Abstract
The quantitative polymerase chain reaction (QPCR) assay allows measurement of DNA damage in the mitochondrial and nuclear genomes without isolation of mitochondria. It also permits measurement of relative mitochondrial genome copy number. Finally, it can be used for measurement of DNA repair in vivo when employed appropriately. In this manuscript we briefly review the methodology of the QPCR assay, discuss its strengths and limitations, address considerations for measurement of mitochondrial DNA repair, and describe methodological changes implemented in recent years. We present QPCR assay primers and reaction conditions for five species not previously described in a Methods article: Caenorhabditis elegans, Fundulus heteroclitus, Danio rerio, Drosophila melanogaster, and adenovirus. Finally, we illustrate the use of the assay by measuring repair of ultraviolet C radiation-induced DNA damage in the nuclear but not mitochondrial genomes of a zebrafish cell culture.
Keywords: quantitative PCR assay, mitochondrial DNA damage, mitochondrial DNA repair
1. Introduction to the QPCR assay
1.1 Theory/Rationale
QPCR, also called long amplicon PCR (LA-PCR) and long extension PCR (LX-PCR), measures gene-specific damage by quantifying the decrease in amplification of DNA extracted from sample cells of organisms following genotoxin exposure. This assay is based on the principle that induced DNA damage impedes the progression of the DNA polymerase when analyzed by PCR, resulting in a decrease in the amount of PCR product (Fig. 1) (1). Thus, the amount of PCR amplification is inversely proportional to the amount of DNA damage on a given template. The amplification of long (typically 10-15 kb) targets of genomic DNA serves to enhance the sensitivity of the assay by increasing the probability that the polymerase will encounter damaged DNA. The PCR signal from a treated sample is compared to that from equal amounts of untreated control DNA, allowing the quantification of relative DNA damage. Damage can then be expressed as lesion frequencies per 10 kb DNA by application of the Poisson distribution. DNA from control samples is defined as undamaged for the purposes of this assay. Amplification of a small mitochondrial target (usually ~200 bp) permits both normalization to mtDNA copy number, and a relative measurement of mtDNA copy number changes.
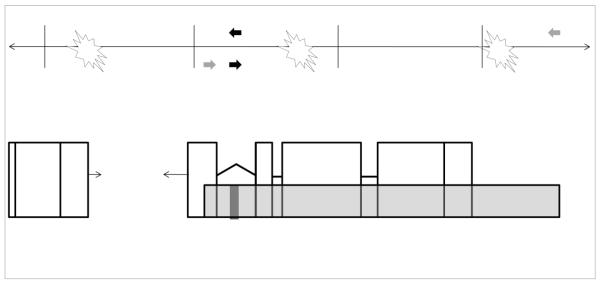
Figure 1. Schematic of the rationale of the QPCR assay for detecting damage in the nuclear genome.
The continuous line ( ↔ ) represents a segment of the nuclear genome and perpendicular hashmarks along the DNA segment mark 10 kb intervals. Open boxes beneath the DNA segment depict genetic coding regions (exons) that are interrupted by interspersed introns (thick black lines). The long amplicon (10-15 kb) is represented as a transparent grey bar that amplifies the majority of a selected gene as well as a portion of the upstream DNA sequence. The bar representing the small amplicon (~200 bases) is shaded black. The filled arrows below and above the DNA segment indicate the position of the corresponding forward and reverse primers for the long (grey) and short (black) amplicon. Lesions are represented as stars that would inhibit or block the progression of the DNA polymerase used in the QPCR assay, thus reducing the amplification of the long product under quantitative conditions. For the smaller amplicon, the relatively close placement of the primers creates a small target size and reduces the probability that such lesions will be detected. Amplification of the short product is not inhibited except by very high levels of damage.
1.2 Advantages and Limitations
1.2.1 Advantages
A major strength of the QPCR assay is its ability to detect a wide range of DNA damage. QPCR can be used to detect DNA adducts, strand breaks and any other type of modification that significantly stalls the progression of the DNA polymerase. Neither the prior knowledge of the nature of the induced lesions nor the use of several tools to detect multiple lesion types is required. Relatedly, damage at many sites along the genome can be detected as opposed, for example, to assays that detect only alterations to specific bases. The QPCR assay also simultaneously allows a relative determination of mitochondrial genome copy number without additional reactions or assays, which is helpful since many treatments can result in changes in mtDNA copy number.
Because the QPCR assay is based on genome-specific primers, its use eliminates the necessity to isolate different types of genomic DNA prior to analysis. Thus, direct and simultaneous comparison of induced damage and repair in both mitochondrial and nuclear genomes can be carried out without differential treatment (during isolation) of mtDNA and nucDNA. Relatedly, the QPCR can be easily modified to evaluate induced damage in different regions of the genome (2, 3), which may be of interest since some genotoxins cause more damage in some areas of the genome (4), and some DNA repair pathways are also differentially active in different parts of the genome (5).
Furthermore, since the primers used in the assay are also species-specific, the presence of DNA from other genomes and/or species in a sample will not affect measurements. For example, the presence of DNA from the OP50 strain of Escherichia coli bacteria on which C. elegans is normally fed will not affect the amplification of the C. elegans nuclear or mitochondrial QPCR targets. (However, contaminating DNA may affect the template quantitation and thus skew results if not handled properly – see QPCR assay, section 2.3).
Finally, a significant advantage is the small amount of genomic DNA (nanogram) required by this assay. We have obtained reliable results using as little as 1 ng of input DNA from DNA extracted as described below (Section 2.1.1 and 2.1.2) (unpublished), and in lysates of individual nematodes (Section 2.1.3) containing ~100 pg DNA (6). Thus, for example, the QPCR assay can be used with small tissue samples and white blood cells from blood draws (7).
1.2.2 Limitations
The extent to which the QPCR assay is able to efficiently detect DNA damage that is not usually considered to be polymerase-blocking, such as 8-hydroxydeoxy-guanosine (8-OHdG), has not been rigorously tested. Notably, however, oxidative damage generated by hydrogen peroxide is routinely detected using this assay (8-12); furthermore, 8-OHdG frequently constitutes only ~10% of oxidative lesions after hydrogen peroxide exposure (13). Additionally, when damage is detected, it is not possible to determine the type of DNA lesion present because of the nondiscriminatory nature of QPCR (as noted above, this may serve as a strength as well as a weakness, depending on the experimental question being asked). Another limitation of QPCR is that this method provides a relative quantification of DNA damage rather than an absolute quantification, since control samples are defined as “undamaged” for the purpose of lesion calculations. However, given the detection limit of this assay (~1 lesion in 105 nucleotides (14)), it is unlikely that a well-chosen control sample would have detectable damage. The assay also provides a relative, rather than absolute, measurement of mtDNA copy number. Absolute quantification compares the PCR of a standard curve to that of the treated sample to determine an absolute value for the input copy number. Our method of QPCR relates the PCR signal of treated DNA to that of another sample, such as the untreated control. These results provide a determination of the relative changes of mtDNA number caused by a damage-inducing agent.
1.3 Examples of chemicals causing damage detectable with the QPCR assay
The QPCR assay has been used to detect mitochondrial and/or nuclear DNA damage induced by many agents. Examples include: rotenone (inducing mtDNA but not nucDNA damage in human cells) (15), methyl methanesulfonate (MMS) (causing nucDNA damage in Saccharomyces cerevisiae) (16), decarbomoyl mitomycin C (inducing mtDNA and nucDNA damage in human cancer cells) (17), menadione (resulting in mtDNA damage in murine myoblasts) (18), benzo[a]pyrene (producing mtDNA and nucDNA damage in killifish) (19, 20), hydrogen peroxide (causing mtDNA but not nucDNA damage in human cells) (9), UV (nucDNA damage induced in mice, C. elegans, adenovirus and zebrafish) (3, 21, 22) (this manuscript, Section 3), zidovudine (3′-azido-3′-deoxythymidine or AZT), lamivudine ((-)2′,3′-dideoxy-3′-thiacytidine or 3TC) and Combivir (zidovudine and lamivudine) (inducing mtDNA in mice) (23), photoactivated hypericin (24), cisplatin, chlorambucil, and psoralen (2), lipopolysaccharide (25), and a complex mixture of environmental contaminants (20).
1.4 Species for which QPCR assay conditions have been identified
The QPCR assay has been applied to many species. Previous publications have detailed conditions for use of the QPCR assay for samples from Homo sapiens, Mus musculus, Rattus norvegicus, Saccharomyces cerevisiae, and Escherichia coli (2, 14, 26, 27). Here, we detail primers and conditions for Caenorhabditis elegans, Fundulus heteroclitus, Danio rerio, Drosophila melanogaster, and adenovirus; the Danio rerio and Drosophila melanogaster primers have not previously been published. We also provide minor corrections to the previously published S. cerevisiae primers (26, 28). We have previously published a protocol for the adaptation of this assay to new species; it is particularly simple to do so for the mitochondrial genome (29).
2. Protocol
Protocols for this assay have been published previously (2, 14, 26). Here, we focus on updates and changes made to the assay in recent years.
2.1 DNA Extraction
2.1.1 Method for DNA isolation: overview
QPCR measures induced DNA damage by amplifying a large segment of DNA. It is therefore critical to isolate high molecular weight, high quality DNA to be used for long-range PCR. Special care should be taken to avoid nicking, shearing, and introducing oxidative damage to the DNA template, as these lesions reduce amplification efficiency. As such, phenol extraction is not suggested because phenol leads to DNA oxidation which may interfere with sensitive downstream applications such as PCR (30). We routinely isolate total DNA using the Qiagen Genomic-tip 20/G and Qiagen DNA Buffer Set (Qiagen, cat. nos. 10223 and 19060, respectively) per the manufacturer’s protocol. The Qiagen protocol for the isolation of genomic DNA from cultured cells results in the removal of mitochondria and the loss of mtDNA. Thus, it is imperative that the Qiagen Tissue Extraction Protocol be used for the analysis of mtDNA.
2.1.2 Method for DNA isolation from cell culture
This method is suitable for the extraction of DNA from cultured cells and soft, easily digested tissue. Briefly, cells or homogenized tissues are incubated in a lysis buffer containing Triton X-100, the denaturant guanidine HCl and Proteinase K (Qiagen cat. no. 19133). Total DNA contained in the extracts is bound to Qiagen Genomic-tip 20/G columns, washed and eluted using standard anion-exchange chromatography techniques which employ varying salt conditions to elute purified DNA. To increase DNA yields, we evenly aliquot each eluate into two 1.7 or 2 mL eppendorf tubes, add 0.7 mL (70%) ethanol to each aliquot and centrifuge samples at 9,500 g. Additionally, DNA can be allowed to precipitate at −20° C prior to centrifugation to further increase yields. DNA integrity can be evaluated using agarose gel electrophoresis (29).
2.1.3 Method for DNA isolation via liquid nitrogen grinding
C. elegans have a cuticular integument that presents a significant barrier in traditional chemical lysis and DNA extraction. For samples obtained from such organisms with a hard, cartilaginous exterior, additional steps may be needed for effective DNA isolation. We routinely employ liquid nitrogen grinding to break up the nematode exterior for DNA extraction. Similarly, for Drosophila melanogaster, batches of flies were frozen and ground in liquid nitrogen prior to digestion and extraction using the Qiagen Genomic Tips protocol. Liquid nitrogen grinding is also an important step for harder tissues such as muscle, and may even be helpful for softer tissues such as liver (discussed below). Below, we described this method as applied to C. elegans, but it can be easily adapted to any other sample.
To isolate DNA, relatively large batches (3000-5000 individuals) of nematodes are collected, washed in K medium and centrifuged (2200 g, 2 min). The resulting pellet (which should be less than 0.5 mL) of nematodes is flash frozen by dripping the pellet/K medium mixture into liquid nitrogen using a Pasteur pipette. We use the liquid nitrogen-cooled mortar (Scienceware liquid nitrogen-cooled mortar cat. no. 37260-0000) for this step. The pellets (frozen droplets) are stored at −80°C until processing.
Prior to grinding the C. elegans samples, the base of the liquid nitrogen-cooled mortar is filled with dry ice and liquid nitrogen is added to the mortar to cool it and the pestle. Once the majority of liquid nitrogen has evaporated, we add approximately six pellets to the pre-cooled mortar and begin grinding. Pellets are ground into a fine powder, ensuring that there are no large chunks. A squeaking sound is heard towards the completion of grinding. It is necessary to use caution to prevent pellets from escaping the mortar during grinding. Using a spatula and/or forceps, the powder is scooped in the pre-aliquoted G2 buffer containing RNase as specified by the Qiagen Genomic DNA Handbook, Sample Preparation and Lysis Protocol for Tissue. The Handbook is followed for the subsequent elution of purified total DNA.
Although tissue samples such as livers from killifish and zebrafish are soft and easily digested and can be treated via homogenization as detailed in the Qiagen Genomic Sample Preparation and Lysis Protocol for Tissue, we often employ liquid nitrogen grinding prior to following the protocol so as to facilitate DNA extraction. This method may be more time intensive, but it speeds the passage of the DNA through the columns, improves yield (although some of the sample may be lost while grinding), and most importantly improves the quality of the DNA recovered. For DNA analysis from fish samples the desired tissue is dissected from sacrificed animals, weighed, and flash frozen in 20% glycerol and stored at −80°C until use. Tissue samples (15-30 mg) are then ground into a fine powder using the protocol for C. elegans grinding.
2.1.4 Method for DNA isolation via lysis
We have also adapted the C. elegans QPCR method to allow DNA amplification from a small number (from 1 to approximately 10) of nematodes (6), removing the need for DNA isolation and quantification. This method, which we term Individual Worm PCR, provides the benefit of allowing individual worm analysis and the determination of inter-individual variability rather than simply allowing the investigation of a batch effect of treatments. Additionally, using this protocol nematodes can be analyzed via QPCR immediately following lysis, reducing the preparation time prior to the QPCR step from the approximately 1.5 days required to isolate and quantify the DNA in the traditional batch method to approximately 1.5 h. A disadvantage of the lysis method is the minute amount of DNA obtained, which prevents the analysis of DNA integrity by agarose gel electrophoresis prior to QPCR and limits the number of QPCRs that can be run.
For the Individual Worm QPCR, using a worm pick, add 1-6 worms to PCR tubes containing lysis buffer at a ratio of 1 worm to 10 μL buffer for early-stage larvae and 1 worm in 20 or 30 μl for gravid adults. The lysis buffer is composed of 1x rTth XL PCR buffer and 1 mg/ml proteinase K. To prepare lysis buffer we add 300 μl of 3.3x rTth XL PCR buffer (XL PCR kit, Applied Biosystems) and 50 μl of Proteinase K (20 mg/ml; Qiagen cat. no. 19133), to 650 μl of Sigma W3500 dH2O (scale as necessary). Samples are immediately frozen at −80°C for at least 10 minutes (we do not allow the worms to remain in the lysis buffer at room temperature for longer than 5 minutes) and are stored at −80° C until we are ready to proceed.
To lyse worms, thawed samples are vortexed for 5 s, heated (using a thermocycler) for 55 minutes at 65°C (the optimal temperature for Proteinase K) and then for 15 minutes at 95°C (to deactivate the Proteinase K), then cooled to 4°C. We proceed immediately to QPCR or store samples at either 4°C (1-2 weeks) or −80°C.
2.1.4 Method for DNA isolation from adenovirus
Eischeid et al. (22) found that viral DNA of sufficiently high quality could be extracted from purified preparations of adenovirus using the QIAamp DNA blood mini kit (Qiagen, Valencia, CA). Adenovirus was propagated in host cells and subsequently isolated and concentrated via polyethylene glycol precipitation.
2.2 Primers for amplification of large and small products
The number of mitochondria per cell and the total DNA content in each mitochondrion can be highly variable depending on cell and tissue type. Therefore, it is necessary to normalize QPCR results for mitochondrial copy number to ensure that changes in amplification levels are true reflections of DNA damage and not artifacts resulting from fluctuations in the amount of mitochondrial DNA present in each sample. This normalization is carried out via amplification of a small mitochondrial target and rests on the premise that the amount of damage present is low enough that it will not inhibit amplification of such a small fragment. Thus, the degree of amplification of the small fragment represents only the amount of DNA. Total nuclear DNA content does not fluctuate, and so under most circumstances the use of a small nuclear product is not necessary. Under some circumstances, however, it may be useful to compare the amplification of a short nuclear fragment between samples to verify the starting amount of template DNA. For example, the use of a small nuclear amplicon may be especially helpful when amplifying DNA directly from a minute number of lysed C. elegans, to confirm that the desired number of nematodes were picked. Alternatively, the short nuclear product is not necessary (for samples lacking contaminating DNA) if the template DNA was extracted by the batch procedure, quantified with PicoGreen and diluted to equal concentrations for all samples, as such samples do not require normalization.
Primers for the generation of short (approximately 200 bp) and long (9 to 14 kb) products for multiple species are presented in Table 1. Additional primers have been developed for various nuclear targets in C. elegans, allowing for the analysis of DNA damage and repair rates in differentially transcribed regions of the genome (3). Additional primers were also described for adenovirus (22), including short products that permit QPCR-based quantification of input DNA, and additional ~1 kb products that may facilitate adaptation to other Adenovirus types. Although primers that generate products approximately 10 kb in length are typically used in long amplicon QPCR, the primers listed in Table 1 for the 1 kb product were sufficient for the detection of UV-induced DNA damage in adenovirus (22) because of the very high doses of UV that are used to inactivate adenovirus. The zebrafish nuclear primers are placed in the genomic neighborhood of the aryl hydrocarbon receptor 2 gene.
Table 1.
Primers used for QPCR assay
| Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Reference |
|---|---|---|
| C. elegans | ||
| nuclear target - 13.7 kb | ||
| AGTCGTTGAACGCAGTGGTGTCAT | CAGTCTTTCTTCGACGCATTCAACG | (3) |
|
| ||
| nuclear target - 9.3 kb | ||
| TGGCTGGAACGAACCGAACCAT | GGCGGTTGTGGAGTGTGGGAAG | (3, 38) |
|
| ||
| nuclear target - 225 bp | ||
| TCCCGTCTATTGCAGGTCTTTCCA | GACGCGCACGATATCTCGATTTTC | (3, 38) |
|
| ||
| mitochondrial target - 10.9 kb | ||
| CCATCAATTGCCCAAAGGGGAGT | TGTCCTCAAGGCTACCACCTTCTTCA | (3) |
|
| ||
| mitochondrial target - 195 bp | ||
| CACACCGGTGAGGTCTTTGGTTC | TGTCCTCAAGGCTACCACCTTCTTCA | (3) |
|
| ||
| D. melanogaster | ||
| nuclear target - 11.5 kb | ||
| GTATTCCTGCGCCAGGAGGATCG | CAGATGCTGGAGCTGCCTTTGGA | |
|
| ||
| nuclear target - 10.3 kb | ||
| GAGGAGCCTTGCGAACAACAGCA | CAATGACAGCTGCGCCTCGAGAT | |
|
| ||
| nuclear target - 152 bp | ||
| CGAGGGATACCTGTGAGCAGCTT | GTCACTTCTTGTGCTGCCATCGT | |
|
| ||
| mitochondrial target - 14.2 kb | ||
| GCCGCTCCTTTCCATTTTTGATTTCC | TGCCAGCAGTCGCGGTTATACCA | |
|
| ||
| mitochondrial target - 151 bp | ||
| GCTCCTGATATAGCATTCCCACGA | CATGAGCAATTCCAGCGGATAAA | |
|
| ||
| Adenovirus type 2 | ||
| 1016 bp fragment | ||
| CAGGAATCGCCCCATCATCGTC | CGCCCGACTTGTTCCTCGTTTG | (22) |
|
| ||
| Fundulus heteroclitus (Atlantic killish) | ||
| nuclear target - 11.5 kb | ||
| CAGCCGCCCGCAAATTCTCA | CAGAATGCGGGCCTTGCTGA | (20) |
|
| ||
| nuclear target - 234 bp | ||
| GCCGCTGCCTTCATTGCTGT | ATGAGCTGGGTGTGCGCTGA | (20) |
|
| ||
| mitochondrial target - 9.4 kb | ||
| TTGCACCAAGAGTTTTTGGTTCCTAAGA CC |
GATGTTGGATCAGGACATCCCAATGGT GCA |
(20, 39) |
|
| ||
| mitochondrial target - 264 bp | ||
| ATCTGCATGGCCAACGCCTA | GGCGGTGCCAGTTTCCTTTT | (20) |
|
| ||
| Danio rerio (zebrafish) | ||
| nuclear target - 10.7 kb | ||
| AGAGCGCGATTGCTGGATTCAC | GTCCTTGCAGGTTGGCAAATGG | |
|
| ||
| nuclear target - 233 bp | ||
| ATGGGCTGGGCGATAAAATTGG | ACATGTGCATGTCGCTCCCAAA | |
|
| ||
| mitochondrial target - 10.3 kb | ||
| TTAAAGCCCCGAATCCAGGTGAGC | GAGATGTTCTCGGGTGTGGGATGG | |
|
| ||
| mitochondrial target - 198 bp | ||
| CAAACACAAGCCTCGCCTGTTTAC | CACTGACTTGATGGGGGAGACAGT | |
|
| ||
| Saccharomyces cerevisiae | ||
| nuclear target - 16.4 kb (forward and reverse primers referred to as 666791 and 683201 respectively) | ||
| ATCATCCCGATTGCTGCCACTAG | CGCTAAAATCCCGTGTATCCCTTG | (26) |
|
| ||
| mitochondrial target - 6.9 kb (forward and reverse primers referred to as 13999 and 20948 respectively) | ||
| GTGCGTATATTTCGTTGATGCGT | GTCACCACCTCCTGATACTTCAA | (26) |
All of the primers presented have already been validated in terms of producing a single band of the expected size. However, it is useful to occasionally verify the size and uniqueness of the product by agarose gel electrophoresis during experimentation, since the plate reader/PicoGreen quantification method only measures total fluorescence of the QPCR product.
Two notes regarding the D. melanogaster primers are warranted. First, the mitochondrial primers amplify the majority of the fly mitochondrial genome (19517 bp total). The portion that is excluded is very AT-rich, so that its exclusion may be somewhat problematic for types of DNA damage that differentially target AT-rich regions. Second, two sets of primers for amplification of a long nuclear product are described, both for the genomic region surrounding the β–tubulin gene. Both sets of primers work with the Oregon strain, but the primers that amplify an 11.5 kb product work with the w1 strain, while those that amplified the 10.3 kb product do not.
We also provide corrections to the previously published S. cerevisiae primers for the amplification of long nuclear (16.4 kb) and mitochondrial (6.9 kb) fragments (26, 28). These corrections are shown in bold print in Table 1 and the genomic target locations corresponding to those previously provided are listed for each primer pair. Other S. cerevisiae primers have also been used in the QPCR assay (16, 26, 28).
2.3 QPCR
To accurately assess the number of induced DNA lesions, it is critical that template DNA be quantified and diluted to the same concentration with minimal variation between samples prior to QPCR. Significant variations in the concentrations of diluted DNA can skew QPCR results. The method used to quantify DNA with PicoGreen dye and dilute DNA to the appropriate concentrations has been detailed (14). We typically amplify 10 ηg of total DNA. It is possible to do this assay with less template; we have found success using as little as 1 ηg of genomic DNA (unpublished). However, we advise including an additional PCR replicate when the amount input DNA is reduced, as the values from resulting quantifications appear to become slightly more variable from PCR to PCR. With each QPCR assay, we also include a 50% control containing 5 ηg (rather than 10 ng) of the untreated DNA control sample (containing half of the volume of the untreated control, balanced by the addition of an equal volume of 1X TE buffer). It is best to prepare a large volume of 50% control dilution, rather than attempting to pipette 50% of the template volume and supplementing with buffer. The number of PCR cycles performed is selected such that the amount of PCR product detected in the 5 ηg reaction is 40 - 60% of that of the 10 ηg untreated control DNA. Only reactions that meet this criterion are considered to represent quantitative PCR reactions. Typical cycling conditions for each primer pair in Table 1 are presented in Table 2.
Table 2.
Conditions for Primers used in QPCR assay
| genome | amplicon size |
cycles | Temp (°C) |
Primer concentrationcon (μm, each) |
MgO(Ac)2 centration (μm) |
|---|---|---|---|---|---|
| C. elegans | |||||
| nuclear | 13.7 kb | 21 | 68 | 0.4 | 1.2 |
| nuclear | 9.3 kb | 20 | 68 | 0.4 | 1.2 |
| nuclear | 225 bp | 23 | 63 | 0.4 | 1.2 |
| mitochondrial | 10.9 kb | 16 | 66 | 0.4 | 1.2 |
| mitochondrial | 195 bp | 18 | 63 | 0.3 | 1.2 |
| D. melanogaster | |||||
| nuclear | 11.5 kb | 22 | 67 | 0.4 | 1.2 |
| nuclear | 10.3 kb | 22 | 67 | 0.4 | 1.2 |
| nuclear | 152 bp | 24 | 65 | 0.4 | 1.2 |
| mitochondrial | 14.2 kb | 17 | 66 | 0.4 | 1.2 |
| mitochondrial | 151 bp | 19 | 61 | 0.4 | 1.2 |
| Adenovirus type 2 | |||||
| 1 kb | 18-19 | 65 | 0.4 | 1.2 | |
| F. heteroclitus (Atlantic killish) | |||||
| nuclear | 11.5 kb | 24 | 68 | 0.4 | 1.1 |
| nuclear | 234 bp | 24 | 62 | 0.4 | 1.2 |
| mitochondrial | 9.4 kb | 16 | 65 | 0.4 | 1.2 |
| mitochondrial | 264 bp | 24 | 62 | 0.4 | 1.2 |
| D. rerio (zebrafish) | |||||
| nuclear | 10.7 kb | 24 | 69 | 0.4 | 1.05 |
| nuclear | 233 bp | 27 | 60 | 0.4 | 1.2 |
| mitochondrial | 10.3 kb | 19 | 68 | 0.4 | 1.2 |
| mitochondrial | 198 bp | 21 | 62 | 0.4 | 1.2 |
When carrying out QPCR, it is exceedingly important to avoid potential sample crosscontamination with PCR products. To ensure a lack of contamination, completed PCR reactions must not be opened in the same room where DNA is extracted and QPCR reactions are set up. A dedicated workstation that is in a physically separate laboratory from where completed reactions are opened and analyzed should be used to set up QPCR reactions. Procedures and other important factors to consider when carrying out QPCR are outlined by Santos et. al (14).
QPCR is performed with the GeneAmp XL PCR system (Applied Biosystems), which uses rTth DNA Polymerase XL enzyme designed to amplify target DNA sequences up to 40 kb. In addition to template DNA, all reactions contain: 1X rTth buffer, 200 μM dNTPs, 1.2 mM MgO(Ac)2 (unless otherwise specified in Table 2), the indicated concentration of forward and reverse primers (see Table 2) and 0.1 mg/mL bovine serum albumin (BSA) in a volume of 40 μL (including Sigma W3500 water). BSA is added to stabilize the polymerase while it carries out the long amplification process. Of note, we find that the quality of BSA varies depending on the source supplying the protein. Therefore we consistently purchase BSA from Roche. We then add 5 μL of template DNA (or 1X TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) buffer for the negative control). All reactions are initiated by a hot start (75 °C, 2 min) prior to the addition of the rTth enzyme. The polymerase is diluted 1:10 in 1X rTth buffer (composed of the supplied 3.3x rTth buffer diluted with Sigma W3500 water) just before starting reactions, and kept at room temperature during the 2 min hot start. After the completion of the hot start, a total of 5 μL (1 Unit) of diluted polymerase is added to each amplification reaction to bring the final volume of each to 50 μL.
Cycling conditions (the number of PCR cycles and the annealing temperature) vary depending on the primers being used (Table 2), as each primer set must be optimized to provide quantitative results and a lack of spurious products. For amplification of long targets, the standard program (following the hot start of 75 °C for 2 min and the addition of the polymerase) includes an initial denaturation step at 94 °C for 1 min followed by a variable number of cycles of at 94 °C for 15 s and a set annealing temperature (see Table 2) for 12 min, with a final extension at 72 °C for 10 min. The thermocycler profile for the amplification of short DNA fragments (following the hot start and the addition of polymerase) includes an initial denaturation step at 94 °C for 1 min, a variable number of cycles of 94 °C for 15 s, a specified annealing temperature (Table 2) for 45 s, and 72 °C for 30 s, with final extension at 72 °C for 10 min. Following QPCR, we maintain reactions between 4 and 8°C.
When amplifying DNA from C. elegans using the batch DNA extraction method (Section 2.1.3), it may become necessary to consider the presence of DNA from E. coli bacteria (the food source). It is our experience that gut bacteria are cleared from healthy, young nematodes in under an hour (it is unclear how long bacterial DNA might persist afterwards). If one has not been very careful to eliminate all OP50 bacteria from the worms prior to DNA extraction (including time to eliminate the bacteria and bacterial DNA from their guts), bacterial DNA will be extracted along with DNA from the nematodes. We have verified that the QPCR C. elegans primers are specific for their targets and do not amplify E. coli DNA (Meyer et al., 2007). Thus, contaminating bacterial DNA will not affect the amplification of the C. elegans nuclear or mitochondrial genome. However, contaminating DNA may affect template quantitation, which may lead to the misinterpretation of results. Furthermore, if the calculated template concentration is unreliable due to the presence of contaminating DNA, it is not possible to get information regarding the mtDNA copy number from the amplification of the small mitochondrial amplicon alone. In that case, the small mtDNA amplicon must be compared to the product of small nuclear amplification, not to the total amount of input template DNA. Finally, it may be necessary to re-optimize the cycle number for samples that are contaminated by bacterial DNA, since the “10 ηg” input will be partly bacterial DNA. Therefore, more cycles may be required. This is especially true in the case of old worms since it is our observation that they do not clear their guts well. In the QPCR analysis of lysed C. elegans DNA (Section 2.1.4), normalization with the short PCR product is crucial.
The cycle numbers listed in Table 2 for the amplification of C. elegans DNA are optimized for the batch method of DNA isolation, but new users should re-optimize cycle numbers since amplification may vary slightly between laboratories depending on thermocycler, etc. When carrying out QPCR on lysates containing very small amounts of DNA, it may be necessary to increase the cycle numbers. For example, for Individual Worm QPCR, we typically carry out 26, 29, 26 and 28 cycles for the small mitochondrial, small nuclear, long mitochondrial and long nuclear unc-2 DNA targets respectively, but this may vary by ratio of worm number to volume of lysis buffer, and the developmental stage of the nematodes.
For proper analysis, each QPCR assay must include the treated DNA sample(s), the untreated positive control(s), 50% input DNA of the untreated positive control (to verify quantitative conditions), and a negative control (used to subtract background fluorescence during quantitation).
2.4 Considerations for use of the QPCR for studies of DNA repair
The QPCR assay has been effectively used for studies of DNA repair many times, by assaying DNA damage at various timepoints after genotoxin exposure. However, particular experimental considerations need to be made in order to study DNA repair using the QPCR assay. In either genome, the illusion of DNA repair can be created by DNA replication-related dilution of damage, since the QPCR assay measures the proportion of damaged templates present. In the nuclear genome, this confounder can be avoided experimentally in a variety of ways, including utilization of time courses and experimental conditions that preclude cell division. A strength of C. elegans as an in vivo model for DNA repair is that adults composed exclusively of nonmitotic, terminally differentiated cells can be used for studies of DNA repair (3). However, the possibility of mitochondrial DNA replication clearly exists even in the absence of cell division. Thus, it is helpful that the QPCR assay measures relative mtDNA copy number as well as damage. A significant amount of mtDNA replication will be reflected by a change in the relative mtDNA copy number, unless replication is accompanied by simultaneous degradation (DNA turnover). If the latter is a concern, then labeling DNA with 5-bromodeoxyuridine (31) may be necessary.
2.5 Calculation of lesion frequency and relative mitochondrial DNA copy number
A protocol for data analysis and DNA quantitation has been previously outlined (2, 14). The assay is based on the quantitative loss (damage) or gain (repair) in fluorescence detected during the PicoGreen analysis of control and treated QPCR products. DNA damage is quantified by comparing the relative amplification of large fragments (approximately 10 kb) of DNA from treated samples to those of controls and normalizing this to the amplification of smaller (approximately 200 bp) fragments. Following QPCR, products are quantified using PicoGreen dye and a fluorescence plate reader in the same manner as the template DNA. The resulting values are converted to relative lesion frequencies per 10 kb DNA by application of the Poisson distribution (lesions/amplicon = −ln(At /Ao); where At represents the amplification of treated samples and Ao is the amplification of untreated controls).
In addition to the determination of the number of lesions and the relative copy number using described quantitation and normalization steps, the concentration of amplified DNA in each QPCR reaction can be calculated using the standard curve. As above, following QPCR the amount of DNA from each reaction is detected using a fluorescence plate reader during PicoGreen analysis. However, we also include a standard curve (the same as that used to quantify template DNA) on each plate. This permits more direct comparisons of amplification efficiencies from reaction to reaction, and facilitates integration of data from large experiments (i.e., those that generate too many samples to be analyzed on a single 96-well plate).
The calculations involved are illustrated in Supplementary data file 1, where the data for the dose-response example in Section 3 is analyzed.
3. Example of Derived Data
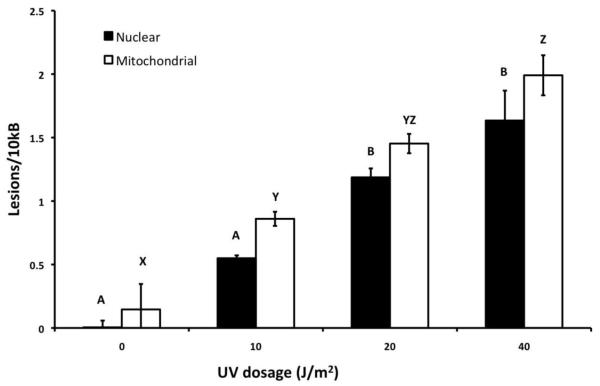
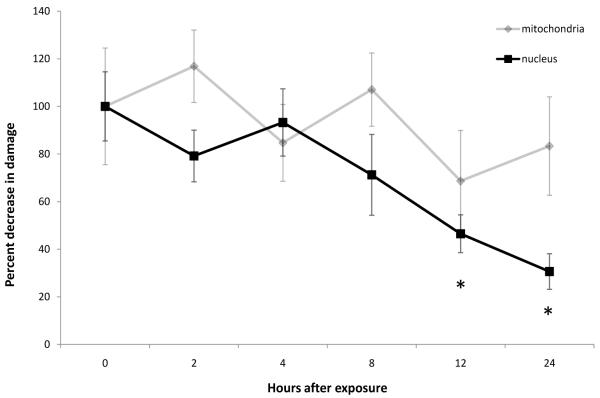
As an example, we present data from an experiment in which the QPCR method was adapted to the zebrafish model. Here, we are able to determine the presence of DNA damage and repair in a zebrafish cell line. Zebrafish embryonic fibroblasts (ZF-4 cell line, ATCC # CRL-2050) were cultured at 28°C (5% CO2) in Dulbecco’s MEM with 15% FBS and 0.1 mg/ml gentamicin as described (32) and exposed to various doses of ultraviolet C radiation (UVC). DNA was then isolated using the Qiagen Tissue Extraction Protocol and Qiagen Genomic tip- 20/G (see Section 2.1.2) and analyzed using QPCR. As shown in Figure 2, dose-dependent damage (p < 0.05 for both mtDNA and nucDNA) was detected with the newly developed primer sets and PCR conditions (see Tables 1 and 2). To test the ability of zebrafish to repair UVC-induced nuclear and mitochondrial DNA damage, cells were treated with 10 J/m2 of UVC and then incubated for various amounts of time to allow DNA repair. DNA was isolated from tissues flash-frozen at each designated time, and mtDNA and nucDNA damage was measured using QPCR. As seen in Figure 3, there was a significant dose-dependent decrease in nucDNA damage (p <0.01), with ~75% of damage repaired after 24h. This is interesting since investigations of nucDNA repair capacity for UV-induced and PAH-induced DNA damage that would normally be repaired by nucleotide excision repair have yielded variable results in several fish species (33-35). NucDNA repair was also observed in zebrafish cell cultures following very high doses of UV using a plasmid repair system (host cell reactivation) to measure DNA repair (36, 37). We did not observe a decrease in damage (i.e., repair) in mtDNA; mtDNA repair in zebrafish was not measurable in previous studies. This method can also be carried out with tissue samples dissected from the zebrafish. Using the same primer sets and cycling conditions would permit the comparison of the DNA damage response in cell culture and whole organisms.
Figure 2. DNA damage in UVC-treated zebrafish cells.
Mitochondrial and nuclear DNA damage was assessed in UVC-treated zebrafish cells. Petri dishes (10 mm diameter) containing roughly 106 cells/ml were drained free of culture medium and treated with 10, 20, and 40 J/m2 of UVC. Total DNA was isolated from cells and analyzed using QPCR. Letters indicate significant differences (p < 0.05) according to Fisher’s least significant differences analysis, carried out after ANOVA.
Figure 3. DNA damage repair in the zebrafish cell line ZF-4.
A petri dish (10 mm diameter) containing roughly 106 cells/ml were drained free of culture medium and treated with 10 J/m2 of UVC. The dishes were immediately filled with new culture media and wrapped in aluminum foil to prevent any exposure to light. Then the plates were kept in normal incubation condition for 0, 2, 4, 8, 12, and 24 hours. After each designated time, DNA was isolated from three random plates per time point, and mtDNA and nucDNA damage was measured using QPCR. * indicates significant differences from 0 hrs (p < 0.01).
4. Summary
The QPCR assay is a well-validated and sensitive protocol that specifically measures DNA damage in various mitochondrial and nuclear DNA regions in a variety of species, thus permitting investigation of mtDNA and nucDNA damage and repair. This is a particularly useful method in investigating mtDNA damage since differential separation of the mitochondrial DNA is not required, and because very small amounts of DNA can be analyzed. Thus, this method lends itself to biomarker-type work in humans, laboratory models, and wildlife species. We believe that this is of particular relevance to the field of environmental health since while defects in mtDNA and mitochondrial function have been implicated in a large number of human diseases, the effects of environmental stressors on mitochondrial DNA are poorly understood.
Supplementary Material
Acknowledgements
This research was supported by NIH P42 ES10356 (to RTD) and NIH R21 NS065468 (to JNM); the funding sources played no role in the research related to this publication, nor in the writing. We thank Elwood Linney for providing the ZF-4 cell lines and for cell culture advice, Bennett Van Houten for support during the adaptation of the assay to D. melanogaster, and Amanda Smith for editing suggestions.
Abbreviations
- QPCR
(quantitative polymerase chain reaction)
- mtDNA
(mitochondrial DNA)
- nucDNA
(nuclear DNA)
- UVC
(ultraviolet C radiation)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ponti M, Forrow SM, Souhami RL, D’Incalci M, Hartley JA. Nucleic Acids Res. 1991;19:2929–33. doi: 10.1093/nar/19.11.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayala-Torres S, Chen Y, Svoboda T, Rosenblatt J, Van Houten B. Methods. 2000;22:135–47. doi: 10.1006/meth.2000.1054. [DOI] [PubMed] [Google Scholar]
- 3.Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van Houten B. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedberg EC, W G, Siede W, Wood RD, Schultz RA, Ellenberger T. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 5.Hanawalt PC, Spivak G. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 6.Boyd WA, Crocker TL, Rodriguez AM, Leung MC, Lehmann DW, Freedman JH, Van Houten B, Meyer JN. Mutat Res. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haugen AC, D PN, Parker JS, Fannin RD, Chou J, Meyer JN, Halweg C, Collins JB, Durr A, Fischbeck K, Van Houten B. PLoS Genetics. 2010 doi: 10.1371/journal.pgen.1000812. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakes FM, Van Houten B. Proc Natl Acad Sci U S A. 1997;94:514–9. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos JH, Hunakova L, Chen Y, Bortner C, Van Houten B. J Biol Chem. 2003;278:1728–34. doi: 10.1074/jbc.M208752200. [DOI] [PubMed] [Google Scholar]
- 10.Sawyer DE, Mercer BG, Wiklendt AM, Aitken RJ. Mutat Res. 2003;529:21–34. doi: 10.1016/s0027-5107(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 11.Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Aging Cell. 2004;3:399–411. doi: 10.1111/j.1474-9728.2004.00124.x. [DOI] [PubMed] [Google Scholar]
- 12.Jarrett SG, Albon J, Boulton M. Free Radic Res. 2006;40:1155–65. doi: 10.1080/10715760600876613. [DOI] [PubMed] [Google Scholar]
- 13.Termini J. Mutat Res. 2000;450:107–24. doi: 10.1016/s0027-5107(00)00019-1. [DOI] [PubMed] [Google Scholar]
- 14.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Methods Mol Biol. 2006;314:183–99. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 15.Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. PLoS One. 2009;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma W, Panduri V, Sterling JF, Van Houten B, Gordenin DA, Resnick MA. Mol Cell Biol. 2009;29:1212–21. doi: 10.1128/MCB.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boamah E, B A, Tomasz M, Myeku N, Figueiredo-Pereira M, Hunter S, Meyer J, Bargonetti J. 2010. in press. [DOI] [PMC free article] [PubMed]
- 18.Trnka J, Blaikie FH, Logan A, Smith RA, Murphy MP. Free Radic Res. 2009;43:4–12. doi: 10.1080/10715760802582183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung D, Cho Y, Collins LB, Swenberg JA, Di Giulio RT. Aquat Toxicol. 2009;95:44–51. doi: 10.1016/j.aquatox.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung D, Cho Y, Meyer JN, Di Giulio RT. Comp Biochem Physiol C Toxicol Pharmacol. 2009;149:182–6. doi: 10.1016/j.cbpc.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmon AB, Ljungman M, Miller RA. J Gerontol A Biol Sci Med Sci. 2008;63:219–31. doi: 10.1093/gerona/63.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eischeid AC, Meyer JN, Linden KG. Appl Environ Microbiol. 2009;75:23–8. doi: 10.1128/AEM.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SS, Santos JH, Meyer JN, Mandavilli BS, Cook DL, Jr., McCash CL, Kissling GE, Nyska A, Foley JF, van Houten B, Copeland WC, Walker VE, Witt KL, Bishop JB. Environ Mol Mutagen. 2007;48:190–200. doi: 10.1002/em.20191. [DOI] [PubMed] [Google Scholar]
- 24.Wielgus AR, Chignell CF, Miller DS, Van Houten B, Meyer J, Hu DN, Roberts JE. Photochem Photobiol. 2007;83:706–13. doi: 10.1562/2006-08-09-RA-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Cardiovasc Res. 2004;64:279–88. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Santos JH, Mandavilli BS, Van Houten B. Methods Mol Biol. 2002;197:159–76. doi: 10.1385/1-59259-284-8:159. [DOI] [PubMed] [Google Scholar]
- 27.Chandrasekhar D, Van Houten B. Mutat Res. 2000;450:19–40. doi: 10.1016/s0027-5107(00)00014-2. [DOI] [PubMed] [Google Scholar]
- 28.Karthikeyan G, Santos JH, Graziewicz MA, Copeland WC, Isaya G, Van Houten B, Resnick MA. Hum Mol Genet. 2003;12:3331–42. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- 29.Meyer JN. Ecotoxicology. (in press)
- 30.Verhagen OJ, Wijkhuijs AJ, van der Sluijs-Gelling AJ, Szczepanski T, van der Linden-Schrever BE, Pongers-Willemse MJ, van Wering ER, van Dongen JJ, van der Schoot CE. Leukemia. 1999;13:1298–9. doi: 10.1038/sj.leu.2401451. [DOI] [PubMed] [Google Scholar]
- 31.Anson RM, Croteau DL, Stierum RH, Filburn C, Parsell R, Bohr VA. Nucleic Acids Res. 1998;26:662–8. doi: 10.1093/nar/26.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassiter CS, Kelley B, Linney E. Gene. 2002;299:141–51. doi: 10.1016/s0378-1119(02)01050-8. [DOI] [PubMed] [Google Scholar]
- 33.Willett KL, Lienesch LA, Di Giulio RT. Comp Biochem Physiol C Toxicol Pharmacol. 2001;128:349–58. doi: 10.1016/s1532-0456(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 34.Weimer TL, Reddy AP, Harttig U, Alexander D, Stamm SC, Miller MR, Baird W, Hendricks J, Bailey G. Toxicol Sci. 2000;57:217–28. doi: 10.1093/toxsci/57.2.217. [DOI] [PubMed] [Google Scholar]
- 35.David WM, Mitchell DL, Walter RB. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:301–9. doi: 10.1016/j.cca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Notch EG, Mayer GD. Aquat Toxicol. 2009;95:273–8. doi: 10.1016/j.aquatox.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Sussman R. Proc Natl Acad Sci U S A. 2007;104:13379–83. doi: 10.1073/pnas.0706157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd WA, Crocker TL, Rodriguez AM, Leung MC, Lehmann DW, Freedman JH, Van Houten B, Meyer JN. Mutat Res. 2009 doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim IC, Kweon HS, Kim YJ, Kim CB, Gye MC, Lee WO, Lee YS, Lee JS. Gene. 2004;336:147–53. doi: 10.1016/j.gene.2004.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.