Abstract
Intramolecular transfer of phosphate during collision-induced dissociation (CID) in ion trap mass spectrometers has recently been described. Because phosphorylation events are assigned to discrete serine, threonine, and tyrosine residues based on the presence of site-determining ions in MS/MS spectra, phosphate transfer may invalidate or confound site localization in published large-scale phosphorylation data sets. Here, we present evidence for the occurrence of this phenomenon using synthetic phosphopeptide libraries, specifically for doubly-charged species. We found, however, that the extent of the transfer reaction was insufficient to cause localization of phosphorylation sites to incorrect residues. We further compared CID to electron-transfer dissociation (ETD) for site localization using synthetic libraries and a large-scale yeast phosphoproteome experiment. The agreement in site localization was >99.5 and 93%, respectively, suggesting that ETD-based site localization is no more reliable than CID. We conclude that intramolecular phosphate transfer does not affect the reliability of current or past phosphorylation data sets.
Keywords: Phosphoproteomics, intramolecular phosphate transfer, phosphopeptide library, collision induced dissociation, electron capture dissociation
Introduction
Recently, Palumbo and Reid described a phenomenon that raises some serious concerns regarding phosphorylation site localization in large scale phosphoproteomic studies1, causing the proteomics community to take notice2–6. The principle concerns are derived from a set of thirty three synthetic phosphopeptides analyzed by ion trap tandem mass spectrometry. Notably, fifteen of the thirty three peptides gave rise to product ions apparently formed following intramolecular phosphate transfer which might lead to ambiguous phosphorylation site assignment. They proposed that phosphoproteomic studies shift toward alternative mechanisms to validate site localization including ETD or ECD (electron capture dissociation) or that experimentally observed CID-based phosphopeptides be compared with independently synthesized phosphopeptide standards.
Here, we evaluated the extent of intramolecular phosphate transfer under typical ion trap CID conditions using two corresponding phosphopeptide libraries (the libraries were based on an example peptide reported to undergo intramolecular phosphate transfer during CID). Each library consisted of 960 singly-phosphorylated peptides containing only two potential phosphate acceptor residues. The libraries were analyzed with an LTQ-Orbitrap mass spectrometer collecting both CID- and ETD-type peptide fragmentation spectra. Using CID, we did find evidence for the appearance of unexpected site-determining ions derived from intramolecular phosphate transfer. However, the rearrangement reaction was only observed for 2+ precursor peptides. One additional fragment ion peak than would be expected by chance was matched. Higher charge states showed no evidence of intramolecular phosphate transfer.
We also evaluated ETD as an alternative fragmentation technique to CID for enhanced identification and site localization. Both libraries and a large-scale yeast phosphorylation experiment were analyzed with same-precursor, back-to-back CID and ETD techniques. In the yeast experiment, CID identified 3 fold more phosphopeptides than ETD from equal numbers of ETD and CID spectra. We found no improvement in the quality of site assignments and limited utility of ETD as a standalone fragmentation technique for large scale phosphoproteomic studies.
RESULTS AND DISCUSSION
How prevalent are gas-phase intra-molecular phosphate transfer reactions?
If intramolecular phosphate transfer reactions occur, then two conditions are likely to be met. First, both species (original and phosphate transfer version) will be present and co-fragmented resulting in the detection of species-specific product ions for both (Figure 1A). Second, when considering only the species formed by phosphate transfer, the number of fragment ion peaks corresponding to incorrect phosphorylation site localization will be greater than what would be expected due to matching ions by random chance.
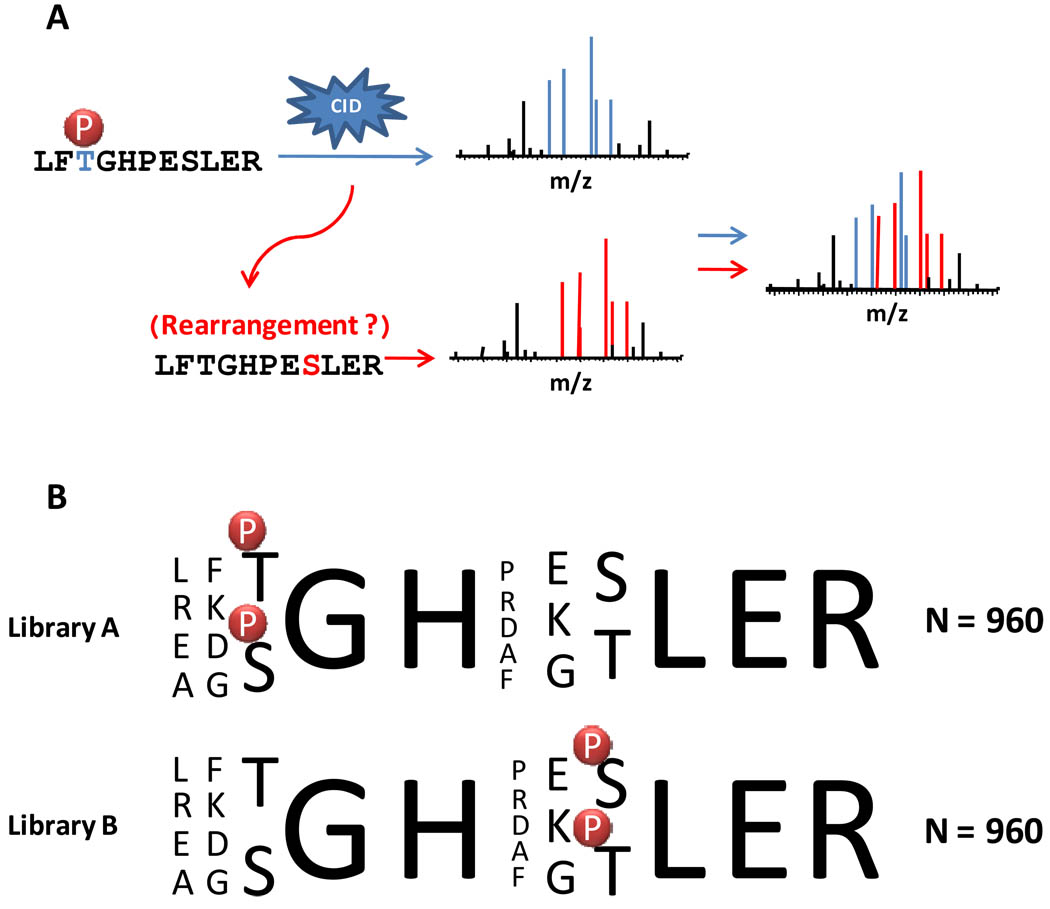
Figure 1.
Schematic for intramolecular phosphate transfer reaction and phosphopeptide libraries used in this study. (A) Intramolecular phosphate transfer has been reported1 as a common occurrence in ion trap MS/MS spectra. When it occurs, the final spectrum may represent a composite of fragment ion peaks from the original and phosphate-transfer species, which could result in mislocalized phosphorylation events. (B) Based on the original sequences used by Palumbo and Reid 1, two libraries were created each with a known site and only one additional potential phosphate-transfer site. Each library was interrogated by LC- MS/MS techniques.
To test this, we began by synthesizing two libraries each containing 960 known-site phosphopeptides. LC-MS/MS analyses of these libraries produced 5417 phosphopeptide identifications at <0.5% FDR (99.5% precision). Among the peptides was the sequence LFtGHPESLER (lower case “t” denotes the phosphorylated residue) which was shown to undergo intramolecular phosphate transfer from the threonine to the serine1. Because only two potential phosphate acceptor sites exist in each peptide, we compared the frequency of site-determining ions for the correct (synthesized) and incorrect (intramolecular phosphate transfer) species of each phosphopeptide at multiple peaks depths versus that which was predicted by the binomial distribution. By examining multiple peak depths (i.e., keeping only N most intense peaks per 100 m/z units, where N=1 to 10) an indication of the abundance of the detected fragment ions can be assessed. Figure 2 illustrates the relationship between charge state, peak depth and site-determining ions predicted by the binomial distribution and that observed for the correct and incorrect sites for the two phosphopeptide libraries. In agreement with Palumbo and Reid1, we did find that for 2+ precursor peptides approximately one additional site-determining ion than would be expected by chance was observed at a peak depth of 10. One ion was predicted to be found by chance and two were observed for the incorrect species. This difference was highly significant (P<10−10 for χ2 test). However, 8 site-determining ions were observed, on average, for the correct site localization. Notably, we did not find evidence of intramolecular phosphate rearrangement for higher charge states (3+ and 4+; Figure 2).
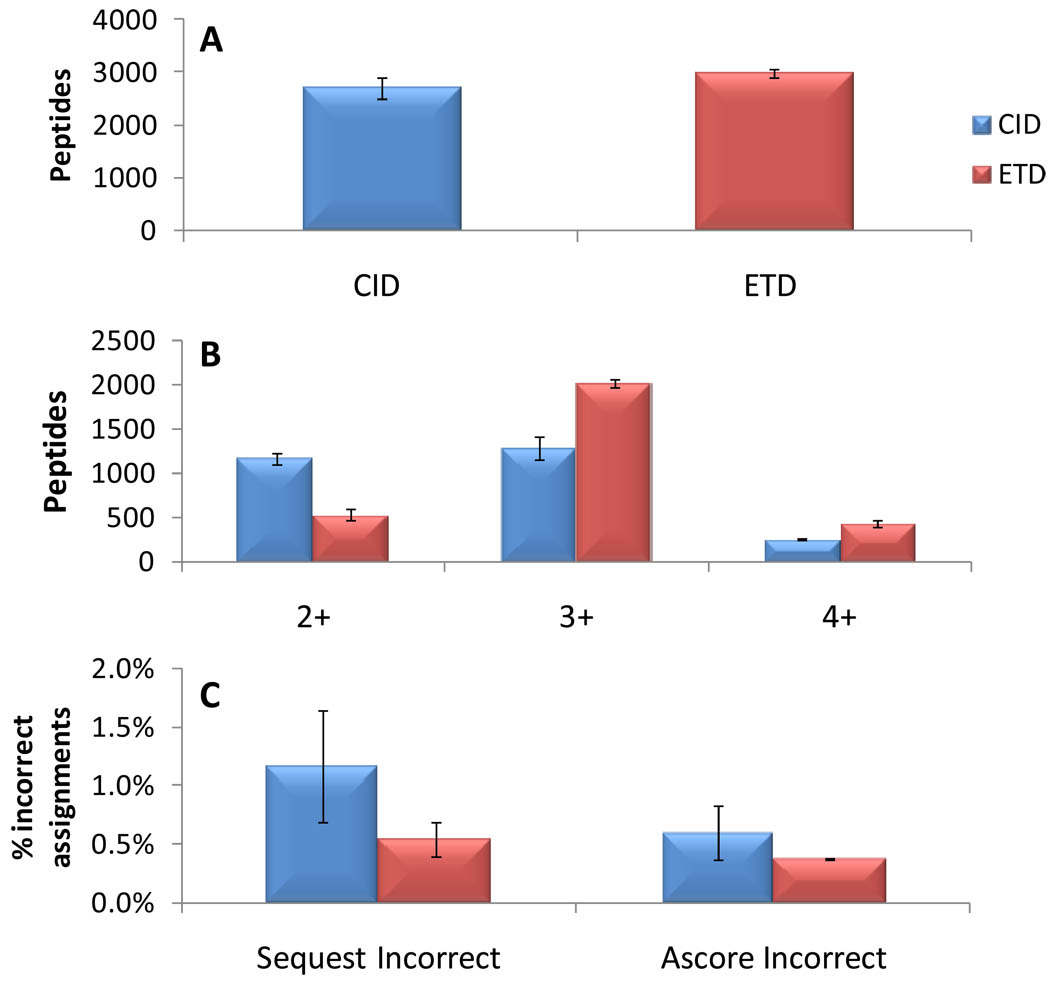
Figure 2.
Evaluation of intramolecular phosphate transfer in ion trap MS/MS spectra. Each library was analyzed in triplicate by LC-MS/MS techniques in an LTQ Orbitrap XL mass spectrometer followed by database searching with Sequest and Ascore for site localization. CID MS/MS spectra (5427) were evaluated individually by keeping only the N most intense peaks (N=1 to 10) in every 100 m/z window. Only site-determining peaks matching to predicted b- or y-type ions from the correct (known) and incorrect (phosphate-transfer species) were counted. Doubly-charged (A), but not triply- or quadruply-charged (B,C) spectra contained evidence of more ions matched than expected by chance. *P<10−10 for both libraries compared to binomial using χ2 test. Data are plotted as the mean from triplicate analysis of the two libraries with error bars at minimum and maximum values.
We next evaluated the ability of any potential intramolecular transfer of phosphate to produce incorrect site localization assignments (Figure 3). From 2506 identified peptides in library A, <1% had both a significant Ascore value (P<0.05) and were assigned to the incorrect (phosphate transfer) position. Nearly identical data were obtained from library B. We conclude that while gas-phase rearrangements of phosphate ions can occur, the extent of the phenomenon is insignificant and does not affect site localization.
Figure 3.
Intramolecular phosphate transfer does not appreciably affect site localization in synthetic peptide libraries. Each library was analyzed in triplicate by LC-MS/MS techniques where back-to-back CID and ETD spectra were collected for each selected phosphopeptide ion. Peptide identifications were filtered to <0.5% FDR. Data are shown as the mean of both libraries (triplicate analysis summed) with error bars at the minimum and maximum values. (A) ETD identified 10.1 % more peptides from these highly basic libraries. (B) CID and ETD are suited to different populations of charge states. (C) The fraction of incorrect site assignments either due to chance or to intramolecular phosphate transfer is very small. ETD is not significantly more reliable than CID at site localization in these known-site synthetic peptide libraries.
Does ETD provide a viable alternative to CID for phosphoproteomics?
The principle challenge facing phosphoproteomics is the vast complexity and large number of phosphopeptides present in a phospho-enriched sample. Peptide sequencing is influenced by the rate at which product ion spectra can be collected and their quality. ETD spectra, while requiring longer scan times on average, can be of very high quality, sometimes with near-complete ion series7. In addition, the ion-ion reactions giving rise to ETD spectra would not be expected to contain molecular rearrangements8. Therefore, ETD has been proposed as a more reliable fragmentation technique1.
We first evaluated ETD fragmentation using our libraries and found no evidence of intramolecular phosphate transfer from the few doubly-charged peptides identified (data not shown). Notably, ETD fragmentation is poor for double-charged precursors where we saw the rearrangement with CID fragmentation. We next compared back-to-back CID/ETD fragmentation of identical precursor ions for each library. Using the mean from both libraries, ETD fragmentation resulted in 2983 identifications compared to 2709 from CID (Figure 3). Higher charge-state species (z = 3 or 4) yielded more matches from ETD spectra. Finally, for greater than 99.5% of all cases with a significant Ascore value (P<0.05), ETD and CID spectra agreed as to the site localization. We conclude that ETD and CID spectra both provide reliable indications of site localization and are not influenced by any phosphate transfer.
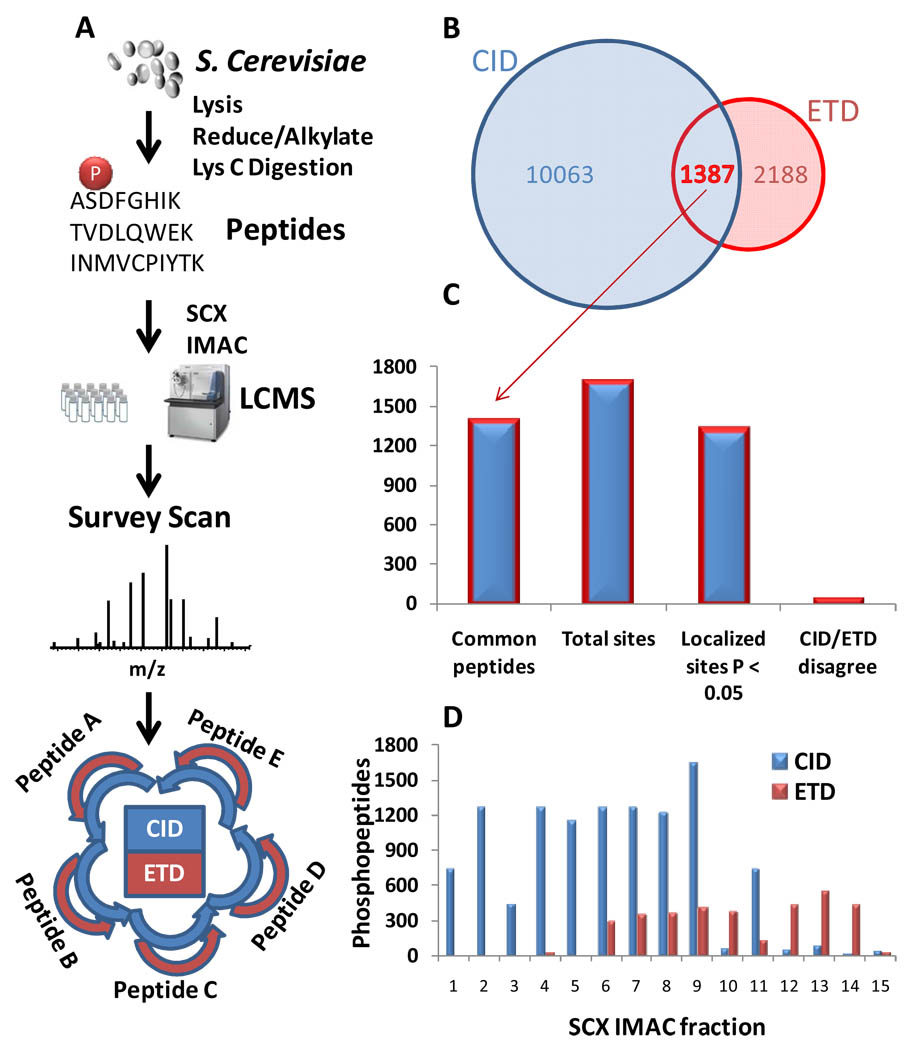
The phosphopeptide libraries used here, while large, may not accurately reflect the true similarities and differences between CID- and ETD-based fragmentation of phosphopeptides typically observed in an actual phosphoproteomic dataset. Therefore, we performed a large-scale yeast phosphoproteome analysis using back-to-back CID and ETD fragmentation of the same precursor ions (Figure 4). Ten mg of yeast was proteolyzed with endoproteinase Lys-C and separated into 15 fractions by strong cation exchange (SCX) chromatography. Each fraction was enriched by immobilized metal ion affinity chromatography (IMAC) and then analyzed by LC-MS/MS techniques. From 15 fractions, CID and ETD identified 11,450 and 3,575 phosphopeptides, respectively (Figure 4B; supplementary table 1). ETD identifications were highly charge-state dependent (data not shown) and were enriched in later SCX fractions (Figure 4D). Importantly, where both CID and ETD localized a site with confidence (P<0.05), the site assignment by the AScore algorithm agreed for 93% of the cases.
Figure 4.
Comparison of CID and ETD fragmentation in a large-scale yeast phosphorylation analysis. (A) Workflow for phosphoproteomic evaluation. Yeast phosphopeptides were enriched via the SCX-IMAC approach10. Back-to-back ETD and CID spectra were acquired for each selected phosphopeptide ion in 15 SCX fractions. (B) CID is considerably more efficient than ETD for large scale phosphoproteomics at a 1% FDR. (C) Among overlapping peptides (1387), the vast majority of localized sites (P<0.05) agreed between CID and ETD (93.2 %). (D) Phosphopeptides identified in each SCX fraction. ETD can be leveraged to identify more phosphopeptides in late eluting SCX fractions.
Conclusions
ETD has been reported to have high value for phosphopeptide sequencing7,9. This study examined both CID and ETD fragmentation of phosphopeptides using back-to-back MS/MS spectra from the same precursor ions. We were encouraged by the results from the library experiments where ETD identified 10% more phosphopeptides than CID. However, in the context of an entire yeast phosphorylation analysis, the success rate was less than 1/3 that of CID. We attribute the better performance in the former to the highly basic nature of the peptide libraries examined. The yeast experiment represented a more realistic challenge. While the numbers of identified peptides by ETD was significantly smaller than CID, the overlap was also small. More than 2,000 phosphopeptides were only detected by ETD and the majority was in the later SCX fractions where the most basic peptides eluted. These findings support the need for selectively collecting ETD spectra when they are most likely to be successful. Indeed, a decision tree8 for collecting CID or ETD spectra has already been reported. Based on these data, CID remains a very effective fragmentation technique for phosphopeptides which could not be directly replaced by ETD.
In agreement with Palumbo and Reid1, we found evidence that some phosphopeptides undergo intramolecular phosphate transfer resulting in the detection of unexpected site-specific fragment ions in ion-trap MS/MS spectra. However, only doubly-charged precursors formed measureable amounts of the transfer species. Furthermore, the transfer species had no effect on site localization precision because only a fraction of ions underwent the reaction. We also found that ETD and CID fragmentation agreed as to site localization for >93% of phosphopeptides from both synthetic libraries and a large-scale yeast experiment. We conclude that intramolecular transfer of phosphate does not affect the reliability of site localization as reported in published large-scale phosphorylation data sets.
METHODS
Peptide library synthesis
Two singly-phosphorylated libraries, each consisting of 960 peptides, were synthesized by Cell Signaling Technology (Danvers, MA). The libraries were based on the following sequences Library A: [LREA][FKDG][TS]GH[PRDAF][EKG][st]LER and Library B: [LREA][FKDG][ts]GH[PRDAF][EKG][ST]LER, where lower case s/t indicate the site of phosphorylation. Contained within these libraries is the peptide used by Palumbo and Reid1 as an example showing evidence for gas-phase rearrangement (LFtGHPESLER). Other entries in the libraries were based on this general peptide scheme.
Yeast whole cell lysate phosphoproteome analysis
Wild type Saccharomyces cerevisiae (BY4742) was used as a model organism for generating a complex mixture of unmodified and modified proteins. Yeast lysate was proteolyzed and fractionated as described previously10. Cells were grown to mid-log phase (OD600 ~ 0.45) in minimal media and collected by centrifugation at 4°C for 10 min, washed with cold water and re-centrifuged. The cell pellet was frozen and disrupted by bead beating in lysis buffer (2 × 90 sec bursts at 4°C). Lysis buffer consisted of 8M urea, 75 mM NaCl, 50mM Tris, pH 8.2, two tablets of protease inhibitor cocktail (complete mini, Roche) per 10 mL of lysis buffer, 50mM NaF, 50mM β-glycerophosphate, 1mM sodium orthovanadate, 10mM sodium pyrophosphate, 1 mM phenylmethanesulphonylfluoride (PMSF).
Cell lysate was assayed for protein concentration by the Bradford method. Ten mg of protein lysate was then reduced for 25 min at 56°C (with 5 mM dithiothreitol), alkylated at room temperature in the dark (in 14 mM iodoacetamide), and quenched with 5 mM dithiothreitol. The mixture was diluted with water to 4M Urea. Proteins were digested overnight at 37°C with 40 µg endoproteinase Lys-C. The sample was centrifuged and the supernatant was collected and lyophilized.
Lyophilized peptides were reconstituted with 1 mL 0.1% trifluoroacetic acid for desalting on a 200 mg tC18 SepPak cartridge (Waters). The cartridge was conditioned with 3 mL acetonitrile, 1 mL (7:3 acetonitrile:0.5% acetic acid), 1 mL (4:6 acetonitrile: 0.5% acetic acid), 3 mL 0.1% trifluoroacetic acid. The peptide mixture was applied to the column. Salts and impurities were washed with 3 mL 0.1% trifluoroacetic acid and 0.25 mL 0.5% acetic acid. Peptides were then eluted into a clean Eppendorf tube with 0.8 mL of 4:6 acetonitrile: 0.5% acetic acid and lyophilized.
Peptides were then fractionated by strong cation exchange (SCX) on a semi-preparative HPLC column (polySULFOETHYL A, 9.4-mm inner diameter 200 mm length, 5-mm particle size, 200 Å pore size (PolyLC)). Mobile phases consisted of (A) 7 mM KH2PO4, pH 2.65, 30% Acetonitrile (vol/vol); (B) 7 mM KH2PO4, 350 mM KCl, pH 2.65, 30% Acetonitrile (vol/vol); (C) 50 mM K2HPO4, 500 mM NaCl, pH 7.5. A gradient from 0% to 35% solvent B over 35 min, followed by a 5 min flush with solvent C, was used to fractionate the peptides. Fifteen fractions spanning the length of the gradient were collected and lyophilized. Peptides were again desalted on Sep Pak cartridges as described above but with 50 mg of tC18 material rather than 200 mg. Volumes of conditioning, loading, washing and elution buffer were adjusted accordingly, and fractions were again lyophilized.
Phosphopeptides were enriched over IMAC resin (Sigma, St. Louis MI) and desalted on C18 Empore 3M (3M, St. Paul, MN) stage tips. Each SCX fraction was resuspended in 120 µL of IMAC binding buffer (25 mM formic acid, 40 % acetonitrile) and incubated in 10 µL of a 50% slurry of IMAC beads for 1 hour at 21°C. The slurry was then poured over the stage tip previously condition with 20 µL of methanol, 20 µL of 50% acetonitrile, 0.5% acetic acid and 2 × 20 µL of 1% formic acid.
Non-phosphopeptides were eluted with 2×50µL IMAC binding buffer. The stage tip was equilibrated with 40 µL of 1% formic acid. Phosphopeptides bound to the IMAC resin were eluted twice with 70 µL of 500 mM K2HPO4, pH 7 and retained on the C18 Empore material. Phosphate salts were removed with 40 µL of 1% formic acid. Enriched phosphopeptides were eluted directly into HPLC vial inserts with 40 µL of 50% acetonitrile 0.5% acetic acid. Each fraction of enriched phosphopeptides was lyophilized and resuspended with 10 µL of 5% formic acid for LC-MS analysis.
Mass Spectrometry
Data were collected using an LTQ Orbitrap XL mass spectrometer with an additional ETD ion source, coupled to an LC system. Column tips were hand-pulled from 150 µm ID fused silica capillary. Microcapillary LC columns were prepared by packing 15 cm of C18 reverse-phase material (Magic C18AQ, Michrom BioResources, Auburn CA). A Famos autosampler (Dionex) with 10 µL loop and 2.4-µL injection needle was plumbed to two Agilent 1100 pumps operating between 80 and 300 µL/min and setup with a flow restrictor to provide an in-column flow rate of 0.5–1 µL/min. The LC program consisted of a 120-min method with 15-min loading time, 83.5-min gradient from 3% to 25% mobile phase B and 100 min of MS data collection. For each scan cycle, one high resolution survey scan acquired in the Orbitrap mass analyzer at 60,000 resolution, 106 automatic gain control (AGC) target and 1,000 ms maximum ion accumulation time, was followed by linear ion trap (LTQ) product ion scans (back-to-back CID and ETD). The 5 most intense precursor ions of appropriate charge state and minimum signal threshold of 3,000 were selected for fragmentation. CID scans were obtained with 120 ms maximum accumulation time, 2.0 Da isolation width and 30 ms activation at 29% normalized collision energy. ETD scans were obtained with 120 ms maximum accumulation time, 2.0 Da isolation width and 60 ms activation. Data were collected with a dynamic exclusion duration of 20 s. Precursor ions with a charge of 1+ or unassigned charge states were excluded.
Database searches and data filtering
All searches were performed using Sequest. For phosphopeptide libraries, an artificial database with 960 entries was assembled and reversed for target/decoy filtering to less than 0.5% false discovery rate (FDR) with multiple parameters including XCorr, dCN, and ppm. Filtered data from the two libraries were used to assess site assignment error rates associated with intramolecular phosphate transfer.
Phosphorylation-site localization
Two methods were used to assign phosphorylation sites. Sequest alone makes an attempt to localize modification sites. In addition, we employ Ascore, an algorithm for probability based phosphosite localization11. Ascore exclusively considers site-determining ions for each STY and assigns modification site(s) based on the difference between site-determining ions for alternate possibilities. If the difference between the number of site determining ions is statistically large, then one can confidently assign the modification site. Notably, not all sites are confidently assigned with p-values < 0.05 (Ascore <13). However, a typical large scale phosphoproteomic study yields thousands of confidently assigned sites12.
Raw data availability
All raw data are available at the Tranche web site (https://trancheproject.org) including .RAW files (6) from triplicate analyses of Library A and Library B using back-to-back ETD/CID and 15 .RAW files for the yeast experiment. Peptide identifications from the libraries and yeast experiment with hyperlinks are available as Supplementary Table 1 and Supplementary Table 2, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded in part by grants from the NIH (HG3456 and GM67945) to S.P.G. We thank Edward Huttlin for statistical advice and critical reading of the manuscript.
References
- 1.Palumbo AM, Reid GE. Evaluation of gas-phase rearrangement and competing fragmentation reactions on protein phosphorylation site assignment using collision induced dissociation-MS/MS and MS3. Anal Chem. 2008;80(24):9735. doi: 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Farha M, et al. Proteomics: from technology developments to biological applications. Anal Chem. 2009;81(12):4585. doi: 10.1021/ac900735j. [DOI] [PubMed] [Google Scholar]
- 3.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 2009;44(6):861. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 4.Edelson-Averbukh M, Shevchenko A, Pipkorn R, Lehmann WD. Gas-phase intramolecular phosphate shift in phosphotyrosine-containing peptide monoanions. Anal Chem. 2009;81(11):4369. doi: 10.1021/ac900244e. [DOI] [PubMed] [Google Scholar]
- 5.Sweet SM, et al. Large scale localization of protein phosphorylation by use of electron capture dissociation mass spectrometry. Mol Cell Proteomics. 2009;8(5):904. doi: 10.1074/mcp.M800451-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers LD, Foster LJ. Phosphoproteomics-finally fulfilling the promise? Mol Biosyst. 2009;5(10):1122. doi: 10.1039/b905580k. [DOI] [PubMed] [Google Scholar]
- 7.Chi A, et al. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104(7):2193. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5(11):959. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coon JJ, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci U S A. 2005;102(27):9463. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beausoleil SA, et al. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 12.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.