Abstract
Optical coherence tomography (OCT) can provide new insight into disease progression and therapy by enabling nondestructive, serial imaging of in vivo cancer models. In previous studies, we have shown the utility of endoscopic OCT for identifying adenomas in the azoxymethane-treated mouse model of colorectal cancer and tracking disease progression over time. Because of improved imaging speed made possible through Fourier domain imaging, three-dimensional imaging of the entire mouse colon is possible. Increased amounts of data can facilitate more accurate classification of tissue but require more time on the part of the researcher to sift through and identify relevant data. We present quantitative software for automatically identifying potentially diseased areas that can be used to create a two-dimensional “disease map” from a three-dimensional Fourier domain OCT data set. In addition to sensing inherent changes in tissue that occur during disease development, the algorithm is sensitive to exogeneous highly scattering gold nanoshells that can be targeted to disease biomarkers. The results of the algorithm were compared to histological diagnosis. The algorithm was then used to assess the ability of gold nanoshells targeted to epidermal growth factor receptor in vivo to enable functional OCT imaging.
Keywords: optical coherence tomography, azoxymethane, mouse, colon, nanoshells, epidermal growth factor, three dimensional imaging, image processing
Introduction
Optical coherence tomography (OCT) is rapidly acquiring acceptance for several biomedical applications, the most notable being assessing retinal diseases of the eye, because no other imaging technology can provide noncontact cross-sectional images of the retina with comparable resolution. A growing application for OCT involves studying diseases of the gastrointestinal (GI) tract. Clinical applications for OCT in the Gl tract are reviewed by Brand et al.1 and Shen and Zuccaro.2 Notable among these applications include detecting Barret’s esophagus,3, 4, 5, 6 esophageal cancer,3, 4 ulcerative colitis,7 inflammatory bowel disease,2 and colonic polyps and adenocarcinoma.8, 9 For patients, a biopsy is still required for diagnosis, but OCT may provide information about subsurface structure and guide biopsy locations.8, 10, 11, 12
Nonclinical applications for OCT include research using animal models.13, 14, 15Because OCT is nondestructive and does not use ionizing radiation, it can be used to monitor disease progression in animal models over time and evaluate treatment or drug effectiveness. Nondestructive imaging with OCT improves on current research practices, which involve euthanizing animals at various time points to track disease statistically.
Previous work in our group has demonstrated the ability of OCT to detect adenomas in vivo in a mouse model of colorectal cancer.14 Mice in the study were treated with a carcinogen, azoxymethane (AOM), which causes the spontaneous development of aberrant crypt foci (ACF), and ultimately adenomas (polyps) in the colon.16 A blinded panel analyzed OCT images of AOM-treated mouse colon and detected 95% of histologically confirmed adenomas. Only one normal location was misclassified as adenoma out of 38 total adenomas identified and correlated with histology. Criteria were also developed for identifying other abnormalities, including gastrointestinal intraepithelial neoplasia (GIN), mucosal lymphoid aggregates, and submucosal lymphoid aggregates.
The current paper extends this work by developing quantitative image metrics to distinguish adenomas, GINs, and normal tissue. These metrics allow classification of the tissue with minimal or no human observation, increasing data throughput and minimizing human errors that can be due to inexperience or lack of training. The improvement in data throughput can be exploited by collecting and analyzing three-dimensional data sets, which can be impractical to analyze manually. High-density data sets minimize sampling errors and, in studies of distal colorectal cancer in the mouse colon, enable all adenomas and GINs to be imaged.
Computer analysis of OCT images, or quantitative OCT, is being studied for a number of applications, including determining blood glucose levels in skin,17, 18 classifying atherosclerotic lesions,19 and detecting and∕or grading precancerous lesions.5, 9, 13, 20, 21 A recent review article by Bazant-Hegemark and Stone22 summarizes these and other applications for quantitative OCT, specifically in dense tissues (such as the colon). One prominent feature in OCT images is light attenuation, which can be quantified by fitting a line to averaged A-scans. For detecting or grading precancerous legions, light attenuation tends to increase with disease.9, 20, 21 Another prominent feature of disease is loss of organization, exploited by Pfau et al.9 for distinguishing normal, hyperplasia, and dysplasia in human colon, albeit qualitatively, and by Korde et al.21 for identifying actinic keratosis in human skin.
Quantitative OCT for detecting colorectal precancers includes work by Pfau et al.9 and Qi et al.23 Pfau et al.9 noted that backscattering, measured by the average gray-scale value in a 60×60 pixel square, was reduced with statistical significance in dysplastic and hyperplastic polyps as compared to normal colon in human subjects. The endoscopist in the study also scored the tissue organization in real time, and the score was found to be a strong predictor of dysplastic polyps. Qi et al.23 identified quantitative measures for identifying ACF, a condition found to precede dysplasia in the colon, in human subjects. By accurately segmenting the colonic crypts, crypts with ACF were identified as being less straight and significantly less parallel than normal crypts.
In the mouse colon, crypt segmentation is difficult to perform, even manually, due to the small size of the crypts as compared to those in human subjects. Therefore, in this work, colon tissue layer properties are evaluated, including gray scale and attenuation in the mucosal layer and underlying submucosal layer visibility.
In addition to detecting adenomas and GINs, the same quantitative metrics are also sensitive to scattering by OCT contrast agents, such as gold nanoshells. Nanoshells are composed of a silica core and gold outer shell and can be engineered to exhibit plasmon resonance at OCT wavelengths in the near infrared.24 Their relative scattering and absorption properties can also be tuned independently of spectral response by varying particle size. Generally speaking, large particles exhibit higher scattering and lower absorption than smaller particles. Having control over the relative amounts of scattering and absorption allows nanoshells to cater to both enhancing image contrast in OCT25, 26, 27 and enabling photothermal therapy.28 Furthermore, nanoshells can be targeted to specific biomarkers, such as HER2 (also known as epidermal growth factor receptor or EGFR), for functional OCT imaging and targeted photothermal therapy.28, 29 Previous papers demonstrate targeted nanoshell enhancement and photothermal therapy on in vitro cell cultures.
Work by Agrawal et al.26 and Kah et al.27 quantitatively demonstrates the effect of nanoshells on OCT images, specifically noting that nanoshells increase μs, which both enhances the surface signal and increases signal attenuation. Although Agrawal et al.26 notes that both effects could be used as a means of contrast enhancement, Kah et al.27 focuses on optimizing the surface signal enhancement while maintaining sufficient imaging depth (i.e., limiting signal attenuation) through choice of nanoshell concentration. Furthermore, Kah et al.27 examines intravenous injection in a mouse xenograft tumor model in vivo and notes that functionalizing gold nanoshells with EGFR increases the fractional concentration of nanoshells in tumors.
By utilizing the sensitivity of the quantitative metrics proposed in this work, nanoshells targeted to EGFR are studied for the first time in vivo in the AOM-treated mouse model. Because of having a highly scattering background and significant inherent scattering variability between adenomas, GINs, and normal areas, the selectivity of nanoshells for EGFR-rich regions can be difficult to assess manually. The proposed metrics are capable of measuring slight changes in tissue optical properties and can statistically evaluate the performance of targeted nanoshells in this model.
Materials and Methods
Endoscopic Spectral Domain OCT
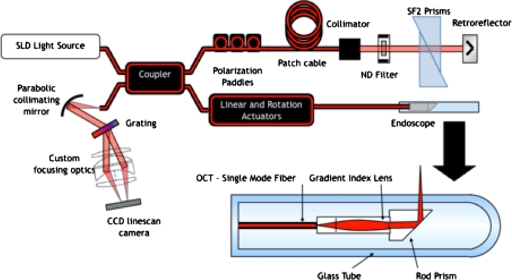
The imaging system is an endoscopic, spectral domain OCT system built for in vivo mouse colon imaging (shown in Fig. 1). The light source is a superluminescent diode (Superlum Broadlighter, Moscow, Russia) centered at 890 nm with a 150-nm full-width-at-half-maximum (FWHM) bandwidth. On the basis of this bandwidth, the theoretical point spread function in air is ∼2.3 μm and in tissue 1.7 μm assuming a refractive index of 1.4. The source optical power is split by a 50:50 coupler of Corning HI780 fiber (AC Photonics, Santa Clara California). The reference arm is attenuated using an neutral density filter with 0.6 optical density, and SF2 right-angle prisms (Thorlabs, Newton, New Jersey) are used for dispersion compensation.
Figure 1.
Imaging system is an endoscopic, spectral domain OCT system. The light source is a superluminescent diode (890 nm, 150 nm FWHM) and is coupled into a fiber-based Michelson interferometer. The detector is a custom built spectrometer that images light from 800 to 1000 nm.
The sample arm consists of a 2-mm-diam endoscope. The endoscope fiber length is ∼2 m with focusing optics at the tip. The optics consist of a 1-mm-diam, 1.435-mm-long glass spacer, angled at 8 deg to limit backreflections, followed by a 1-mm-diam, 1.906-mm-long GRIN lens (GRINTECH GmbH, Jena, Germany). The focusing optics produce a numerical aperture of 0.1 (1∕e2 point), resulting in a FWHM spot diameter of 5 μm at focus. A side-firing endoscope is accomplished using a prism with aluminized face oriented at 48 deg relative to the incident light. The tip optics are enclosed in a 2-mm-diam glass tube. Light exits the tube 3 deg from the normal to limit light coupling from the tube interfaces.
The detector consists of a custom-built CCD-based spectrometer. Light exits the detection fiber and is collimated by an off-axis parabolic mirror (Edmund Optics, Barrington, New Jersey). Collimated light is dispersed by a volume phase holographic grating with 1200 line pairs per millimeter, 830 nm blaze (Wasatch Photonics, Logan, Utah). Light is incident at 31 deg and exits at 26–43 deg (800–1000-nm wavelength). A custom focusing optic, Cooke triplet design using off-the-shelf lenses, focuses the 17 deg field of view onto a 2048-pixel silicon CCD array (Atmel AT71SM2CL2014-BAO, San Jose, California). Data are collected using a frame grabber, Meteor2-CL∕32 from Matrox Imaging (Dorval, Quebec). Roll-off inherent to Fourier domain systems is affected by pixel size as well as optical diffraction and aberrations introduced by the grating and imaging optics. In practice with this detector, the signal drops by ∼6 dB at 0.85-mm imaging depth in air.
The system axial point-spread function is affected by the source bandwidth imaged at the detector as well as system and media dispersion. The detector images over 200 nm of the source bandwidth, retaining the theoretical 2.3 μm. System dispersion is corrected in system calibration, similar to Ref. 30, and is summarized as follows. Images are collected from a single scatterer at two different depths. A minimum of 1000 A-scans are collected from each depth and averaged to approximate the background signal. Because of phase instabilities, the average signal does not retain phase information about the scatterers, only the background signal. The background is subtracted from each signal and then the Hilbert transform is performed to calculate the phase. In the absence of system dispersion, the phase of each scatterer should be linear with respect to wavenumber; however, system dispersion introduces a nonlinear phase component at each wavenumber so that the resultant phase is nonlinear. Because the system dispersion phase component is common to both scatterer signals, the subtraction of these two phases eliminates the system dispersion phase component. This resultant phase should now be linear with respect to wavenumber. Linear interpolation is performed to approximate even sampling with respect to wavenumber. After resampling the phase, the system dispersion phase component can be measured by subtracting the linear portion of the phase. The known system dispersion phase component can now be removed for all subsequent measurements by multiplying the measured signal by exp(−i*phasesystem) at each pixel. The resultant axial point spread function in practice is under 3 μm in air.
Sensitivity and dynamic range were measured using the method by Tumlinson et al.31 for endoscopic systems. The sensitivity is 88 dB. The dynamic range was measured for multiple OCT images of mouse colon and found to vary between 44 and 68 dB.
Gold Nanoshell Fabrication and Functionalization
Highly scattering gold nanoshells were produced as described previously.28 The nanoshells were ∼120 nm diam with a resonant peak near 800 nm. Nanoshells were coated in polyethyleneglycol and functionalized with epidermal growth factor receptor antibody. Both functionalized (EGFR-targeted nanoshells) and unfunctionalized (pegylated nanoshells) were imaged in mouse colon.
Imaging Protocol
A total of 11 A∕J mice were imaged in this study. Of the 11, nine were treated with AOM to induce colorectal cancer. AOM (10 mg∕kg) was administered once a week for five weeks, starting at age seven weeks. The other two mice were treated with saline of equivalent volume with the same schedule (control).
Twelve to 24 h prior to imaging, mice were separated without food and given Pedialyte in place of water. Immediately prior to imaging, mice were anesthetized with 2.5% Avertin delivered intraperitoneally. The distal colon was gently flushed with 3–10 mL of warm phosphate buffered saline (PBS). The endoscope was lubricated with a water-based lubricant and inserted ∼35 mm inside the colon. For all mice, OCT images were collected in eight positions, 30-mm long and spaced 45 deg apart, without any contrast agent. Subsequently, highly scattering gold nanoshells targeted to EGFR fabricated at Rice University, 0.2 mL with 1×1010 nanoshells∕mL (2×109 nanoshells), were intraluminally applied in the colons of five AOM-treated mice and one control mouse. After 20 min, the colon was flushed with 10 mL PBS. Untargeted, pegylated gold nanoshells (Rice University) were similarly applied in the colons of three AOM-treated mice and one control mouse. Protocols were approved by the University of Arizona Institutional Animal Care and Use Committee.
Eight AOM mice (AOM 1–8) and two control mice (control 1–2) were used to test the quantitative metrics for distinguishing tissue types and detecting gold nanoshells. An additional AOM mouse (AOM 9) was used to demonstrate the ability of the algorithm to detect disease in a three-dimensional data set. The data set was acquired by taking 120 OCT images, 20-mm long, spaced 3 deg apart in the colon. Imaging procedures per each animal are outlined in Table 1.
Table 1.
Animal information.
| Alphanumeric label | Imaged without contrast | Imaged with nanoshells | |||
|---|---|---|---|---|---|
| 8 images | 120 images | EGFR targeted | Pegylated | ||
| AOM 1 | ˟ | ˟ | |||
| AOM 2 | ˟ | ˟ | |||
| AOM 3 | ˟ | ˟ | |||
| AOM 4 | ˟ | ˟ | |||
| AOM 5 | ˟ | ˟ | |||
| AOM 6 | ˟ | ˟ | |||
| AOM 7 | ˟ | ˟ | |||
| AOM 8 | ˟ | ˟ | |||
| AOM 9 | ˟ | ||||
| Control 1 | ˟ | ˟ | |||
| Control 2 | ˟ | ˟ | |||
| Totals | 11 | 10 | 1 | 6 | 4 |
Histology and Immunohistochemistry
After imaging, mice were euthanized and 30 mm of colon was excised. Colons were fixed with Poly∕LEM Fixative (Polysciences) for 12 h, processed, embedded routinely in paraffin, and sectioned 6-μm thick. Sectioned tissue was stained with hematoxylin and eosin (H&E) or EGFR immunostain as described below.
After antigen retrieval, sections were washed, blocked with goat serum, and incubated overnight with primary rabbit polyclonal anti-EGFR (1005):sc-03 (Santa Cruz Biotechnology). Endogeneous peroxidase activity was blocked by immersion in 3% hydrogen peroxide in water followed by a 1-h incubation at room temperature with Biotinylated Anti-Rabbit IgG (H+L). Sections were incubated for 10 min at room temperature with streptavidin-HRP (Dako). Complexes were visualized (brown) with 3,3′-diaminobenzidine (Dako), and the sections were counterstained with 0.5% methyl green.
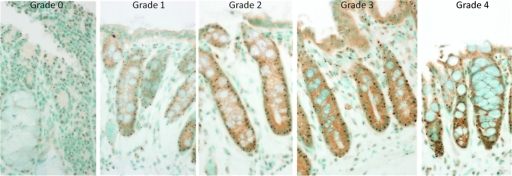
Disease was positively identified using H&E slides and correlated with disease features in OCT as described by Hariri et al.14 Diseased areas identified in the H&E slides were adenomas and GIN; all other areas were classified as normal. EGFR immunostained slides were used to quantify EGFR expression in adenomas, GINs, and selected normal regions. Each region was given a grade between 0 and 4, judged by eye, representing the average color hue and saturation. The darker the brown was in the slides, the higher the EGFR grade was, as shown in Fig. 2.
Figure 2.
Normal colon sections with EGFR immunostaining are shown, ranging from less (grade 0) to more (grade 4) EGFR. Note the increasing brown color saturation from grades 0 to 4. (In gray-scale print, increasing brown saturation appears as a darkening around the crypts and apical cell layer.) (Color online only.)
Image Processing and Quantitative Metrics
OCT image properties associated with normal, GIN, and adenoma were previously determined in a study of 18 AOM-treated mice.14 These imaging features are listed in Table 2 as well as illustrated in Fig. 3.
Table 2.
Imaging properties of normal, GIN, and adenoma.
| Normal | GIN | Adenoma | |
|---|---|---|---|
| Image properties | Clearly visible boundaries | Visible M∕SM boundary | No visible layers |
| Mild attenuation through mucosa | Moderate to high attenuation in mucosa | High attenuation in mucosa | |
| Thickened mucosa | Thickened mucosa |
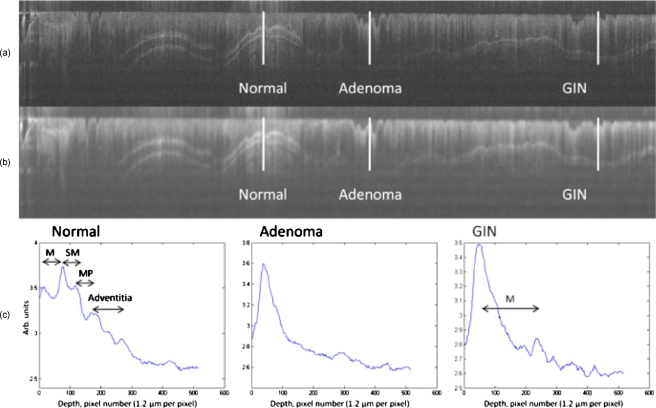
Figure 3.
OCT imaging of mouse colon clearly distinguished normal from diseased tissue. (a) A 22×1.5 mm logged OCT image of AOM-treated mouse colon. (b) The same OCT image after median filtering with a large (21×21 pixel, 125×25 μm) kernel. (c) Plots corresponding to the logged backscattered intensity as a function of depth. The position and length of the plots are indicated by the white lines in the OCT images labeled “Normal,” “Adenoma,” and “GIN.” Anatomical features are indicated on the Normal and GIN plots. In both plots, the mucosal (M) surface and mucosa (M)/submucosa (SM) boundary are visible. The Normal plot also clearly shows the boundaries between the submucosa (SM), muscularis propria (MP), and adventitia. Because of mucosal thickening and heightened attenuation with depth, these boundaries are not visible in the GIN. The adenoma displays no clearly discernable boundaries. Note also the marked mucosal thickening between the normal and GIN tissues.
To differentiate tissue based on the image features described in Table 2, specifically boundary visibility, attenuation in the mucosa, and mucosal thickening, three image properties were measured: (i) Average gray-scale value in the mucosa after logarithmic transformation, (ii) slope of a line fitted to log-transformed gray-scale values in the mucosa, and (iii) slope standard deviation from the tissue surface to ∼360-μm deep, which encompasses the average depth of normal mouse colon to the muscularis propria∕adventitia boundary. These three metrics can be described as zero-, first-, and second-order properties, respectively. The gray-scale value and slope in the mucosa quantify the amount of attenuation in the mucosa, while the slope standard deviation is affected by boundary visibility. Mucosal thickening decreases boundary visibility in at least two ways; first, it pushes the boundaries further from the system focus and, second, it increases the number of scatterers ahead of the boundaries. In effect, slope standard deviation is affected by boundary visibility, which is affected by mucosal thickening.
Sections 2E1, 2E2, 2E3 describe the specific steps in the automated disease detection algorithm, including image preprocessing, calculating the zero-, first-, and second-order image metrics, and assessing the efficacy of each metric for identifying disease.
Preprocessing
Image preprocessing includes identifying the surface of the colon and minimizing noise. The surface of the colon was manually traced for all images, although in future studies the surface can be detected automatically using standard edge detection methods, such as Sobel edge detection. To reduce single pixel noise and smooth speckle, a median filter was performed using a large kernel, 21×21 pixels or 125×25 μm.32 A rectangular filter was chosen to preserve horizontal features. The exact dimensions were determined empirically, balancing smoothing the A-scan with retaining boundary information. Then, a logarithmic transformation was performed in order to better visualize features as well as linearize tissue attenuation.
Residual mucous and cell debris could cause some ambiguity in determining the surface of the colon. In order to be less sensitive to errors in surface tracing, the maximum value in the first 25 pixels, or 30 μm below the segmented tissue surface, was assumed to be the actual start of the colon tissue. On the basis of work identifying physical contrast mechanisms in mouse colon performed by Tumlinson et al.33 the maximum in this range likely corresponds to the base of the apical cell layer.
Calculating Metrics
All three metrics were calculated for each A-scan. The average grayscale value and slope were calculated over a relatively short depth, 75 pixels or 90 μm from the tissue surface, in order to limit the measurement to the mucosal layer. The equation for this metric is shown below where i is the pixel number and A(i) is the gray-scale value at pixel i
The slope was determined by fitting a line to pixels within the first 90 μm using Matlab’s (The Mathworks, Inc.) “robustfit” function with the default settings. The algorithm uses least-squares regression and iteratively reweights the residuals with a bisquare weighting function. This algorithm was compared to simply averaging the difference between adjacent pixels as well as other available Matlab functions, including “polyfit” and “regress,” and was found to be the least sensitive to surface drawing errors and residual noise.
The slope standard deviation was calculated over a larger window, 300 pixels or 360 μm, in order to ensure that all visible tissue boundaries were encompassed. The difference in gray-scale value between pixels spaced 10 apart was calculated over the entire depth, and the standard deviation of these slopes was calculated, as shown in the equation below where i is the pixel number and A(i) is the gray-scale value at pixel i.
This metric is sensitive to sharp changes in slope, which occur at hyperintense interfaces, such as the mucosal∕submucosal boundary,
For normal areas, the gray-scale value in the mucosa is expected to be high, the slope is expected to be flat or near zero, and the slope standard deviation is expected to be high. In diseased areas, the opposite is expected: low gray-scale value, more negative slope, and low slope standard deviation. After adding nanoshells, increased surface backscattering is expected, resulting in increased gray-scale values, more negative slopes, and decreased slope standard deviations. Table 3 summarizes the metrics, expected results for tissue type, and physical meanings.
Table 3.
Quantitative metric predictions for different tissue types and tissue with nanoshells.
| Normal | GIN | Adenoma | Nanoshells | Physical Meaning | |
|---|---|---|---|---|---|
| Gray-scale value | High | Medium | Low | Higher | Increases with increased backscattering in mucosal layer |
| Slope | Low (near zero) | More negative | Most negative | More negative | Becomes more negative with increased attenuation in mucosal layer |
| Slope standard deviation | High | Medium | Low | Lower | Increase with boundary visibility |
Metric Analysis
To assess the correlation between the quantitative metrics and disease, p-values using the z-test were calculated as well as the sensitivity and specificity to disease at different metric value thresholds.
Adenomas and GINs were identified in OCT images manually using criteria developed by Hariri et al.14 Each adenoma and GIN identified was verified in H&E histology slides. The rest of the colon was classified as normal. Normal regions were segmented in 667 A-scan (4 mm) pieces, which was the average size of the observed adenomas in this study. In cases where a left-over segment was <667 A-scans, a minimum segment size of 167 A-scans (1 mm) was required for the segment to be included in the analysis. In each adenoma, GIN, or normal segment, the average of each metric for every A-scan was calculated.
A-scans that were too far from the endoscope focus were excluded in the analysis. The endoscope focus was located ∼0.2 mm from the endoscope outer glass; therefore, A-scans with the colon surface located deeper than 0.2 mm were excluded.
To calculate p-value, the average and standard deviation of the metrics were calculated for each tissue type (adenoma, GIN, and normal). The number of samples, N, was taken as the number of adenomas, GINs, or normal segments. The z-test was performed between adenoma and GIN values, adenoma and normal values, and GIN and normal values. This analysis was performed for images without nanoshells because nanoshells significantly change OCT image properties.
The z-test was also performed between adenomas pre- and postnanoshells, GINs pre- and postnanoshells, and normals pre- and postnanoshells to demonstrate sensitivity to nanoshells. To test for nanoshell-enhancement correlated with EGFR expression, each tissue type (adenoma, GIN, and normal) was subdivided into EGFR grade levels. To maintain a sufficient N, grades 0 and 1 were combined and renamed “low grade,” and grades 3 and 4 were combined and renamed “high grade.” In keeping with the new naming convention, grade 2 was renamed “medium grade.” The z-test was performed for each tissue type subdivided by EGFR grade pre- and postnanoshells.
Results
Distinguishing Adenoma, GIN, and Normal Regions
Correlating OCT and histology revealed 140 adenomas, 32 GINs, and 269 normal segments. One saline-treated mouse, control 2, was not classified as normal due to being diagnosed with diffuse mucosal edema. Using the surface depth and segment size criteria, 5 adenomas, 4 GINs, and 42 normal segments were excluded. The included tissue segments totaled 135 adenomas, 28 GINs, and 227 normal segments.
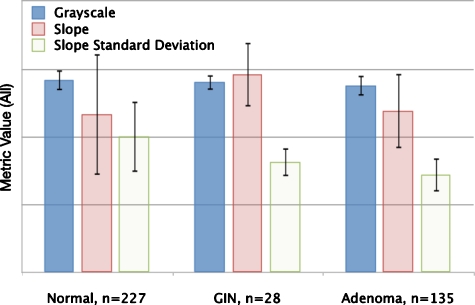
The best metric for distinguishing adenomas, GINs, and normals was the slope standard deviation metric. As predicted, this metric was highest for normals and decreases for GINs and adenomas, and it was statistically significantly different between the three tissue types with p<0.0002 in all cases. At one threshold value, this metric was 89% sensitive and 70% specific for identifying adenomas and GINs from normals. At a lower threshold value, this metric distinguished adenomas from GINs with 70% sensitivity and 76% specificity.
Average gray scale and slope were less predictive. Average gray scale decreased as predicted from normals to GINs to adenomas. This difference was statistically significant for differentiating adenomas from normals and adenomas from GINs but not for differentiating normals from GINs. The difference in slope was statistically significant between normals and GINs, with GINs have steeper slopes than normals. The slope for adenomas was steeper than the slope for normals, as predicted, but less steep than the slope for GINs, contrary to prediction. The metrics are shown graphically for each tissue type in Fig. 4.
Figure 4.
Metric values (on an arbitrary scale) for three different tissue types: Normals, GINs, and Adenomas. From Normal to GIN to Adenoma, the gray-scale and slope standard deviation decrease, as anticipated. From Normal to GIN and from Normal to Adenoma, the slope becomes steeper, as anticipated, but from GIN to Adenoma, the slope becomes less negative.
Utility for 3D Imaging
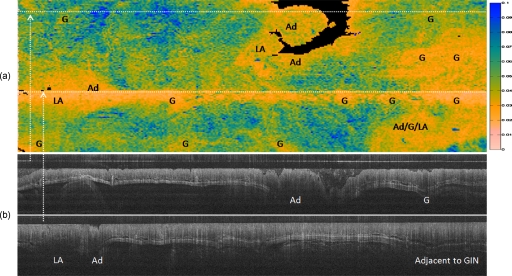
The slope standard deviation metric was calculated for a three-dimensional data set from one AOM-treated mouse, shown in Fig. 5. This metric was mapped to a gray-scale value, where dark gray (blue online) indicated the highest slope standard deviation or most “normal” and white (pink online) indicated the lowest slope standard deviation or most “diseased.” A-scans with the colon surface deeper than 0.2 mm, which could not be classified, were mapped to black. The image is overlaid with histologically verified features, specifically adenoma (Ad), GIN (G), and lymphoid aggregate (LA). LAs are normal immune tissue in the colon. As the image suggests, LAs can be confounders for adenomas and GINs.
Figure 5.
(a) Two-dimensional representation (“disease map”) of the mouse colon, 20 mm long in which left is rostral and right is caudal, and in which the top and bottom wrap around to complete the cylindrically shaped colon ∼6 mm in circumference. The metric plotted is the slope standard deviation, ranging from 0 (diseased) to 0.1 (normal) arbitrary units. Locations of adenomas (Ad), GINs (G), and LA, verified both in OCT and histology, are labeled. Note that the white (pink online) areas, which indicate a low slope standard deviation or “disease,” correspond with the locations of adenomas, GINs, and lymphoid aggregates. Although lymphoid aggregates are considered confounders for disease in OCT, note the colocalization of lymphoid aggregates and adenomas, which is typical in the AOM-treated mouse model. Dark gray (blue online) areas indicate high slope standard deviation or “normal.” Black areas indicate that the colon surface fell below the minimum depth, 0.2 mm, from the endoscope window and could not be classified. The logged OCT images (20 mm long by 0.3 mm deep) at the bottom were taken at locations indicated by the dotted lines in the “disease map” at the top. (Color online only.)
Identifying Nanoshells
Data from EGFR-targeted nanoshells and untargeted pegylated nanoshells were first combined in order to show sensitivity to nanoshells overall regardless of specificity to EGFR. For all tissue types, average gray scale increases as predicted with p<0.05 between pre- and postnanoshell images. Slope and slope standard deviation, however, did not have a consistent response to nanoshells for all three tissue types.
Visualizing EGFR with Targeted Nanoshells
The data were graded using EGFR immunostained slides and each tissue type was subdivided into low, medium, and high grade. The number of GINs after subdividing into grades was too small to analyze; therefore, only normals and adenomas were considered for this analysis. To grade normals, normal sections were correlated with histology based on surrounding features, including adenomas, GINs, and lymphoid aggregates. Some normal sections were excluded from analysis for lack of an identifiable landmark.
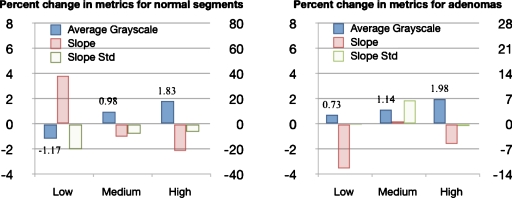
Figure 6 shows the percent change in the metrics after adding nanoshells for each EGFR grade, where the percent change was calculated as the value postnanoshells (xpost) minus the value prenanoshells (xpre) divided by the value prenanoshells times 100%, (xpost−xpre)∕xpre*100%. Figure 7 shows representative OCT images before and after adding nanoshells. For both normals and adenomas, the average gray scale increases postnanoshells and the percent change in gray scale increases with EGFR grade, suggesting higher nanoshell density with higher EGFR expression. The percent change in slope and slope standard deviation postnanoshells decreases with EGFR grade in normals, which suggests that other image effects may be present besides increased attenuation. The percent change in slope and slope standard deviation for adenomas showed no trend with EGFR grade.
Figure 6.
Charts show the percent change in image metrics post-EGFR-targeted nanoshells. The average gray-scale metric is plotted using the left axis, with values indicated. The slope and slope standard deviation metrics are plotted using the right axis. For the normal segments, gray scale increases and slope becomes less negative with EGFR-grade. The slope standard deviation decreases with nanoshells, as anticipated, but the percent change decreases with EGFR-grade. For adenomas, grayscale increases with EGFR-grade, while slope and slope standard deviation do not correlate with EGFR-grade. Note that for both normal segments and adenomas, the percent change in average gray scale increases with EGFR-grade postnanoshells.
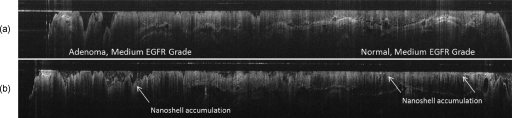
Figure 7.
Logged OCT images (30 mm long by 0.3 mm deep) are in the same colon location in the same mouse (a) pre- and (b) post nanoshells. Note the increase in surface signal in the postnanoshell image at the positions indicated by the arrows.
Image metrics for untargeted pegylated nanoshells do not show a consistent response for all three tissue types nor show a trend with EGFR grade for any of the metrics, suggesting nonspecific adherence to tissue.
Discussion
The quantitative metrics outlined in this paper demonstrate computer classification of tissue type (adenoma, GIN, normal) and functional OCT imaging with highly scattering gold nanoshells. The utility of these metrics to create en face disease maps and to identify regions with concentrated targeted nanoshells can enhance the development of drugs and therapies for colorectal cancer in the AOM-treated mouse model.
The metrics were developed to reflect expected changes in optical properties with disease progression in the colon and with nanoshells. Areas of AOM-induced abnormalities (ACF, GIN, adenoma) tend to appear as hypointense regions in OCT images. As disease develops in AOM-treated mice, the nuclear to cytoplasmic ratio in cells increases, and the nucleus becomes larger. Arifler et al.34 examines nuclear changes in dysplasia compared to normal cells, using finite-difference time-domain (FDTD) modeling and images of cervical biopsies of dysplasia. The results show significant increases in nuclear size and refractive index heterogeneity in cervical dysplasia biopsies as compared to normal cells. FDTD modeling and confocal microscopy demonstrate increased backscattering in dysplastic cells due to these nuclear changes.
Increased backscattering results in increased attenuation in the mucosa. In this work, a slight decrease in gray scale in the mucosa is observed as the tissue progresses from normal to GIN to adenoma, consistent with heightened attenuation. Furthermore, an increase in attenuation is observed for GINs and adenomas as compared to normals, as expected. Contrary to prediction, however, attenuation is greater for GINs than adenomas, with the difference in slope being statistically significant (p<0.05). Further study is necessary to elucidate this reversal; however, it may be due to cellular or architectural changes in adenomas. For example, in dysplastic cells, Arifler et al.34 noted a higher concentration of enlarged nuclei in the basal∕parabasal portion of the tissue epithelium as compared to the superficial portion and measured the highest backscattering from the basal∕parabasal portion. Increased scattering from deeper areas of the mucosa may offset attenuation due to scattering at the surface.
The slope standard deviation, which quantifies the visibility of the underlying layers, is not impacted by the nominal slope. This metric most successfully differentiates the three tissue types: normal, GIN, and adenoma. Perhaps the most useful application for this metric is the ability to visualize a three-dimensional data set as a two-dimensional, en face disease map. This utility allows a researcher to quickly visualize the entire mouse colon, nondestructively and over multiple time points. It also enables the researcher to quickly locate relevant OCT images to verify the presence of disease.
Using the metrics to generate a two-dimensional disease map can be extended to functional OCT imaging of a biomarker such as EGFR. The most consistent quantitative metric for nanoshells detection is increased grayscale. This effect is consistent with quantitative measurements made by Kah et al.27 in a xenograft tumor model, which demonstrates that signal enhancement (increased gray scale) dominates as compared to signal attenuation (slope) in the first 200 μm. Slope standard deviation could have been confounded by increased surface brightness, by magnifying small slope variations. The results of this study suggest specificity of nanoshells targeted to EGFR, with change in average grayscale trending with increased EGFR grade. Quantitative metrics enable the measurement of this effect which is subtle to visualize by eye in the OCT images.
Acknowledgments
The research was supported in part by NIH R01 CA109835 as well as the Achievement Rewards for College Scientists, The University of Arizona Technology Research Initiative Fund Graduate Fellowship, and Philanthropic Educational Organization scholarships.
References
- Brand S., Poneros J. M., Bouma B. E., Tearney G. J., Compton C. C., and Nishioka N. S., “Optical coherence tomography in the gastrointestinal tract,” Endoscopy 32, 796–803 (2000). 10.1055/s-2000-7714 [DOI] [PubMed] [Google Scholar]
- Shen B. and Zuccaro G. J., “Optical coherence tomography in the gastrointestinal tract,” Gastrointest Endosc Clin. N. Am. 14, 555–571 (2004). 10.1016/j.giec.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Jackle S., Gladkova N., Feldchtein F., Terentieva A., Brand B., Gelikonov G., Gelikonov V., Sergeev A., Fritscher-Ravens A., Freund J., Seitz U., Schröder S., and Soehendra N., “In vivo endoscopic optical coherence tomography of esophagitis, Barrett’s esophagus, and adenocarcinoma of the esophagus,” Endoscopy 32, 750–755 (2000). 10.1055/s-2000-7705 [DOI] [PubMed] [Google Scholar]
- Evans J. A., Poneros J. M., Bouma B. E., Bressner J., Halpern E. F., Shishkov M., Lauwers G. Y., Mino-Kenudson M., Nishioka N. S., and Tearney G. J., “Optical coherence tomography to identify intramucosal carcinoma and high-grade dysplasia in Barrett’s esophagus,” Clin. Gastroenterol. Hepatol. 4, 38–43 (2006). 10.1016/S1542-3565(05)00746-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Sivak M. V., Isenberg G., Willis J. E., and Rollins A. M., “Computer-aided diagnosis of dysplasia in Barrett’s esophagus using endoscopic optical coherence tomography,” J. Biomed. Opt. 11, 044010 (2006). 10.1117/1.2337314 [DOI] [PubMed] [Google Scholar]
- Chen Y., Aquirre A. D., Hsiung P. L., Desai S., Herz P. R., Pedrosa M., Huang Q., Figueiredo M., Huang S. W., Koski A., Schmitt J. M., Fujimoto J. G., and Mashimo H., “Ultrahigh resolution optical coherence tomography of Barrett’s esophagus: preliminary descriptive clinical study correlating images with histology,” Endoscopy 39, 599–605 (2007). 10.1055/s-2007-966648 [DOI] [PubMed] [Google Scholar]
- Adler D. C., Zhou C., Tsai T. H., Schmitt J., Huang Q., Mashimo H., and Fujimoto J. G., “Three-dimensional endomicroscopy of the human colon using optical coherence tomography,” Opt. Express 17, 784–796 (2009). 10.1364/OE.17.000784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackle S., Gladkova N., Feldchtein F., Terentieva A., Brand B., Gelikonov G., Gelikonov V., Sergeev A., Fritscher-Ravens A., Freund J., Seitz U., Soehendra S., and Schrodern N., “In vivo endoscopic optical coherence tomography of the human gastrointestinal tract—toward optical biopsy,” Endoscopy 32, 743–749 (2000). 10.1055/s-2000-7711 [DOI] [PubMed] [Google Scholar]
- Pfau P. R., Sivak M. V. J., Chak A., Kinnard M., Wong R. C., Isenberg G. A., Izatt J. A., Rollins A., and Westphal V., “Criteria for the diagnosis of dysplasia by endoscopic optical coherence tomography,” Gastrointest. Endosc. 58, 196–202 (2003). 10.1067/mge.2003.344 [DOI] [PubMed] [Google Scholar]
- Fujimoto J. G., Brezinski M. E., Tearney G. J., Boppart S. A., Bouma B. E., Hee M. R., Southern J. F., and Swanson E. A., “Optical biopsy and imaging using optical coherence tomography,” Nat. Med. 1, 970–972 (1995). 10.1038/nm0995-970 [DOI] [PubMed] [Google Scholar]
- Tearney G. J., Brezinski M. E., Bouma B. E., Boppart S. A., Pitris C., Southern J. F., and Fujimoto J. G., “In vivo endoscopic optical biopsy with optical coherence tomography,” Science 276, 2037–2039 (1997). 10.1126/science.276.5321.2037 [DOI] [PubMed] [Google Scholar]
- Herz P., Chen Y., Aguirre A., Fujimoto J., Mashimo H., Schmitt J., Koski A., Goodnow J., and Petersen C., “Ultrahigh resolution optical biopsy with endoscopic optical coherence tomography,” Opt. Express 12, 3532–3542 (2004). 10.1364/OPEX.12.003532 [DOI] [PubMed] [Google Scholar]
- Cobb M. J., Chen Y., Bailey S. L., Kemp C. J., and Li X., “Non-invasive imaging of carcinogen-induced early neoplasia using ultrahigh-resolution optical coherence tomography,” Cancer Biomark. 2, 163–173 (2006). [DOI] [PubMed] [Google Scholar]
- Hariri L. P., Qiu Z., Tumlinson A. R., Besselsen D. G., Gerner E. W., Ignatenko N. A., Povazay B., Hermann B., Sattmann H., McNally J., Unterhuber A., Drexler W., and Barton J. K., “Serial endoscopy in azoxymethane treated mice using ultra-high resolution optical coherence tomography,” Cancer Biol. Ther. 6, 1753–1762 (2007). [DOI] [PubMed] [Google Scholar]
- Xiong H., Zeng C., Guo Z., Zhong H., Wang R., Liu S., and He Y., “Potential ability of hematoporphyrin to enhance an optical coherence tomographic image of gastric cancer in vivo in mice,” Phys. Med. Biol. 53, 6767–6775 (2008). 10.1088/0031-9155/53/23/008 [DOI] [PubMed] [Google Scholar]
- Bissahoyo A., Pearsall R. S., Hanlon K., Amann V., Hicks D., Godfrey V. L., and Threadgill D. W., “Azoxymethane is a genetic background-dependent colorectal tumor initiator and promoter in mice: effects of dose, route, and diet,” Toxicol. Sci. 88, 340–345 (2005). 10.1093/toxsci/kfi313 [DOI] [PubMed] [Google Scholar]
- Esenaliev R. O., Larin K. V., Larina I. V., and Motamedi M., “Noninvasive monitoring of glucose concentration with optical coherence tomography,” Opt. Lett. 26, 992–994 (2001). 10.1364/OL.26.000992 [DOI] [PubMed] [Google Scholar]
- Kuranov R. V., Sapozhnikova V. V., Prough D. S., Cicenaite I., and Esenaliev R. O., “In vivo study of glucose-induced changes in skin properties assessed with optical coherence tomography,” Phys. Med. Biol. 51, 3885–3900 (2006). 10.1088/0031-9155/51/16/001 [DOI] [PubMed] [Google Scholar]
- van der Meer F. J., Faber D. J., Baraznji Sassoon D. M., Aalders M. C., Pasterkamp G., and van Leeuwen T. G., “Localized measurement of optical attenuation coefficients of atherosclerotic plaque constituents by quantitative optical coherence tomography,” IEEE Trans. Med. Imaging 24, 1369–1376 (2005). 10.1109/TMI.2005.854297 [DOI] [PubMed] [Google Scholar]
- Turchin I. V., Sergeeva E. A., Dolin L. S., Kamensky V. A., Shakhova N. M., and Richards-Kortum R., “Novel algorithm of processing optical coherence tomography images for differentiation of biological tissue pathologies,” J. Biomed. Opt. 10, 064024 (2005). 10.1117/1.2137670 [DOI] [PubMed] [Google Scholar]
- Korde V. R., Bonnema G. T., Xu W., Krishnamurthy C., Ranger-Moore J., Saboda K., Slayton L. D., Salasche S. J., Warneke J. A., Alberts D. S., and Barton J. K., “Using optical coherence tomography to evaluate skin sun damage and precancer,” Lasers Surg. Med. 39, 687–695 (2007). 10.1002/lsm.20573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazant-Hegemark F. and Stone N., “Towards automated classification of clinical optical coherence tomography data of dense tissues,” Lasers Med. Sci. 24, 627–638 (2009). 10.1007/s10103-008-0615-6 [DOI] [PubMed] [Google Scholar]
- Qi X., Pan Y., Hu Z., Kang W., Willis J. E., Olowe K., Sivak M. V., and Rollins A. M., “Automated quantification of colonic crypt morphology using integrated microscopy and optical coherence tomography,” J. Biomed. Opt. 13, 054055 (2008). 10.1117/1.2993323 [DOI] [PubMed] [Google Scholar]
- Lin A. W., Lewinski N. A., West J. L., Halas N. J., and Drezek R. A., “Optically tunable nanoparticle contrast agents for early cancer detection: model-based analysis of gold nanoshells,” J. Biomed. Opt. 10, 064035 (2005). 10.1117/1.2141825 [DOI] [PubMed] [Google Scholar]
- Barton J. K., Halas N. J., West J. L., and Drezek R. A., “Nanoshells as an optical coherence tomography contrast agent,” Proc. SPIE 5316, 99–106 (2004). 10.1117/12.529235 [DOI] [Google Scholar]
- Agrawal A., Huang S., Wei Haw Lin A., Lee M. H., Barton J. K., Drezek R. A., and Pfefer T. J., “Quantitative evaluation of optical coherence tomography signal enhancement with gold nanoshells,” J. Biomed. Opt. 11, 041121 (2006). 10.1117/1.2339071 [DOI] [PubMed] [Google Scholar]
- Kah J. C., Olivo M., Chow T. H., Song K. S., Koh K. Z., Mhaisalkar S., and Sheppard C. J., “Control of optical contrast using gold nanoshells for optical coherence tomography imaging of mouse xenograft tumor model in vivo,” J. Biomed. Opt. 14, 054015 (2009). 10.1117/1.3233946 [DOI] [PubMed] [Google Scholar]
- Loo C., Lowery A., Halas N., West J., and Drezek R., “Immunotargeted nanoshells for integrated cancer imaging and therapy,” Nano Lett. 5, 709–711 (2005). 10.1021/nl050127s [DOI] [PubMed] [Google Scholar]
- Loo C., Hirsch L., Lee M. H., Chang E., West J., Halas N., and Drezek R., “Gold nanoshell bioconjugates for molecular imaging in living cells,” Opt. Lett. 30, 1012–1014 (2005). 10.1364/OL.30.001012 [DOI] [PubMed] [Google Scholar]
- Makita S., Fabritius T., and Yasuno Y., “Full-range, high-speed, high-resolution 1-μm spectral-domain optical coherence tomography using BM-scan for volumetric imaging of the human posterior eye,” Opt. Express 16, 8406–8420 (2008). 10.1364/OE.16.008406 [DOI] [PubMed] [Google Scholar]
- Tumlinson A. R., Povazay B., Hariri L. P., McNally J., Unterhuber A., Hermann B., Sattmann H., Drexler W., and Barton J. K., “In vivo ultrahigh-resolution optical coherence tomography of mouse colon with an achromatized endoscope,” J. Biomed. Opt. 11, 064003 (2006). 10.1117/1.2399454 [DOI] [PubMed] [Google Scholar]
- Schmitt J. M., Xiang S. H., and Yung K. M., “Speckle in optical coherence tomography,” J. Biomed. Opt. 4, 95–105 (1999). 10.1117/1.429925 [DOI] [PubMed] [Google Scholar]
- Tumlinson A. R., Hariri L. P., Drexler W., and Barton J. K., “Scatter sensitive microscopy techniques to identify contrasting mucosal structures in ultrahigh-resolution optical coherence tomograms of mouse colon,” Proc. SPIE 6851, 68510A (2008). 10.1117/12.763968 [DOI] [Google Scholar]
- Arifler D., Guillaud M., Carraro A., Malpica A., Follen M., and Richards-Kortum R., “Light scattering from normal and dysplastic cervical cells at different epithelial depths: finite-difference time-domain modeling with a perfectly matched layer boundary condition,” J. Biomed. Opt. 8, 484–494 (2003). 10.1117/1.1578640 [DOI] [PubMed] [Google Scholar]