Abstract
Accurate volume estimation in PET is crucial for different oncology applications. The objective of our study was to develop a new fuzzy locally adaptive Bayesian (FLAB) segmentation for automatic lesion volume delineation. FLAB was compared with a threshold approach as well as the previously proposed fuzzy hidden Markov chains (FHMC) and the Fuzzy C-Means (FCM) algorithms. The performance of the algorithms was assessed on acquired datasets of the IEC phantom, covering a range of spherical lesion sizes (10–37mm), contrast ratios (4:1 and 8:1), noise levels (1, 2 and 5 min acquisitions) and voxel sizes (8mm3 and 64mm3). In addition, the performance of the FLAB model was assessed on realistic non-uniform and non-spherical volumes simulated from patient lesions. Results show that FLAB performs better than the other methodologies, particularly for smaller objects. The volume error was 5%–15% for the different sphere sizes (down to 13mm), contrast and image qualities considered, with a high reproducibility (variation <4%). By comparison, the thresholding results were greatly dependent on image contrast and noise, whereas FCM results were less dependent on noise but consistently failed to segment lesions <2cm. In addition, FLAB performed consistently better for lesions <2cm in comparison to the FHMC algorithm. Finally the FLAB model provided errors less than 10% for non-spherical lesions with inhomogeneous activity distributions. Future developments will concentrate on an extension of FLAB in order to allow the segmentation of separate activity distribution regions within the same functional volume as well as a robustness study with respect to different scanners and reconstruction algorithms.
Keywords: Algorithms; Bayes Theorem; Computer Simulation; Fuzzy Logic; Humans; Image Processing, Computer-Assisted; methods; Markov Chains; Neoplasms; radionuclide imaging; Normal Distribution; Positron-Emission Tomography; methods; Reproducibility of Results
Keywords: Oncology, PET, segmentation, volume determination
I. Introduction
Positron Emission Tomography (PET) is now a widely used tool in the field of oncology, especially in applications such as diagnosis, and more recently radiotherapy planning [1] or response to therapy and patient follow-up studies [2]. On the one hand, accurate activity concentration recovery is crucial for correct diagnosis and monitoring response to therapy. On the other hand, applications such as Intensity-Modulated Radiation Therapy (IMRT) treatment planning using PET also require accurate shape and volume determination of the lesions of interest, in order to reduce collateral damage to healthy tissues and to ensure maximum dose delivered to the active disease. Various methodologies used for the determination of volume of interest (VOI) have been proposed. On the one hand, segmentation methods requiring a manual delineation of the boundaries of the object of interest have been established as laborious and highly subjective [2]. Alternatively, the performance of already available automatic algorithms is hampered by the low resolution and associated partial volume effects (PVE), as well as low contrast and signal to noise ratios generally characterizing PET images.
Most of the previously proposed work dealing with VOIs determination in PET use thresholding, either adaptive, based on a priori Computed Tomography (CT) knowledge [3], or a fixed threshold using values derived from phantom studies (from 30 to 75% of maximum local activity concentration value) [1], [2], [3]. Thresholding is however known to be significantly susceptible to noise and contrast variations, leading to variable VOIs determination as shown in recent clinical studies [4]. As far as automatic detection of lesions from PET datasets is concerned, different methodologies have been previously proposed including edge detection [5], watersheds [6], fuzzy C-Means [7] or clustering [8]. The performance of these algorithms is also sensitive to variations in lesion-to-background contrast and/or noise levels. In addition, past work has in its majority considered the ability of such automatic methodologies for the detection of lesions (sensitivity), and not for their performance in terms of accuracy in the specific VOI determination task. Finally, all of the afore-mentioned algorithms have additional drawbacks associated with necessary pre- or post-processing steps. For example in the case of the watershed algorithm, a pre-processing step using a filtering pass is required to smooth the image, and a post-processing step is necessary to fuse the regions resulting from the over-segmentation of the algorithm. Such a need for user-dependent initializations, pre- and post-processing steps, or additional information like CT or expert knowledge render the use of these algorithms more complicated and the outcome dependent on choices made by the user in relation to these necessary steps.
Bayesian based image segmentation methods are automatic algorithms allowing noise modelling and have shown to be less sensitive to noise than other segmentation approaches due to their statistical modelling [9]. They offer an unsupervised estimation of the parameters needed for the image segmentation and limit the user’s input to the number of classes to be searched for in the image. Reconstructed images require no further pre- or post-processing treatment (such as for example filtering) prior to the segmentation process. Instead, image noise is considered as additional information (a parameter in the classification decision process) to be taken into account rather than to be filtered or ignored. They have only been previously used in PET imaging in the form of Hidden Markov Fields (HMF) [10] and more recently we have investigated the performance of hidden Markov chains (HMC) for volume determination, a faster model that was in addition extended to include fuzzy modelling, Fuzzy HMC (FHMC) [11]. Although FHMC was shown to provide overall superior results relative to the threshold reference methodology, independent of lesion contrast and image signal-to-noise ratio, it is unable to correctly segment objects <2 cm in diameter. This is mainly due to the 3D Hilbert-Peano path [12] used to transform the 3D volume into an 1D chain, since voxels defining small objects may find themselves far away from each other on the chain, thus being misidentified by the algorithm as noise and becoming not significant enough to form a class apart from the background.
Consequently, the main objectives of this study were to improve the segmentation of small objects by (a). developing a fuzzy local adaptive Bayesian (FLAB) model, and (b) comparing the performance of this new algorithm with that of the thresholding methodologies currently used in clinical practice as well as the Fuzzy C-Means (FCM) and the previously proposed FHMC algorithms. In addition, as a secondary objective we have also investigated the use of the Pearson’s system [13] in order to potentially improve the noise modelling used in the algorithm, instead of simply assuming a Gaussian distribution.
Different imaging conditions were considered in this study in terms of statistical quality, as well as lesion size and source-to-background (S/B) ratio. The images were reconstructed using an iterative algorithm, since this type of reconstruction algorithms form today’s state of the art in whole body PET imaging in routine clinical oncology practice [14]. In addition, the new FLAB algorithm was evaluated using simulated images of non homogeneous and non spherical tumors derived from tumors of patients undergoing radiotherapy.
II. Material and Methods
A. FLAB model
The FLAB model is an unsupervised statistical methodology that takes place in the Bayesian framework. Let T be a finite set corresponding to the voxels of a 3D PET image. We consider two random processes Y = (yt)t∈T and X = (xt)t∈T. Y represents the observed image and takes its values in ℝ whereas X represents the “hidden” segmentation map and takes its values in the set {1, …, C}, with C being the number of classes. The segmentation problem consists of estimating the hidden X from the available noisy observation Y. The relationship between X and Y can be modeled by the joint distribution P(X,Y), which can be obtained using the Bayes formula:
| (1) |
P(Y|X) is the likelihood of the observation Y conditionally with respect to the hidden ground-truth X, and P(X) is the prior knowledge concerning X. The Bayes rule allows the determination of the posterior distribution of X with respect to the observation Y: P(X|Y). Contrary to the FHMC model [11], we do not assume here that a Markov process can model the prior distribution of X, thus simplifying its expression.
The fuzzy measure
The general idea behind the implementation of a fuzzy model within the Bayesian framework was previously introduced in [15], [16] and was used for a local Bayesian segmentation scheme in [15]. Its implementation is based on the incorporation of a finite number of fuzzy levels Fi in combination with two homogeneous (or “hard”) classes, in comparison to the standard implementation where only a finite number of hard classes are considered. This model allows the coexistence of voxels belonging to one of two hard classes and voxels belonging to a “fuzzy level” depending on its membership to the two hard classes. While the statistical part of the algorithm models the uncertainty of the classification, with the assumption being that the voxel is identified but the observed data is noisy, the fuzzy part models the imprecision of the voxel’s membership, with the assumption being that the voxel may contain both classes. One way to achieve this extension is to simultaneously use Dirac and Lesbegue measures, considering that X in the fuzzy model takes its values in [0,1] instead of Ω = {1, …, C}. We define therefore a new measure ν =δ0 + δ1 + ζ on [0,1], given that δ0 and δ1 are the Dirac measures at 0 and 1, and ζ is the Lesbegue measure on the fuzzy interval ]0,1[. This approach is adapted for the segmentation of PET images since they are both noisy and of low resolution. The “noise” aspect when considering Bayesian models is the way the values of each class to be found in the image are distributed around a mean value. The noise model used, whose respective mean and variance are to be determined by the estimation steps, can therefore be adapted to image specific characteristics. Finally, the fuzzy measure facilitates a more realistic modelling of the objects’ borders transitions between foreground and background, allowing in such a way to indirectly account for the effects of blurring associated with low resolution PET images.
Distribution of X (a priori model)
Using ν =δ0+δ1+ζ as a measure on [0,1], the a priori distribution of each xt can be defined by a density h on [0,1], with respect to ν. If we assume that X is a stationary process and that the distribution of each xt is uniform on the fuzzy class, this density can be written as:
| (2) |
where, h satisfies the following normalization condition:
Using this simple modelling for the prior distribution leads to ignoring the spatial relationship of each voxel with respect to its local neighborhood. Although it is possible to include such spatial information using the contextual framework [15], the use of such modelling leads to an increase in the number of parameters to be handled, and in practice, no more than one or two neighbors can be actually taken into account. Hence, the contextual approach is not of interest since we aim to explore all the information available in the 3D volume around each voxel, i.e. at least 26 neighbors (8-connectivity extended in three dimensions). As an alternative, the adaptive framework [15] can be used. In this adaptive modelling, the spatial information is inserted into the estimation step of the algorithm (see section parameters estimation).
Distribution of Y (observation or noise model) and the Pearson’s system
In order to define the distribution of Y conditional on X, let us consider two independent random variables Y0 and Y1, associated with the two “hard” values 0 and 1, whose densities f0and f1 are characterized by means and variances (μ0, σ02) and (μ1, σ12) respectively. The mean and variance of each fuzzy level Fi are derived from the ones estimated in the two hard classes as follows:
| (3) |
where, εi is the value associated to a fuzzy level Fi. For the case of two fuzzy levels and were used according to results previously published [11].
The assumption that the noise for each class of the observed data can fit a Gaussian distribution was considered as a first approximation as with the previous implementation of the FHMC algorithm [11]. In this work we propose the study of the Pearson’s system that contains seven other distributions. In this context, instead of using a Gaussian distribution, an additional step is introduced to detect which laws best fit the actual distribution of the voxels in the image, for each class considered at a given iteration of the estimation step of the algorithm. The theory behind the Pearson’s system has been previously detailed in [17] and a description of its use in mixture estimation and statistical image segmentation is given in [13]. Here, we briefly describe the Pearson’s system in our particular context.
A distribution density f on ℝ belongs to the Pearson’s system if it satisfies:
| (4) |
Different shapes of distributions as well as the parameters determining a given distribution are provided by the variations of the coefficients a, c0, c1 and c2. For m =1, 2, 3 and 4, let us consider the first four statistical moments of a partition Yp of Y defined by:
| (5) |
We also define two parameters γ1 and γ2 as follows:
| (6) |
where is called “skewness” and γ2 is called “kurtosis”. The coefficients a, c0, c1 and c2 are related to (5) and (6) by equations that can be found in the appendix (section A.1). Given , the eight distribution density families {f1,…, f8} contained in the system of Pearson can now be defined by a set of conditions using λ, γ1 and γ2 (see appendix, section A.2). These eight distribution density families are illustrated in figure 1. Finally, the protocol used for the determination of which density family best fits each measured distribution can be found in section A.3 of the appendix.
Fig. 1.
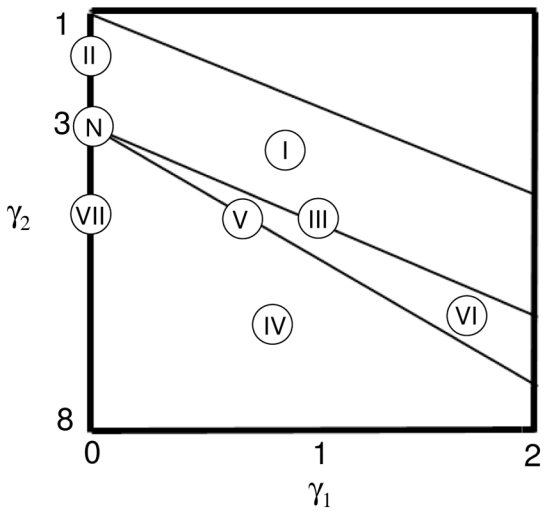
The eight distribution families in the graph of Pearson, function of gamma1 and gamma 2 [17]. I for Beta I, II for type II, III for Gamma, IV for type IV, V for Inverse Gamma, VI for Beta II, VII for type VII and N for Normal.
Parameters estimation
The different parameters necessary to be estimated for the segmentation process are:
| (7) |
Both a priori (A) and noise (B) parameters are unknown and may vary from one image to another. An iterative procedure called Stochastic Expectation Maximization (SEM) [18], a stochastic version of the EM algorithm [19], is used for the estimation of these parameters. This is achieved by sampling a realization of X according to its posterior distribution P(X|Y) and computing empirical values of the parameters of interest using this realization. The stochastic nature of this procedure makes it less sensitive to the initial guess of the parameters using the K-Means [20] than deterministic procedures like the EM algorithm. The system of Pearson can be used as an additional step (inside each iteration of the algorithm) in order to determine the type of distribution to use. The posterior distribution d (with respect to class c for a given voxel t used at iteration q) for sampling the posterior realization is given by:
| (8) |
where, fq−1(yt|c) is a density whose distribution type is chosen using the Pearson system and whose mean and variance were estimated at iteration q−1, and, pt,cq−1 is the prior probability of voxel t belonging to class c estimated at iteration q−1.
In the adaptive framework priors are re-estimated using a local neighbouring window with priors pt,c depending on the position t of the voxel in the image. Although in the 2D case, a window centred on the voxel of interest is used [15], for our application we use a 3D ”cube” centred on each voxel. The size of the estimation “cube” was experimentally determined for the specific application of PET imaging, since it depends on the size of the objects of interest (10–50mm in diameter) relative to the reconstructed voxel size (2×2×2 or 4×4×4 mm3). An estimation cube should from one hand be small enough to yield good local characteristics [15], while on the other hand it should not be too large with respect to the size of the object of interest. Considering this, we tested two different estimation “cube” sizes; namely covering 3×3×3 and 5×5×5 voxels.
It is worth noting that only the priors are concerned by the use of the adaptive framework. Noise parameters are estimated the same way as in the blind context [15]. The detailed description of the SEM algorithm in our context is given in the appendix (section B.1).
Segmentation
In order to perform segmentation on a voxel by voxel basis, we need to use a criterion to classify each voxel as either part of the background or the functional VOI. For this purpose we use the maximum posterior likelihood (MPL) method as suggested by [15]. To compute a solution, the MPL method requires the parameters defining the a priori model (priors of each class for each voxel) as well as the noisy observation data model (mean and variance of each class), estimated using SEM. The MPL computes the posterior density and selects for each voxel the class that maximizes it, using the procedure described below.
Let us consider d(ε|yt) given by (8) computed using the parameters estimated by the SEM estimation algorithm. Using d(F|yt) =1 − d(0|yt) − d(1|yt), the decision rule assigning the class c or fuzzy level Fi to the voxel t knowing the observed value yt is given by the following procedure:
For each voxel, let ct = arg maxn∈{0,1,F} d(n|yt). If ct ∈ {0,1}, then assign the hard class 0 (ct = 0) or 1 (ct = 1) to the voxel t. Else if ct belongs to the fuzzy domain (ct = F), use ct = arg maxn∈]0,1[d(n|yt) to determine its exact value using the quantitation of the fuzzy interval into fuzzy levels (see section 2.1.3) and assign one of the fuzzy levels to the voxel. In our implementation of FLAB, each ct can take four different values: 0, and 1.
B. Alternative approaches used for comparison
Thresholding
Various thresholding methodologies have been proposed in the past for functional volume determination [2], [3], [4]. For comparison purposes with the developed methodology, threshold at 42% of the maximum value inside the lesion was chosen for VOI determination, based on suggestions from previous publications [2], [3]. The methodology was implemented through region growing using the voxel of maximum intensity in the object of interest as a seed. Using a 3-D neighborhood (26 neighbors) the region is iteratively increased by adding neighboring voxels if their intensity is superior or equal to the selected threshold value. The results derived using this method will be denoted from here onwards as T42.
Fuzzy C-Means
The Fuzzy C-Means algorithm was introduced in [21]. It was suggested for PET image segmentation in [7]. For the purpose of this study it was implemented using the following objective function O:
| (9) |
where e ≥ 1 is a weighting exponent and mi are the centre values of the classes. The weighting exponent e controls the fuzzy aspect of the image and is usually set to 2 (hard segmentation is represented by e = 1). The algorithm converges to the value at which the objective function has a local maximum. The results derived using this method will be denoted from here onwards as FCM.
C. Validation studies
Datasets
Acquisitions of the IEC image quality phantom [22], containing six different spherical lesions of 10, 13, 17, 22, 28 and 37 mm in diameter (figure 3(a)) were carried out in list-mode format using a Philips GEMINI PET/CT scanner. The spatial resolution of this system is 4.9 mm full width at half maximum (FWHM) at the center of the field of view [23]. Partial volume effects are therefore expected to be significant even for the largest sphere. The 28 mm diameter sphere was not considered in this study since it was replaced by a hand-made plastic sphere whose diameter was not known precisely. Different parameters were considered covering a large spectrum of configurations allowing assessment of the influence of different parameters susceptible to affect the functional VOI determination. The statistical quality of the images was varied by considering 1, 2 or 5 minutes list-mode time frames. Two different signal-to-background (S/B) ratios (4:1 and 8:1) were considered, by introducing 7.4kBq/cm3 in the background and 29.6 or 59.2kBq/cm3 respectively in the spheres. Two different voxel sizes (2×2×2 or 4×4×4 mm3) were used in the reconstruction of each of the different statistical quality datasets using the 3D RAMLA algorithm, with specific parameters previously optimized for clinical use [14]. Visual illustration of the acquired images is given in figure 2. In addition, an estimation of the FLAB algorithm’s reproducibility was performed by considering five different 1 minute list-mode time frames acquired consecutively and reconstructed using 8 mm3 voxels.
Fig. 3.
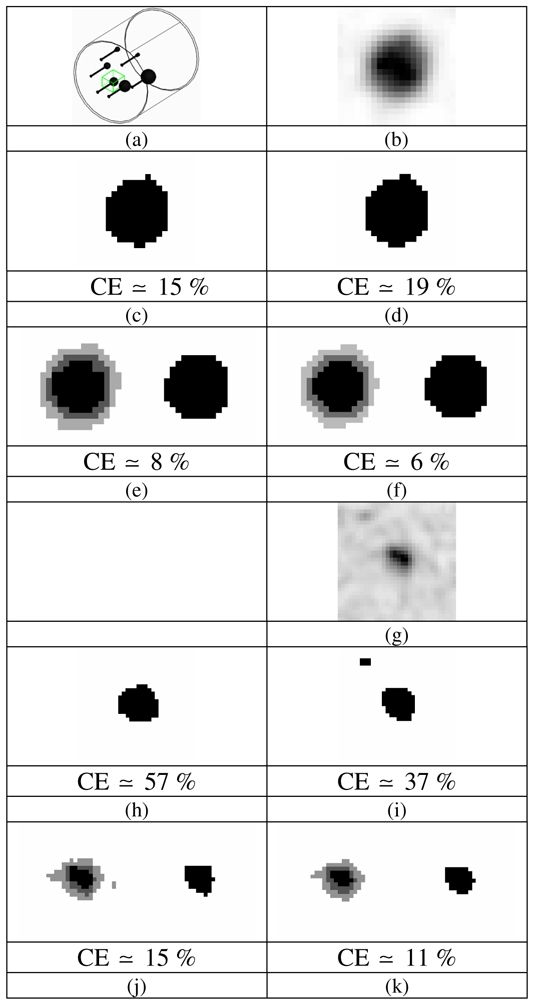
(a) Graphical representation of the IEC phantom and illustration of the 3D box selection for the 22 mm sphere and examples of segmentation maps (only central slice is shown); (b–f) for the 22 mm sphere (8:1 contrast, 5 min acquisition) and (g–k) for the 17 mm sphere (4:1 contrast, 2 min acquisition) with corresponding volume errors (computed on the whole volume): (b & g) PET ROI, (c & h) T42 map, (d & i) FCM map, (e & j) FHMC and (f & k) FLAB maps with 2 fuzzy levels (light and dark grey voxels). Both images are extracted from 8 mm3 voxel size reconstructions.
Fig. 2.
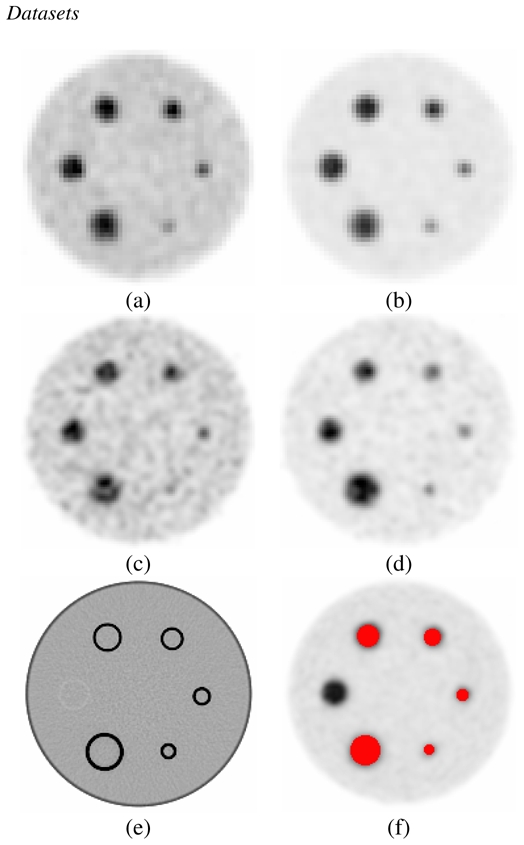
Different images used in the segmentation study; (a) ratio 4:1, 2 min acquisition time, 64 mm3 voxels, (b) ratio 8:1, 2 min, 64 mm3, (c) ratio 4:1, 2 min, 8 mm3, (d) ratio 8:1, 2 min, 8 mm3, (e) CT acquisition, (f) voxel-by- voxel ground-truth generated using CT image on the PET image. Note the 28 mm sphere is in plastic and not clearly seen (since its real diameter was unknown this sphere was excluded from any analysis in this work).
Finally, to test the algorithm against more clinically realistic conditions of tumor shapes, we simulated three lesions with non spherical shapes and inhomogeneous activity distributions. These lesions were generated using real lung tumor images from three patients undergoing 18FDG PET scans for radiotherapy treatment planning purposes. A ground-truth was drawn by a nuclear medicine physician (on a slice-by-slice basis) based on the reconstructed patient images. In the case of the first tumor, the simulated contrast between the region of the highest activity concentration and the rest of the tumor was around 2.2:1 whereas in the case of the second tumor, it is closer to 1.4:1. Finally, the third tumor is almost homogeneous. The overall contrast between the whole tumor and the background was 6:1 and 5:1 for the first and second tumors respectively and less than 2:1 for the third one. In terms of lesion size, the largest lesion “diameter” was 4.1 cm, 2.9 cm and 1.5cm for the first, second and third lesion respectively. These lesions were subsequently placed within the lungs of the NCAT phantom [24]. No respiratory or cardiac motion was considered. Normal organ FDG concentration was assumed for the simulation [25], with the maximum activity concentration in the lesions being four times the mean activity concentration in the lungs. The NCAT emission and attenuation maps were finally combined with a model of the Philips PET/CT scanner previously validated with GATE [26]. A total of 45 million coincidences were simulated corresponding to the statistics of a standard clinical acquisition over a single axial field of view of 18 cm [26]. Images were subsequently reconstructed from the list mode output of the simulation using 8 mm3 voxels. As well as using all of the simulated true coincidences, images were reconstructed for each lesion using only 40% and 20% of the overall detected coincidences in order to evaluate the accuracy of the segmentation algorithms at different noise levels (similar to the IEC phantom study using 5, 2 and 1 min acquisitions for the image reconstruction). Visual illustration of these simulated tumor images (central slice), with their ground-truth drawn from the corresponding patient tumors are displayed in figures 7, 8 and 9(a–c). Each segmentation algorithm considered was applied to the lesion and the segmentation map was compared with the ground-truth. Note that in this framework, the ground-truth does not need to be accurate with respect to the true patient image. What is important is that we are able to compare the segmentation obtained on the simulated image with the ground-truth used in the simulation. The corresponding segmentation maps (central slice) for each algorithm can be found in figures 7, 8 and 9(d–g).
Fig. 7.

(a) Real tumour used as model, (b) voxelized ground-truth (manually drawn) and its binary version, and (c) simulated tumour. Segmentation binary maps obtained using (d) T42, (e) FCM, (f) FHMC and (g) FLAB are shown. Image is 34×34 voxels with 8 mm3 voxels.
Fig. 8.
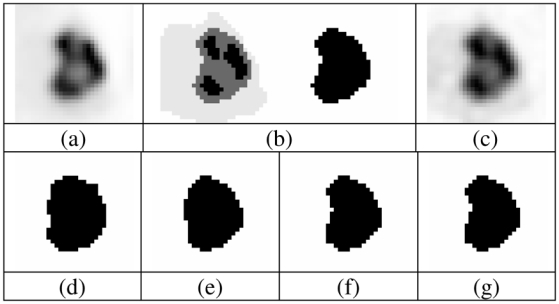
(a) Real tumour used as model, (b) voxelized ground-truth (manually drawn) and its binary version, and (c) simulated tumour. Segmentation binary maps obtained using (d) T42, (e) FCM, (f) FHMC and (g) FLAB are shown. Image is 30×30 voxels with 8 mm3 voxels.
Fig. 9.
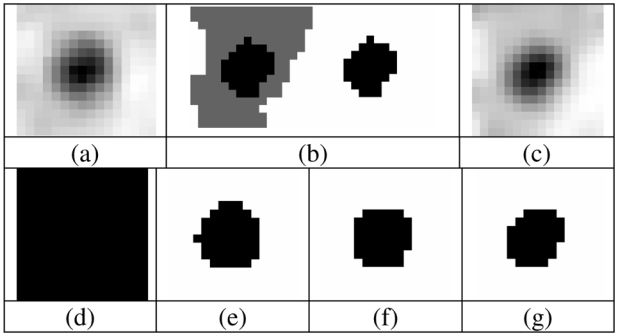
(a) Real tumour used as model, (b) voxelized ground-truth (manually drawn) and its binary version, and (c) simulated tumour. Segmentation binary maps obtained using (d) T42, (e) FCM, (f) FHMC and (g) FLAB are shown. Image is 16×16 voxels with 8 mm3 voxels
Analysis
As our goal is not lesion detection in the whole body image but the estimation of a lesion’s volume with the best accuracy possible, we assume that the lesion has been previously identified by the clinician and automatically or manually placed in a 3-D “box” well encompassing the object (see (figure 3(a)). Although no significant impact on the segmentation results was observed through small changes in placement or size of the box, certain conditions must be respected. Evidently it should be large enough to contain the entire extent of the object of interest and a significant number of background voxels so the algorithm is able to detect and estimate the parameters of the background class. On the other hand it should be small enough in order to avoid including neighboring tissues with significant uptake that would end up being classified as functional VOI, requiring manual post-processing. However, the shape of this box does not have to be perfectly cubic or with specified dimensions (contrary to the FHMC case [11]), and as a result it could be drawn accordingly to exclude structures in the background that are of no interest.
Subsequently, the images of the selected area were segmented in two classes (functional VOI and background) using each of the methods under evaluation (T42, FCM, FHMC and FLAB). In the FHMC and FLAB cases, considering the optimization results obtained in [11], two fuzzy levels were considered in the segmentation process and the functional volumes were defined using the first hard class and the first fuzzy level. A voxel-to-voxel ground-truth was generated for the phantom dataset using the CT image registered with the PET reconstructed image (see figure 2(e) and 2(f)). Classification errors (CE) were then computed on a voxel-by-voxel basis following the definition used in [11]:
| (10) |
PCE stands for positive classification errors, including voxels of the background that are classified as belonging to the object of interest, and NCE stands for negative classification errors including voxels of the object that are classified as belonging to the background. These classification errors essentially occur on the boundaries of the objects of interest because of activity “spill in” and “spill out”. If the segmentation results in PCEs and NCEs of equal amounts, the computed VOI would be very close to the true known volume whereas the shape and position of the object would be incorrect (this essentially occurs for objects >2 cm, while for smaller objects the errors are essentially PCE). As shown in equation 10, the total number of PCEs and NCEs is considered with respect to the number of voxels defining the sphere (VoS). Although the size of classification errors can be bigger than 100%, in the case where a large number of background voxels in the selected area of interest are misclassified as belonging to the sphere, maximum classification errors considered in this paper where limited to 100%, since any such values represent complete failure of the segmentation process. Although the combination of PCE and NCE into CE leads to a loss of information as far as the direction of the bias is concerned, classification errors represent more pertinent information than overall volume errors, which reflect neither accurate magnitude nor direction of the bias for a segmented volume. For comparison purposes overall volume errors (with respect to the known volume of the sphere) were also computed and shown in figure 6.
Fig. 6.
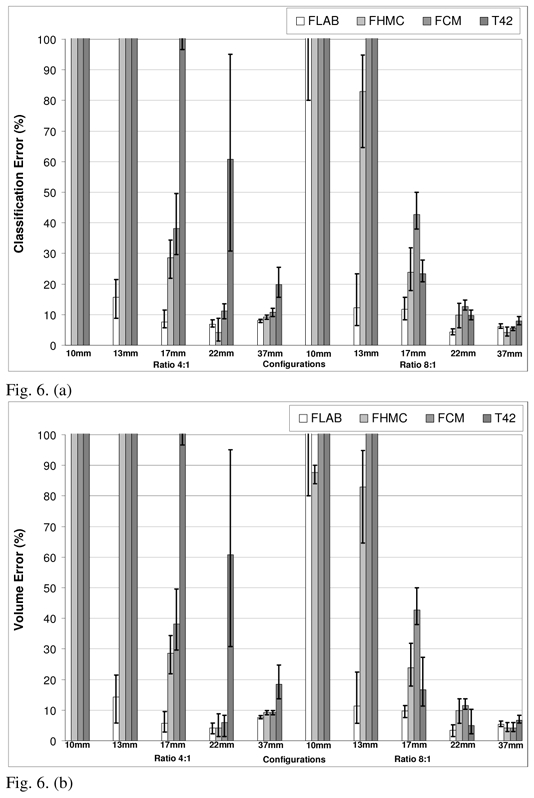
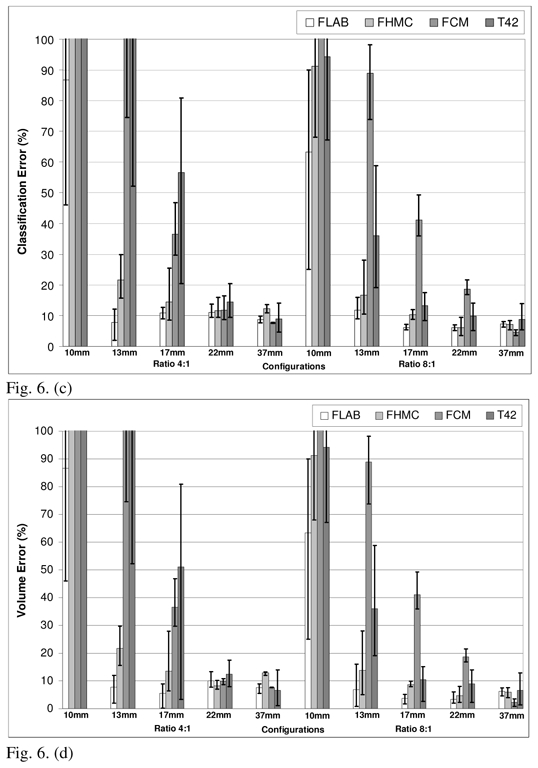
Comparison of performances for FLAB, FHMC, FCM and T42 on data reconstructed with (a) classification errors and (b) volume errors, for 64mm3 and (c) classification errors and (d) volume errors, for 8 mm3 voxels. The top of the error bar is the result concerning the worst statistical quality images (1 min acquisition), the medium one concerns the medium quality (2 min acquisition), and the lowest one corresponds to the superior statistical quality (5 min acquisition).
As far as the simulated tumors are concerned, both overall volume errors (with respect to the known volume of the ground-truth) and CE were computed. Since all the algorithms under investigation in this study perform binary segmentations (i.e. able to distinguish between tumor tissue and background only), no evaluation was performed of their ability to distinguish different regions within a given tumor.
III. Results
Different segmentation maps obtained using each of the methods under evaluation (FHMC, FLAB, T42 and FCM) are presented in figure 3(c–f) for a slice centered on the 22 mm sphere considering a “good quality” image (8:1 contrast and 5 min acquisition) (fig. 3(a)) to visually illustrate the variations of the segmentation maps obtained. Segmentation results in the case of a “lower quality” image (4:1 contrast and 2 min acquisition) and a smallest sphere (17 mm) (fig. 3(g)) are presented in figure 3(h–k). Both images are representative of the 8mm3 voxel size reconstructions.
In the different figures shown in this section the CE are given for all five spheres (10, 13, 17, 22 and 37 mm) and for both contrast ratios (4:1 on the left part of each figure, 8:1 on the right part) considered. The error bars in the figures represent the different results obtained for each of the 3 different levels of image statistical quality considered. The top of the error bar is the result concerning the worst statistical quality images (1 min acquisition), the medium one concerns the medium quality (2 min acquisition), and the lowest one corresponds to the superior statistical quality (5 min acquisition). The only exception is figure 5 where the error bars represent the variability of the FLAB segmentation results considering the application of the algorithm on multiple images of 1 minute acquisitions (five independent realizations).
Fig. 5.
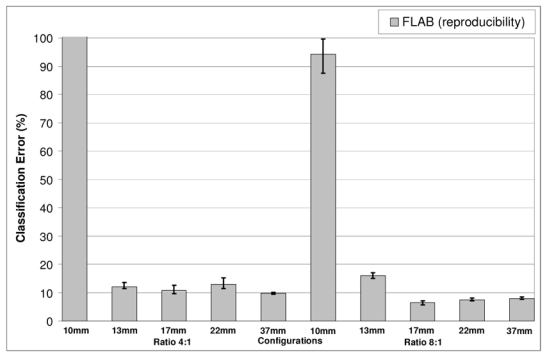
Study of FLAB reproducibility using five different 1 minute list-mode time frames (reconstructed with 8mm3 voxel size). The error bars represent the variability of the FLAB segmentation results considering the application of the algorithm on multiple images of 1 minute acquisitions (five realizations)
Figure 4 contains the results on the optimization of the algorithm for the specific application of lesion segmentation in PET images. Considering the selected volume of interest around a lesion, the Pearson’s system systematically led to the detection of Beta I distributions for both the background and the lesion activity distributions (although with different parameters). However, the parameters γ1 and γ2 (see section 2.1.3, eq. 6) placed the estimated distributions very close to the Gaussian one in the Pearson graph (as it can be seen in figure 1, the surface matching Beta I distribution (I) is in contact with the point defining the Normal distribution (N)). Consequently only small changes in the volume estimation results were consistently obtained using the Beta I instead of a Gaussian distribution (figure 4(a)). Considering these results the Gaussian distribution was kept in the final implementation of the algorithm for the description of both the background and lesion activity distributions.
Fig. 4.
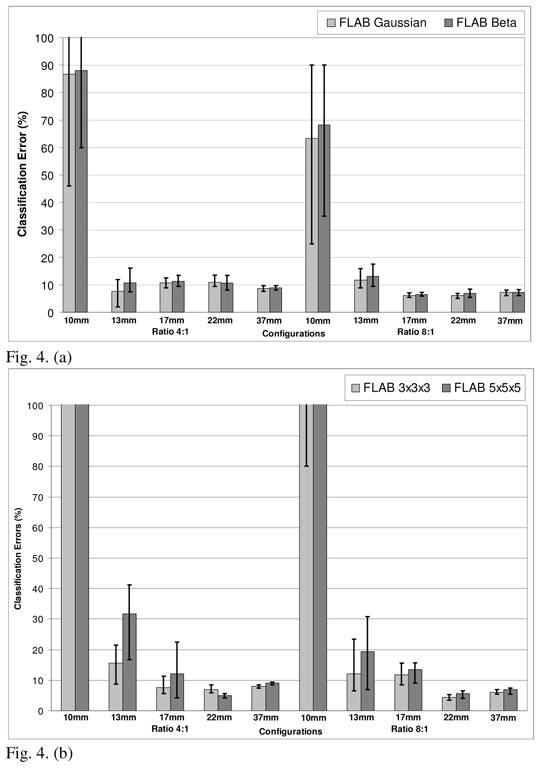
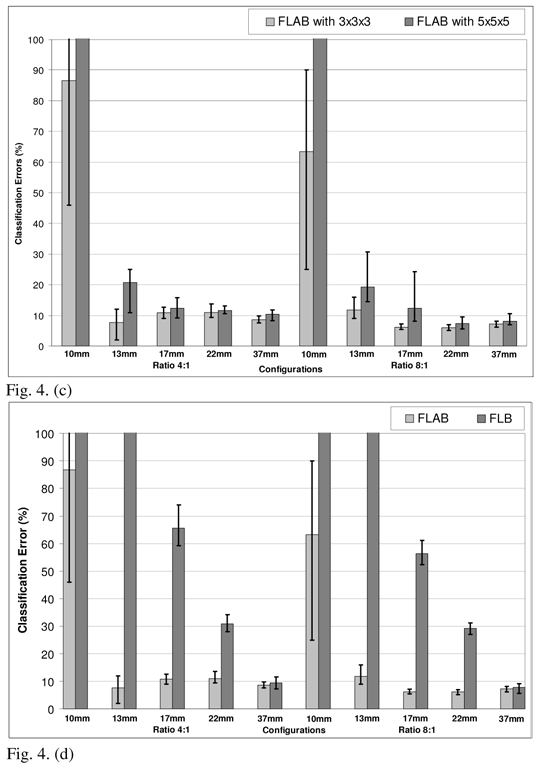
Optimization of the FLAB algorithm. Classification errors for (a) Beta I distributions (detected using the Pearson’s system) or Gaussian distributions (for the 8mm3 voxel size); (b) 3×3×3 or 5×5×5 voxels for the estimation cube (for the 64mm3 voxel size); (c) 3×3×3 or 5×5×5 voxels for the estimation cube (for the 8mm3 voxel size); (d) with (FLAB) or without (FLB) adaptive estimation of priors (for the 8mm3 voxel size). The top of the error bar is the result concerning the worst statistical quality images (1 min acquisition), the medium one concerns the medium quality (2 min acquisition), and the lowest one corresponds to the superior statistical quality (5 min acquisition).
In terms of the size of the estimation “cube” used for the re-estimation of the priors in the adaptive framework, a size of 3×3×3 voxels led to consistently better results across different lesion and voxel sizes as well as S/B contrast and noise configurations as shown in figures 4(b) and 4(c). Finally, figure 4(d) demonstrates the impact in terms of the improved results through the use of the adaptive estimation, for the 8mm3 configuration. In this figure the FLAB segmentation results are compared to the results without adaptive estimation (FLB for Fuzzy Local Bayesian, using the same fuzzy levels implementation), where priors are the same for all the voxels of the image and are computed using the entire image instead of using only the local neighbourhood of each voxel. As is demonstrated by this figure, the inclusion of the adaptive estimation significantly improves the segmentation results throughout the different lesion sizes and contrast configurations considered.
Results in relation to the FLAB algorithm’s reproducibility can be seen in figure 5. In this particular figure, error bars represent the variation of the segmentation results (mean and variance) using the five different images obtained from the consecutive 1 minute acquisitions. A variation of <4% in the segmented volumes was obtained from the application of the algorithm on the five different images for all spheres except from the 1 cm sphere which the algorithm consistently failed to correctly segment. This segmentation failure is most probably the cause of this larger variability observed for the segmented volume of the 1 cm sphere.
Figure 6 presents the classification errors and corresponding overall volume errors relative to the CT-based ground-truth obtained using each approach, for both 64 and 8 mm3 voxel sizes (fig. 6(a)–(b) and 6(c)–(d) respectively). Globally, volume errors are very closely linked to classification errors: when the segmentation results in strictly NCE, the volume error (underestimation) is equal to the CE. When the segmentation results in only PCE, the volume error (overestimation) is also equal to the CE. And when both NCE and PCE occur, the volume error is inferior to the CE (it essentially occurs for medium-sized spheres). FLAB led to superior results in comparison to all the other methodologies on the whole dataset. The proposed algorithm gives good results (on average between 5 and 20% CE) independently of the contrast ratio and for all spheres except from the 1 cm one for which a minimum error of 25% was obtained for the most favorable configuration evaluated (8:1 contrast and a 5 min. acquisition). The use of a reconstruction voxel size of 8mm3 allowed an improvement in the segmented volume errors from 10–25% to 5–15% for lesions between 1cm and 2cm.
As shown in figure 6, T42 gave errors <20% for the three biggest spheres with the 8:1 contrast and 64 mm3 voxel size, while for a 4:1 contrast T42 did not manage to accurately segment any of the spheres. By reducing the reconstruction voxel size to 8mm3 an improvement was obtained in the results of the T42 with errors <15% for the three larger spheres and a contrast 8:1, while errors of <20% were obtained for the 22mm and 37mm spheres with a 4:1 contrast ratio. In the case of the FCM algorithm errors of <20% and >40% were seen for lesions larger and smaller than 2 cm respectively. No substantial differences were seen in these results from the reduction in the reconstruction voxel sizes from 64 mm3 to 8 mm3. Finally, FLAB performed better in comparison to the previously developed fuzzy Bayesian approach (FHMC) for all different lesion sizes and statistical image qualities considered with a larger magnitude effect (improvements of over 100% in the errors) observed in the spheres with a diameter <2 cm. Relative to the FLAB results globally larger improvements in the accuracy of the segmented volumes were observed for the FHMC algorithm with a reduction in the reconstructed voxel size. On the other hand, in percentage terms the dependence of the algorithm results to the statistical quality of the images was similar for both the FLAB and FHMC results.
Figures 7, 8 and 9 show visual illustration of the segmentation maps obtained on the simulated tumors. Figure 10 contains the results for both classification errors (NCE+PCE divided by the number of voxels defining the tumor ground-truth volume) and volume errors (with respect to known overall volume of the tumor) for each approach.
Fig. 10.
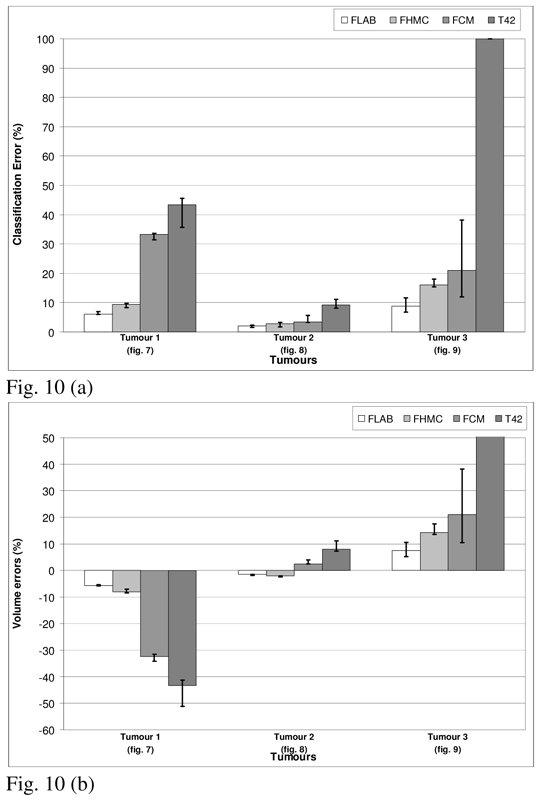
Segmentation results for the three simulated tumours. (a) Classification errors and (b) overall volume errors. The top of the error bar is the result concerning the worst statistical quality images (20% of detected coincidences), the medium one concerns the medium quality (40%), and the lowest one corresponds to the superior statistical quality (100%).
The results for the first and third tumors (fig. 7) show the largest differences between the four algorithms. In the case of the first tumor, this difference can be attributed to the non-uniform activity distribution (the contrast between the region of highest activity and the rest of the tumor is around 2.2:1) relative to the second tumor (closer to 1.4:1). Consequently, the segmentation results of T42 and FCM lead to large under evaluation (−30 to −50%) of the true volume of the first tumor since they limit themselves to the highest activity area, whereas in the case of the second tumor they are unable to differentiate between the two regions, hence recovering the entire tumor (less than 10% error for all methods). On the other hand, the third tumor despite being uniform is small with a low tumor to background ratio (1.5 cm in “diameter” and contrast <2:1). As a result, thresholding using 42% of maximum value fails completely (the region growing never stops and expands into the entire selection box) and FCM despite qualitatively satisfying results leads to a large over evaluation (from 10 to 40% volume error depending on the image statistical quality) of the volume. As far as FHMC and FLAB are concerned, they are both able to recover the whole tumor in all cases with volume errors between 2% and <20% (see fig. 10). While FLAB in comparison with the FHMC performed better in terms of both the misclassification and the overall volume errors, FHMC results were less competitive with decreasing tumor sizes as seen also in the IEC phantom results (fig. 10). Finally, the variability of the results (demonstrated by the error bars in figure 10) considering the different noise levels was higher for FCM and T42, illustrating their lower robustness to noise in comparison to the fuzzy statistical approaches.
IV. Discussion
Over the past few years there has been an increasing interest in clinical applications such as the use of PET for IMRT planning, for which an accurate estimation of the functional volume is indispensable. Unfortunately, accurate manual delineation is impossible to achieve due to high inter- and intra-observer variability [2] resulting from the noisy and low resolution nature of the PET images. Current state of the art methodologies for functional volume determination involve the use of adaptive thresholding based on anatomical information or phantom studies. Thresholding however is known to be sensitive to contrast variation as well as noise [2,4], since it does not include any explicit modelling of noise or spatial relationship. In addition, proposed adaptive thresholding methodologies require a priori knowledge of the tumour volumes currently obtained by CT images, based on the non-valid assumption that the functional and anatomical volumes are the same [3]. In addition, proposed correction methodologies accounting for the effects of background activity levels depend on lesion contrast and background noise as well as being imaging system specific [4]. On the other hand, previously developed automatic algorithms have also shown dependence on the level of noise and lesion contrast, most frequently requiring pre- or post-processing steps and variable initialization parameter values depending on image characteristics rendering their use complicated and their performance highly variable.
We have previously developed and assessed the performance for functional volume segmentation of a modified version of the Hidden Markov Chains algorithm (FHMC) [11]. In this algorithm a number of fuzzy levels have been added to introduce the notion of imprecision allowing this way to account for the effects of low image spatial resolution in addition to the noise modelling (which is part of the standard HMC framework). Although the algorithm was shown to accurately segment functional volumes (errors <15%) for lesions >2cm throughout different contrast and noise conditions, it was unable to accurately segment lesions <2cm. The main reason behind the failure of FHMC concerning the segmentation of such small lesions was the small number of voxels associated with the object of interest in combination to image noise levels, and the Hilbert-Peano path [12] used to transform the image into a chain. The spatial correlation of such small objects is lost once the image is transformed into a chain, because the voxels belonging to the object may find themselves far away from each other in the chain, thus resulting in transition probabilities that prevent these voxels to form a class differentiated from the background. In addition, it was thought that the assumption of a Gaussian noise distribution in the images to be segmented may have also been partly responsible.
FLAB clearly improved the results of FHMC, essentially due to the adaptive estimation of the priors using the whole 3D neighbourhood of each voxel, as the results of figure 5-c clearly demonstrate. FLAB results obtained on the objects >2 cm were similar to those obtained through the use of FHMC as were their respective robustness with respect to noise levels. Finally, FLAB resulted in faster computation times in comparison with the FHMC.
In addition, highly reproducible results (<4% variability, to compare with the 8 to 20% variability observed on manual segmentation [2]) were obtained for different image contrast ratios and lesion sizes >1cm. We should emphasize here that the performance of the FLAB in comparison to other segmentation algorithms was evaluated in this study on images reconstructed using a specific iterative reconstruction algorithm used today in clinical practise. Since the FLAB segmentation algorithm has been developed in order to better cope with variable noise and contrast characteristics it should be the least affected by such changes introduced as a result of using an alternative reconstruction algorithm [27]. On the other hand, the use of the system of Pearson for the determination of image voxel value distributions did not lead to significant changes or improvements in the results in comparison to the Gaussian assumption. Although this was shown to be the case for the images reconstructed using the specific iterative reconstruction algorithm used here it may not be the case if an alternative reconstruction algorithm is used, where potentially the use of the system of Pearson for the characterisation of the image voxel values distribution may still prove to play a role in the segmentation process and needs to be further investigated.
By comparison the use of T42 led, as expected, to segmented functional volumes greatly dependent on image contrast and noise levels while being comparable to the FLAB results considering medium image statistical quality and lesions >17mm with an 8:1 tumour to background ratio. Finally, the resulting volumes from the application of the automatic segmentation algorithm FCM were less dependent to image statistical quality but consistently failed to segment lesions <2cm.
In this study, as in every other phantom study presented to date in the literature, we have firstly considered the performance of the different algorithms for the segmentation of uniformly filled spherical lesions. To our knowledge there has been no study up to now specifically investigating the functional volume segmentation task for inhomogeneous uptake lesions, for example lesions with necrotic or partially necrotic regions. Although it has not been the major aim of their work, Nestle et al demonstrated some evidence of the issues associated with the use of either fixed or background adjusted thresholding methodologies for lesions with inhomogeneous activity distributions and shapes in the clinical set up for non small cell lung cancer [4]. As it was shown in this study using simulated realistic lesions, the FLAB model is able to successfully deal with non-uniform lesion shapes and variable activity concentrations in contrast with the threshold based or fuzzy C-means segmentation algorithms considered. On the other hand, the binary 2-class modelling (background or lesion) is obviously not adequate to permit the differentiation of multiple regions inside the tumour with largely different activity concentrations, as well as extracting the overall tumour in the case of strong heterogeneity. However, whereas it seems difficult to improve threshold-based segmentation methods in order to allow the identification of regions with variable activity concentration within the same functional volume of interest, the fuzzy model of FLAB may be extended to more than two hard classes to allow modelling a combination of inhomogeneous regions within a given volume. This could further enhance the use of FLAB for functional volume segmentation in future potential clinical applications.
The objectives of this study were to address the issue of functional volume determination and lesion segmentation. The FLAB model, as with any other segmentation algorithms, does not modify the values of the image voxels. As such, the use of the functional volume obtained with the FLAB algorithm, although is the closest to the true volume of the tumor as demonstrated by the results in this study, does not lead to the accurate activity concentration within the lesion. This is as a result of including voxels whose values have been decreased by spill-out from partial volume effects, usually leading to an under-estimation of the activity concentration whose magnitude depends on the size of the lesion [11]. Although the segmented volume should therefore not be used for directly recovering the accurate activity concentration, they can be used in combination with partial volume correction methodologies potentially allowing a more accurate correction in comparison to the use of anatomical volumes [28].
V. Conclusion
A modified version of a fuzzy local Bayesian segmentation algorithm has been developed. The suggested approach combines statistical and fuzzy modelling in order to address specific issues in the segmentation of low resolution noisy PET images. It is automatic, fully 3D and uses adaptive estimation of priors to yield good local spatial characteristics that improve segmentation of small objects of interest. Results obtained with images of the IEC phantom reconstructed with the 3D RAMLA iterative algorithm have shown that it is more effective than the reference thresholding methodology and other previously proposed automatic algorithms such as FHMC or the FCM methods for functional volume determination in PET images. The algorithm has also been tested successfully against realistic simulated tumors, using real patient tumors as model, with non-spherical shape and inhomogeneous activity distributions. Future developments will concentrate on the incorporation into FLAB of three hard classes and three different fuzzy transitions, in order to allow the segmentation within the same lesion of variable activity distributions in the case of highly heterogeneous functional uptake in the tumor volumes. We will also evaluate the use of different noise models in an associated robustness study using acquisitions with different scanner models and reconstruction algorithms.
Acknowledgments
This work was financially supported by a Region of Brittany research grant under the “Renouvellement des compétences” program 1202-2004, the French National Research Agency (ANR) under the contract ANR-06-CIS6-004-03, and the Cancéropôle Grand Ouest under the contract R05014NG.
Appendix
A.1 Relationship between coefficients a, c0, c1 and c2 and equations (5) and (6):
A.2 Definition of the 8 distribution density families:
Beta I and Gaussian distributions with respect to a class c are defined as follows:
| (18) |
| (19) |
where is the Beta function (with Γ the Gamma function).
We also have the following relationships between the parameters α and β, and the mean and variance (μ̂c, denote estimated mean and variance) of class c (this is useful to get the parameters α and β from the estimated means and variances obtained through the SEM algorithm):
A.3 Recipe for identification of the best family to fit distributions of classes:
Let us consider the voxels y1, …, yt and their partitions Q0 and Q1 into two classes. The moments can be estimated from empirical moments, and we use the following to detect which family best fits each distribution:
Consider the partitions Q0, Q1 of (x1, …, xt) defined by i ∈ Q0 ⇔ xi = 0and i ∈ Q1 ⇔ xi =1
-
For each class i use Qi in order to estimate the μm,i empirical moments by:
for m = 2,3,4
For each class i, calculate γ1,i and γ2,i from the estimated μm,i (m=1,2,3,4) according to (6).
For each class i, use γ1,i, γ2,i and rules (appendix A.2) to determine which family its density f belongs to.
B.1 SEM algorithm:
Give an initial value of the parameters ω0 =[pt,00, pt,10, μ00, (σ02)0, μ10, (σ12)0 using K-Means algorithm for the noise and equal probabilities for the priors.
-
At each iteration q, ωq is obtained from ωq−1 and the data (y1,…, yt) using:
-
Choose a distribution for the classes 0 and 1 according to the Pearson system rules (section 2.1.3 and sections A.2 and A.3 of the appendix).
For each yt, compute the a posteriori probabilities dq(0|yt) and dq(1|yt) using (8) and sample a value in the set {0,1,F} according to dq(0|yt), dq(1|yt) and 1 − dq(0|yt) − dq(1|yt) (F representing the fuzzy voxels). Let us denote R = (r1q, …, rTq) the posterior realization obtained through this sampling.
Let Q0q ={t|rtq = 0} and Q1q = {t|rtq = 1};
-
Reestimate the priors using:
where Ct is the estimation cube centred on voxel t and δ(a,b) the Kronecker function.
-
Reestimate the noise parameters using:
-
-
For the means and variances of the fuzzy levels, use (3).
Repeat step 2 until stabilization of the parameters. Stabilization is defined by a criterion of % change in the values of the parameters between two successive iterations (we used 0.1% and the algorithm usually stops before 25 iterations) and a maximum number of iterations if the stabilization criterion is not met (usually 50 iterations).
References
- 1.Jarritt H, Carson K, Hounsel AR, Visvikis D. The role of PET/CT scanning in radiotherapy planning. British Journal of Radiology. 2006;79:S27–S35. doi: 10.1259/bjr/35628509. [DOI] [PubMed] [Google Scholar]
- 2.Krak NC, Boellaard R, et al. Effects of ROI definition and reconstruction method on quantitative outcome and applicability in a response monitoring trial. European Journal of Nuclear Medicine and Molecular Imaging. 2005;32:294–301. doi: 10.1007/s00259-004-1566-1. [DOI] [PubMed] [Google Scholar]
- 3.Erdi YE, Mawlawi O, Larson SM, Imbriaco M, Yeung H, Finn R, Humm JL. Segmentation of Lung Lesion Volume by Adaptative Positron Emission Tomography Image Thresholding. Cancer. 1997;80(S12):2505–2509. doi: 10.1002/(sici)1097-0142(19971215)80:12+<2505::aid-cncr24>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Nestle U, Kremp S, Schaefer-Schuler A, Sebastian-Welch C, Hellwig D, Rübe C, Kirsch CM. Comparison of Different Methods for Delineation of 18F-FDG PET-Positive Tissue for Target Volume Definition in Radiotherapy of Patients with Non-Small Cell Lung Cancer. Journal of Nuclear Medicine. 2005;46(8):1342–8. [PubMed] [Google Scholar]
- 5.Reutter BW, Klein GJ, Huesman RH. Automated 3D Segmentation of Respiratory-Gated PET Transmission Images. IEEE Transactions on Nuclear Science. 1997;44(6):2473–2476. [Google Scholar]
- 6.Riddell C, Brigger P, Carson RE, Bacharach SL. The Watershed Algorithm: A Method to Segment Noisy PET Transmission Images. IEEE Transactions on Nuclear Science. 1999;46(3):731–719. [Google Scholar]
- 7.Zhu W, Jiang T. Automation Segmentation of PET Image for Brain Tumours. IEEE NSS-MIC conference Records. 2003;4:2627–2629. [Google Scholar]
- 8.Kim J, Feng DD, Cai TW, Eberl S. Automatic 3D Temporal Kinetics Segmentation of Dynamic Emission Tomography Image Using Adaptative Region Growing Cluster Analysis. IEEE NSS-MIC conference Records. 2002;3:1580–1583. [Google Scholar]
- 9.Pieczynski W. Modèles de Markov en traitement d’images. Traitement du Signal. 2003;20(3):255–277. [Google Scholar]
- 10.Chen JL, Gunn SR, Nixon MS. Markov Random Field Model for segmentation of PET images. Lecture Notes on Computer Science. 2001;2082:468–474. [Google Scholar]
- 11.Hatt M, Lamare F, Boussion N, Turzo A, Collet C, Salzenstein F, Roux C, Carson K, Jarritt P, Cheze-Le Rest C, Visvikis D. Fuzzy hidden Markov chains segmentation for volume determination and quantitation in PET. Physics in Medicine and Biology. 2007;52:3467–3491. doi: 10.1088/0031-9155/52/12/010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamata S, Eason RO, Bandou Y. A New Algorithm for N-dimensional Hilbert Scanning. IEEE Transactions on Image Processing. 1999;8:964–973. doi: 10.1109/83.772242. [DOI] [PubMed] [Google Scholar]
- 13.Delignon Y, Marzouki A, Pieczynski W. Estimation of Generalized Mixtures and Its Application in Image Segmentation. IEEE Transactions on Image Processing. 1997;6(10) doi: 10.1109/83.624951. [DOI] [PubMed] [Google Scholar]
- 14.Visvikis D, Turzo A, Gouret S, Damine P, Lamare F, Bizais Y, Cheze Le Rest C. Characterisation of SUV accuracy in FDG PET using 3D RAMLA and the Philips Allegro PET scanner. Journal of Nuclear Medicine. 2004;45(5):103. [Google Scholar]
- 15.Caillol H, Pieczynski W, Hillon A. Estimation of Fuzzy Gaussian Mixture and Unsupervised Statistical Image Segmentation. IEEE Transactions on Image Processing. 1997;6(3) doi: 10.1109/83.557353. [DOI] [PubMed] [Google Scholar]
- 16.Salzenstein F, Pieczynski W. Parameter Estimation in hidden fuzzy Markov random fields and image segmentation. Graphical Models and Image Processing. 1997;59(4):205–220. [Google Scholar]
- 17.Johnson NL, Kotz S. Distributions in Statistics: Continuous Univariate Distributions. Vol. 1. New York: Wiley; 1970. [Google Scholar]
- 18.Celeux G, Diebolt J. L’algorithme SEM: un algorithme d’apprentissage probabiliste pour la reconnaissance de mélanges de densités. Revue de statistique appliquée. 1986;34(2) [Google Scholar]
- 19.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B. 1977;39:1–38. [Google Scholar]
- 20.McQueen J. Some methods for classification and analysis of multivariate observations. Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability. 1967;1:281–297. [Google Scholar]
- 21.Dunn JC. A Fuzzy relative of the Isodata process and its use in detecting compact well-separeted clusters. J Cybernet. 1974;31:32–57. [Google Scholar]
- 22.Jordan K. IEC emission phantom Appendix Performance evaluation of positron emission tomographs. Medical and Public Health Research Programme of the European Community. 1990 [Google Scholar]
- 23.Lodge MA, Dilsizian V, Line BR. Performance assessment of the Philips GEMINI PET/CT scanner. Journal of Nuclear Medicine. 2004;45:425. [Google Scholar]
- 24.Segars WP, Lalush DS, Tsui BMW. Modelling respiratory mechanics in the MCAT and spline-based MCAT phantoms. IEEE Transactions on Nuclear Sciences. 2001;48:89–97. [Google Scholar]
- 25.Ramos CD, Erdi YE, Gonen M, Riedel E, Yeung HW, Macapinlac HA, Chisin R, Larson SM. FDG-PET standardized uptake values in normal anatomical structures using iterative reconstruction segmented attenuation correction and filtered back-projection. European Journal of Nuclear Medicine. 2001;28:155–64. doi: 10.1007/s002590000421. [DOI] [PubMed] [Google Scholar]
- 26.Lamare F, Turzo A, Bizais Y, Cheze-Le Rest C, Visvikis D. Validation of a Monte Carlo simulation of the Philips Allegro/Gemini PET systems using GATE. Physics in Medicine and Biology. 2006;51:943–962. doi: 10.1088/0031-9155/51/4/013. [DOI] [PubMed] [Google Scholar]
- 27.Hatt M, Bailly P, Turzo A, Roux C, Visvikis D. PET functional volume segmentation: a robustness study. IEEE NSS-MIC conference Records. 2008 in press. [Google Scholar]
- 28.Boussion N, Hatt M, Visvikis D. Partial volume correction in PET based on functional volumes. Journal of Nuclear Medicine. 2008;49(S1):388. doi: 10.1007/s00259-009-1065-5. [DOI] [PubMed] [Google Scholar]


