M, Takehara K, Sato S. Autoantibody against matrix metalloproteinase-3 in patients with systemic sclerosis. Clin Exp Immunol. 2004;138(2):357–63. doi: 10.1111/j.1365-2249.2004.02615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawaki Y, Ikuta Y, Idobe Y, Kawasaki H. Serum matrixmetalloproteinase-1 in patients with chronic viral hepatitis. J Gastroenterol Hepatol. 1999;14(2):138–45. doi: 10.1046/j.1440-1746.1999.01821.x. [DOI] [PubMed] [Google Scholar]
- Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- Strup-Perrot C, Mathé D, Linard C, Violot D, Milliat F, François A, Bourhis J, Vozenin-Brotons MC. Global gene expression profiles reveal an increase in mRNA levels of collagens, MMPs, and TIMPs in late radiation enteritis. Am J Physiol Gastrointest Liver Physiol. 2004;287(4):G875–85. doi: 10.1152/ajpgi.00088.2004. [DOI] [PubMed] [Google Scholar]
- Chang YC, Yang SF, Tai KW, Chou MY, Hsieh S. Increased tissue inhibitor of metalloproteinase-1 expression and inhibition of gelatinase: A activity in buccal mucosal fibroblasts by arecoline as possible mechanisms for oral submucous fibrosis. Oral Oncol. 2002;38(2):195–200. doi: 10.1016/S1368-8375(01)00045-8. [DOI] [PubMed] [Google Scholar]
- Nishizawa R, Nagata M, Noman AA, Kitamura N, Fujita H, Hoshina H, Kubota T, Itagaki M, Shingaki S, Ohnishi M, Kurita H, Katsura K, Saito C, Yoshie H, Takagi R. The 2G allele of promoter region of matrix metalloproteinase-1 as an essential pre-condition for the early onset of oral squamous cell carcinoma. BMC Cancer. 2007;7:187. doi: 10.1186/1471-2407-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCawley L, Crawford H, King L, Mudgett J, Matrisian L. Aprotective role for matrix metalloproteinase-3 in squamous cell carcinoma. Cancer Res. 2004;64:6965–6972. doi: 10.1158/0008-5472.CAN-04-0910. [DOI] [PubMed] [Google Scholar]