Summary
Retinoic acid (RA) induces differentiation of neuroblastoma cells in vitro and is used with variable success to treat aggressive forms of this disease. This variability in clinical response to RA is enigmatic, as no mutations in components of the RA signaling cascade have been found. Using a large-scale RNAi genetic screen, we identify crosstalk between the tumor suppressor NF1 and retinoic acid induced differentiation in neuroblastoma. Loss of NF1 activates RAS-MEK signaling, which in turn represses ZNF423, a critical transcriptional co-activator of the retinoic acid receptors. Neuroblastomas with low levels of both NF1 and ZNF423 have extremely poor outcome. We find NF1 mutations in neuroblastoma cell lines and in primary tumors. Inhibition of MEK signaling downstream of NF1 restores responsiveness to RA, suggesting a therapeutic strategy to overcome RA resistance in NF1 deficient neuroblastomas.
Keywords: Neurofibromatosis, differentiation, retinoic acid, neuroblastoma, RAS
Introduction
Neuroblastoma is a malignancy of early childhood that arises from the developing autonomic nervous system (Maris et al., 2007). It is the most frequently diagnosed cancer in the first year of life and shows a remarkably heterogeneous clinical behavior. Some tumors regress spontaneously, whereas others progress to highly aggressive metastatic disease with a poor overall survival rate. Amplification of the MYCN oncogene, and/or loss of the chromosomal regions 1p36 and 11q23 correlate with poor disease outcome (Attiyeh et al., 2005; Caron et al., 1996; Seeger et al., 1985). Recurrent gene mutations are relatively rare in neuroblastoma. Somatic and germ line activating mutations in the ALK kinase have been identified in 7% of neuroblastomas, providing the basis for a promising molecular targeted therapy in this subgroup (reviewed in Mosse et al., 2009). PHOX2B and PTPN11 mutations were found in 2% and 3% of neuroblastomas, respectively (Bentires-Alj et al., 2004; van Limpt et al., 2004).
Neuroblastomas frequently transdifferentiate into more benign ganglioneuroblastomas and ganglioneuromas. Progression in the opposite direction is also observed in patients. The vitamin A metabolite retinoic acid (RA) is an important morphogen for the developing nervous system in vivo and capable of differentiating neuronal cells in vitro (Duester, 2008). Various neuroblastoma cell lines cease proliferation, differentiate into neuronal-like cells or undergo apoptosis upon exposure to RA (Sidell et al., 1983). These observations established the basis for the clinical application of RA in the treatment of neuroblastoma. RA is one of the few targeted therapeutics currently used in the clinic for aggressive neuroblastoma, but the benefit is limited. A phase III randomized trial showed that treatment with 13-cis-RA given after completion of intensive chemo-radiotherapy yields a slight but significant improvement in event-free survival in high-risk neuroblastoma (Matthay et al., 2009; Matthay et al., 1999).
The physiological functions of retinoids are primarily exerted through the regulation of specific target genes mediated by retinoid acid receptors (RARs). RARs are nuclear hormone receptors that function as ligand dependent transcription factors (reviewed in Rochette-Egly and Germain, 2009). Their activity requires hetero-dimerization with the retinoid X receptors (RXR) that can also associate with several other nuclear hormone receptors. RAR/RXR heterodimers constitutively bind to retinoic acid response elements (RAREs) in the promoter regions of target genes and actively repress transcription in the absence of ligand. This process involves recruitment of the corepressors NCoR and SMRT and histone deacetylases (HDAC). RA binds to RAR and triggers conformational changes that release the corepressors and in turn promote the assembly of coactivator complexes. Subsequently, transcription of target genes is initiated. Many of the coactivators, including CBP/p300, PCAF and SRC1-3 (NCOA1-3), possess histone acetylase (HAT) activity that promotes transactivation of RAR/RXR. In contrast, ligand-dependent corepressors such as LCoR and PRAME recruit HDACs or PcG proteins to ligand-bound RAR/RXR complexes to repress their activities (Epping et al., 2005; Fernandes et al., 2003). Therefore, co-activators/repressors play crucial roles for the context dependent action of RA and may be important determinants and biomarkers for RA based therapies in the clinic.
Recently, we have identified ZNF423 as a critical co-factor required for RAR/RXR function using a large-scale RNA interference (RNAi) based screen in F9 mouse teratocarcinoma cells, a widely used model system to study RA signaling (Huang et al., 2009). We found that ZNF423 is also crucial for RA mediated growth arrest and differentiation in human neuroblastoma cell lines and its reduced expression was a powerful marker of poor prognosis in neuroblastoma patients, independent of MYCN amplification. However, we have not observed loss of heterozygosity at the ZNF423 locus in primary neuroblastomas, nor could we restore ZNF423 levels in neuroblastoma cell lines with low ZNF423 by DNA demethylating agents. These results indicate that ZNF423 expression is determined by transcriptional regulation, rather than by epigenetic silencing or genetic loss. The signaling pathways that regulate ZNF423 expression could therefore be the critical components governing the RA response.
As pointed out above, the overall response rate to RA in neuroblastoma patients is low, suggesting that only a subgroup of patients benefits from the treatment. Currently, no predictive markers of RA responsiveness are available for clinical use. No alterations in any of the core components of the RA signaling pathway have been described in neuroblastoma tumors, posing a conundrum how to explain the diversity in RA sensitivity. To gain more mechanistic insights in the factors that control RA responsiveness in neuroblastoma, we performed an unbiased large-scale RNAi screen in neuroblastoma cells. We identify here the tumor suppressor NF1 as a major determinant of RA sensitivity in neuroblastoma cells and find genetic alterations in NF1 in primary tumors and neuroblastoma cell lines. We provide proof of concept experiments for a therapeutic strategy to overcome RA resistance in NF1 deficient neuroblastomas by the co-application of RA with MEK inhibitors.
Results
NF1 suppression confers resistance to RA in neuroblastoma
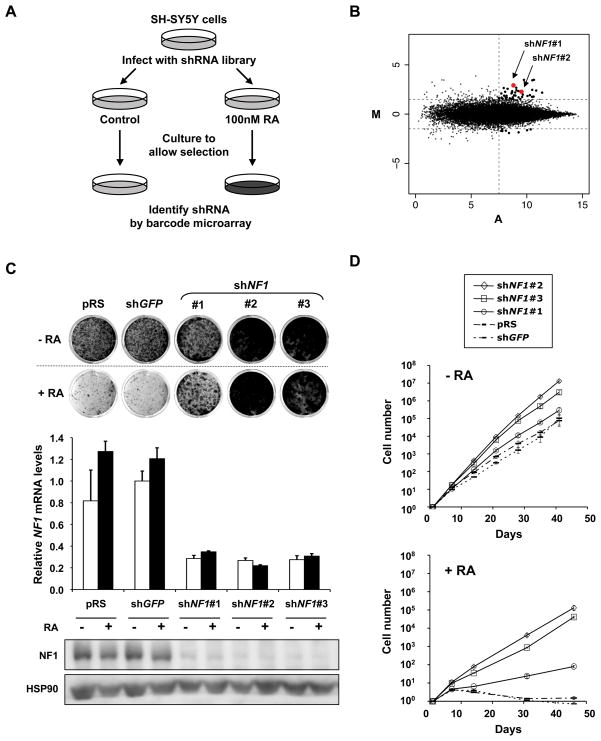
In response to RA the human neuroblastoma cell line SH-SY5Y ceases proliferation and differentiates into neuronal-like cells. To identify novel determinants of RA resistance in neuroblastoma, we performed a large-scale RNAi-based loss-of-function genetic screen using a collection of 24,000 short hairpin (shRNA) vectors targeting 8,000 human genes (Berns et al., 2004). We used a barcoding technology to identify genes whose suppression causes resistance to RA (Brummelkamp et al., 2006). The entire shRNA library was introduced into SH-SY5Y cells by retroviral infection and cells were plated at low density with or without RA (Figure 1A). After four weeks of RA selection, genomic DNA was isolated from both cultures. The stably integrated shRNA cassettes (19-mer bar code sequences) were recovered by PCR. The relative abundance of individual shRNA vectors was quantified by hybridization of the PCR products to microarrays harboring all 24,000 barcode sequences. The barcode screen was carried out in triplicate and the results are shown in Figure 1B. Each dot in the M/A-plot represents one individual shRNA vector in the library. M- and A-values reflect relative enrichment and hybridization signal intensity. Reproducible outliers are generally located in the right upper corner. Low-intensity spots are prone to technical artifacts and thus unreliable. Therefore we restricted our candidate selection by applying M/A cut-off values as indicated in Figure 1B. To rule out ‘off-target’ effects, we only consider a gene identified from the screen as a genuine hit, if at least two independent shRNAs suppress the expression of the target and also confer RA resistance. Within the list of 44 candidate hits fulfilling the cut-off criteria, the only gene that was represented by two shRNAs was the tumor suppressor gene NF1 (neurofibromin 1) and was studied further.
Figure 1. A genome-wide RNAi screen identifies NF1 as a critical determinant of RA sensitivity in neuroblastoma cells.
(A) Schematic outline of the RA resistance barcode screen performed in SH-SY5Y cells. Human shRNA library polyclonal virus was produced to infect SH-SY5Y cells, which were then left untreated (control) or treated with 100 nM all-trans retinoic acid (RA). After 4 weeks of selection, shRNA inserts from both populations were recovered, labeled and hybridized to DNA.
(B) Analysis of the relative abundance of the recovered shRNA cassettes from RA barcode experiment. Averaged data from three independent experiments were normalized and 2log transformed. Among the 44 top shRNA candidates (M>1.5 and A>7.5), two independent shNF1 vectors (in red) were identified.
(C) Validation of independent shRNAs targeting NF1. The functional phenotypes of non-overlapping shNF1 vectors are indicated by the colony formation assay in 100 nM RA. The pRS vector and shGFP were used as controls. The cells were fixed, stained and photographed after 14 days (untreated) or 21 days (RA treatment). The knockdown ability of each of the shRNAs was measured by examining the NF1 mRNA levels by qRT-PCR and the NF1 protein levels by western blotting. Error bars denote standard deviation (SD). See also supplemental figure S1.
(D) RA resistance by NF1 RNAi was dependent on RA signaling. Proliferation curves according to the 3T3 protocol of SH-SY5Y cells expressing shNF1 vectors, shGFP or pRS, in the absence and presence of RA (100 nM). Error bars denote SD.
To validate the results of the screen, we individually introduced the two NF1 shRNAs (#1 and #2) from the library and one newly generated (#3) into SH-SY5Y cells. Vectors targeting GFP (shGFP) or harboring only the empty vector (pRS) served as controls throughout the study. All three NF1 knockdown vectors conferred resistance to RA in long-term colony formation assays and efficiently suppressed NF1 mRNA and protein expression (Figure 1C). We also noted a growth advantage of NF1 knockdown cells in the absence of exogenously added RA. This may result from resistance to low levels of RA present in the culture medium or from a RA-independent proliferation advantage. Therefore, we performed a long-term proliferation assay of SH-SY5Y cells expressing shRNAs against NF1, GFP or control vector (pRS) using the 3T3 protocol (Figure 1D). A slightly increased growth rate of NF1 knockdown cells was indeed detected in the absence of exogenous RA. However, when exposed to RA, shNF1 expressing cells continued to proliferate, while control cells completely ceased proliferation after a latency of about 10 days and the total cell number even declined, indicating that prolonged RA treatment caused cell death of the control cells. Thus, NF1 knockdown cells are clearly rescued from RA mediated differentiation and growth arrest. Identical results were obtained in other neuroblastoma cell lines having different levels of MYCN amplification (SK-N-SH, SJ-NB6, Kelly, HT-230 and LAN-5, Figure S1 and data not shown), indicating NF1 knockdown can cause resistance to RA independent of MYCN amplification status in a range of neuroblastomas.
RAS signaling linked to RA response
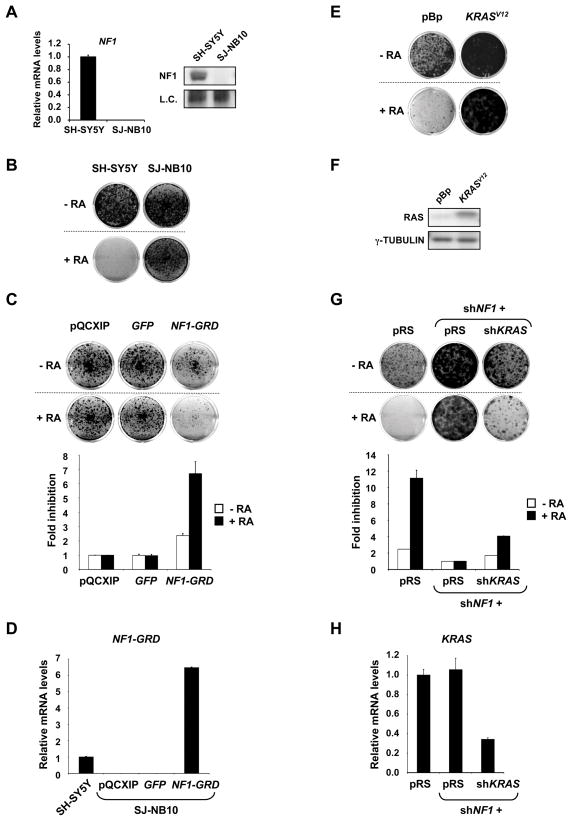
It has been found that the neuroblastoma cell line SJ-NB-10 (NB90-9) harbors a homozygous deletion of some exons of the NF1 gene, suggesting that these cells could be insensitive to RA (The et al., 1993). Figure 2 shows that SJ-NB-10 cells indeed lack NF1 mRNA and protein (Figure 2A) and fail to respond to RA (Figure 2B). NF1 is a large protein that has GTPase activating activity (GAP) and functions as a negative regulator of RAS proteins by promoting their conversion from the active GTP bound to the inactive GDP bound form (Cichowski and Jacks, 2001; Martin et al., 1990). Loss of NF1 expression results in hyperactive RAS signaling and therefore we reasoned that hyperactive RAS signaling could cause RA resistance in this NF1 deficient neuroblastoma cell line.
Figure 2. RAS signaling linked to RA response in neuroblastoma cells.
(A) NF1 deficiency in SJ-NB10 (NB90-9) cells. NF1 mRNA and protein were undetectable by qRT-PCR and western blotting. SH-SY5Y cells were included as a positive control. Error bars denote SD.
(B) SJ-NB10 cells are insensitive to RA. SJ-NB10 and the control SH-SY5Y Cells were grown in the absence or presence of RA (100 nM). Treated and untreated dishes of each cell line were fixed, stained and photographed at the same time. SH-SY5Y cells were harvested after 14 days and SJ-NB10 cells after 28 days.
(C) Enforced-expression of NF1-GRD in SJ-NB10 cells leads to growth inhibition in the absence of exogenous RA and enhanced responses to RA. Cells expressing pQCXIP-vector, -GFP or -NF1-GRD were grown in the absence or presence of RA (100 nM) for 28 days, after which cells were fixed, stained and photographed. Relative cell growth was then measured by crystal violet quantification. Fold inhibition values were normalized to cells infected with pQCXIP vector. Error bars denote SD.
(D) Relative NF1-GRD mRNA levels in SH-SY5Y cells and SJ-NB10 cells expressing pQCXIP-vector, -GFP or -NF1-GRD. Error bars denote SD.
(E) Ectopic-expression of KRASV12 in SH-SY5Y cells leads to RA resistance. Cells expressing pBABE-vector or -KRASV12 were grown in the absence or presence of RA (100 nM). The cells were fixed, stained and photographed after 14 days (untreated) or 21 days (RA treatment).
(F) The RAS protein expression levels in SH-SY5Y cells expressing pBABE-vector or -KRASV12.
(G) Endogenous KRAS is required for the RA resistance driven by NF1 knockdown. SH-SY5Y cells expressing pRS, shNF1 plus pRS or shNF1 plus shKRAS were grown in the absence or presence of RA (1 μM). The cells were fixed, stained and photographed after 14 days (untreated) or 21 days (RA treatment). Relative cell growth was then measured by crystal violet quantification. Fold inhibition values were normalized to cells infected with shNF1 plus pRS, in the absence or presence of RA. Error bars denote SD.
(H) The KRAS mRNA levels in SH-SY5Y cells expressing pRS, shNF1 plus pRS or shNF1 plus shKRAS. Error bars denote SD.
We stably introduced the GAP-related domain of NF1 (NF1-GRD) into the NF1 deficient SJ-NB-10 cells. Expression of the NF1-GRD is widely used to restore NF1 dependent GAP activity and thus inhibition of RAS activity in cells lacking NF1 (Martin et al., 1990). Expression of the NF1-GRD was determined by qRT-PCR using NF1-GRD specific primers (Figure 2D). SJ-NB-10 cells expressing NF1-GRD were exquisitely sensitive to RA and cell growth was impaired by almost 7-fold relative to the control cells. In the absence of RA, however, the inhibition was only 2.5-fold, underscoring the RA specific sensitization effect (Figure 2C).
Our results suggest that hyperactive RAS signaling is responsible for the RA resistance associated with a naturally occurring homozygous NF1 deletion in a human neuroblastoma cell line. To substantiate the role of RAS signaling further, we tested if an activated RAS (KRASV12, Figure 2F) oncogene can cause RA resistance in sensitive SH-SY5Y cells. Similar to NF1 knockdown, cells expressing the KRAS oncogene potently bypassed RA induced growth arrest (Figure 2E).
To investigate the role of endogenous KRAS in the RA resistance caused by NF1 loss, we performed an epistasis analysis. SH-SY5Y cells were simultaneously infected with shRNAs against KRAS and NF1. Suppression of KRAS (Figure 2H) in combination with NF1 impaired the occurrence of RA resistant cells driven by NF1 knockdown, whereas cells co-infected with shNF1 and pRS were highly resistant to RA (Figure 2G). This indicates that KRAS is critically required to mediate the effects of NF1 knockdown on the RA response.
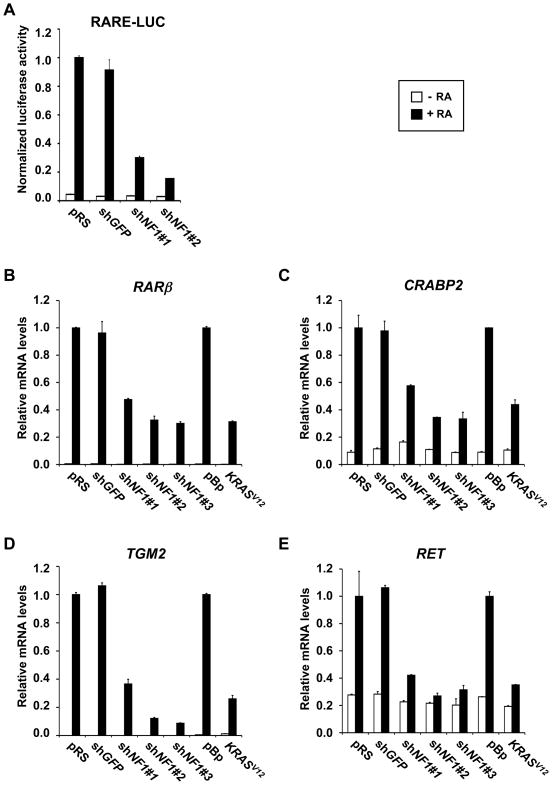
NF1 loss inhibits transcriptional response to RA
RA exerts its biological effects largely through gene regulation. We therefore addressed whether suppression of NF1 interferes with the transcriptional response to RA. ShRNAs targeting NF1 or GFP were co-transfected with a reporter gene containing a consensus Retinoic Acid Response Element (RARE) linked to luciferase (RARE-Luc). Both shRNAs against NF1 inhibited the reporter gene activation by RA (Figure 3A). Moreover, NF1 knockdown also suppressed expression of bona fide endogenous RA target genes, RARβ, CRABP2 and TGM2 (Figure 3B–D). Similarly, exogenous expression of KRASV12 also repressed RA dependent target gene activation. Similar results were obtained in other neuroblastoma cell lines (data not shown). These results further suggest that hyperactive RAS signaling downstream of NF1 blocks the transcriptional response to RA in neuroblastoma cells. Transcriptional induction of the RET tyrosine kinase, a gene associated with neuroblastoma differentiation (Bunone et al., 1995), was consistently blocked in shNF1 and KRASV12 expressing cells, indicating that NF1 suppression not only overrides proliferation arrest, but also differentiation induced by RA (Figure 3E).
Figure 3. NF1 loss inhibits transcriptional response to RA.
(A) NF1 RNAi inhibits activation of a RARE-Luciferase (RARE-Luc) reporter gene by endogenous RAR/RXR in response to 24 hours of 100 nM RA stimulation in SH-SY5Y cells. Normalized luciferase activities shown are ratios between luciferase values and Renilla internal control values. Error bars denote SD.
(B to E) NF1 knockdown or ectopic expression of KRASV12 suppresses transcriptional activation of endogenous RA target genes in response to RA. mRNA expression analysis of RA target genes RARβ (B), CRABP2 (C), TGM2 (D) and RET (E) in SH-SY5Y cells expressing controls, shRNAs targeting NF1 or pBABE- KRASV12 after 100 nM RA stimulation for 7 days. Error bars denote SD.
NF1 loss downregulates the RAR/RXR co-activator ZNF423
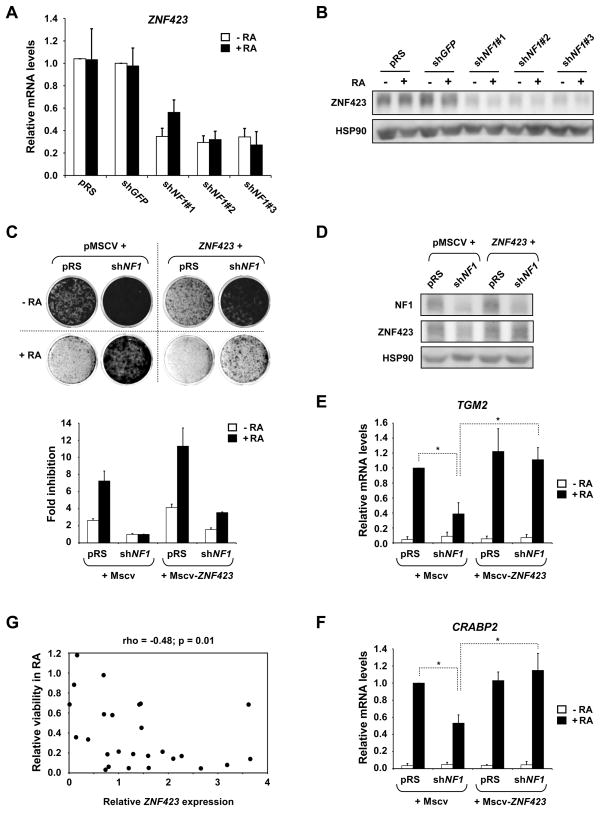
Since co-factors are critical determinants of RAR/RXR transcriptional activity, we asked whether modulation of a co-factor(s) might play a role in the RA resistance caused by NF1 knockdown. Global gene expression analysis in NF1 knockdown cells identified 196 and 167 genes that were greater than 2-fold up- or down-regulated (Table S1). Among these genes, we found the RAR/RXR co-activator ZNF423 to be significantly repressed in shNF1 expressing cells. We have previously shown that ZNF423 physically interacts with RAR/RXR heterodimers and binds to RA responsive promoters (Huang et al., 2009). Suppression of ZNF423 caused RA resistance in multiple neuroblastoma cell lines and reconstitution of ZNF423 levels in ZNF423low neuroblastoma cell lines restored their response to RA. Thus, repression of ZNF423 by loss of NF1 could play an important role in mediating RA resistance. We confirmed down-regulation of ZNF423 mRNA and protein levels in shNF1 expressing cells by qRT-PCR and Western blotting (Figure 4A, B). A reduction in ZNF423 expression by knockdown of NF1 was also seen in additional neuroblastoma cell lines (Figure S2A–D).
Figure 4. NF1 loss suppresses RA response by downregulating the RAR/RXR coactivator ZNF423.
(A and B) NF1 RNAi leads to down-regulation of ZNF423 mRNA and protein levels. SH-SY5Y cells expressing controls and shRNAs targeting NF1 were grown in the absence or presence of RA (100 nM) for 7 days. A) ZNF423 mRNA levels are suppressed in NF1 knockdown cells. (B) ZNF423 protein levels are also reduced n NF1 knockdown cells. See also supplemental Figure S2 and Table S1.
(C) Re-expression of ZNF423 reverses the RA resistance driven by NF1 knockdown. SH-SY5Y cells expressing MSCV control or MSCV-ZNF423 were retrovirally infected with viruses containing pRS or shNF1, and were grown in the absence or presence of 1 μM RA. Cells were then fixed, stained and photographed after 12 days (untreated) or 14 days (RA treatment). Relative cell growth was then measured by crystal violet quantification. Fold inhibition values were normalized to cells expressing both MSCV control and shNF1, in the absence or presence of RA. Error bars denote SD.
(D) The NF1 and ZNF423 protein levels in SH-SY5Y cells described in Figure 4C. Cells were grown in the presence of 1 μM RA for 14 days.
(E and F) Re-expression of ZNF423 restores activation of the RA target genes TGM2 and CRABP2 in NF1 knockdown cells. Error bars denote SD.*p<0.01 (t-test for comparison of three independent biological experiments)
(G) ZNF423 expression and RA responsiveness in neuroblastoma cell lines. ZNF423 mRNA levels from a panel of 26 different neuroblastoma cell lines were determined by qRT-PCR. The relative viability in 100nM RA was determined by the ratio of cell growth of treated versus untreated cultures measured by crystal violet quantification after long-term colony formation assays. A significant association between ZNF423 expression and RA sensitivity was determined by Spearman rank correlation analysis. See also supplemental figure S2.
To address whether suppression of ZNF423 is causal for the RA resistance of NF1 knockdown cells, we re-expressed ZNF423 at physiological levels in shNF1 expressing SH-SY5Y cells (Figure 4C, D). Reconstitution of ZNF423 was capable of partially restoring responsiveness to RA in otherwise insensitive NF1 knockdown cells (Figure 4C). In line with our previous study, ZNF423 expression caused hypersensitivity to RA along with a moderate inhibition of proliferation (Fig. 4C). ZNF423 reconstitution in NF1 knockdown cells also restored expression of bona fide RA target genes, such as TGM2 and CRABP2 (Figure 4E, F). Our results demonstrate that ZNF423 is a major, but possibly not the only, component downstream of NF1 in the control of RA sensitivity in neuroblastoma cells.
ZNF423 predicts RA responsiveness
As NF1 loss causes resistance to RA, at least in part, by suppression of ZNF423, our data raise the possibility that ZNF423 levels might predict sensitivity of neuroblastomas to RA. To begin to address this, we explored the predictive value of ZNF423 expression for RA responsiveness in a panel of 26 different neuroblastoma cell lines. We investigated the sensitivity to RA (100nM) by long-term colony formation assays. The response to RA was determined by the relative growth of treated versus untreated cultures. ZNF423 mRNA levels were measured by qRT-PCR in the absence of RA (Figure 4G and S2E, F). The response to RA ranged from highly sensitive to fully resistant. High expression of ZNF423 was significantly associated with sensitivity to RA (Spearman correlation p=0.01). The range of ZNF423 expression within the cell line panel was comparable to the regulation observed by knockdown of NF1 in SH-SY5Y and other neuroblastoma cell lines (Figure 4A, S2E). This underscores the functional relevance of ZNF423 suppression by knockdown of NF1.
NF1 expression predicts outcome of neuroblastoma
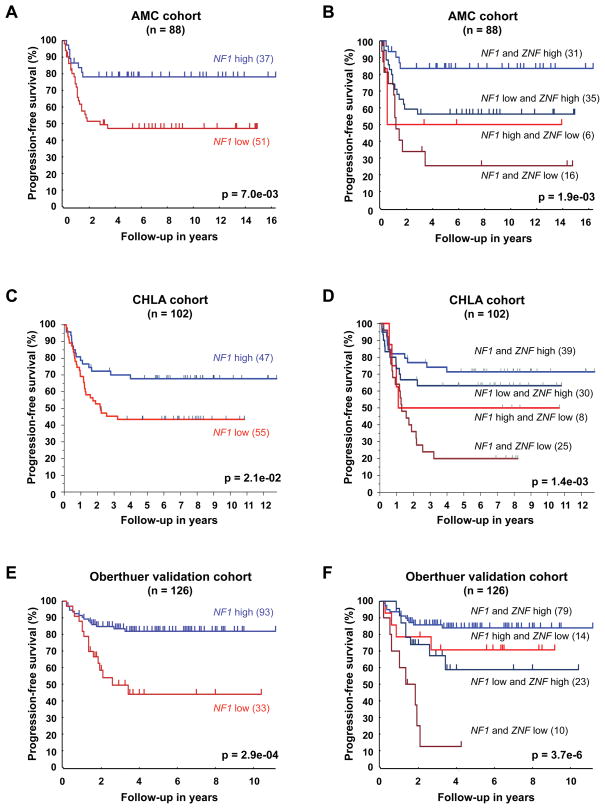
We have shown previously that ZNF423 expression is prognostic for event-free survival of neuroblastoma patients (Huang et al., 2009). Given the functional connection between NF1 and ZNF423, we investigated a possible prognostic significance of NF1 expression in neuroblastoma by microarray analysis in three independent patient cohorts. The first cohort consists of 88 neuroblastoma patients from the Academic Medical Center (AMC) in Amsterdam, Netherlands (Huang et al., 2009). We used this cohort as a “training set” to generate an optimal cut-off value of NF1 expression by a Kaplan scanning approach (see experimental procedures for details). Patients were then classified into NF1 low or high based on their NF1 levels in the primary tumors using this cut-off value and progression-free survival was assessed by Kaplan-Meier analysis. The NF1high subgroup had a significantly better outcome than the NF1low subgroup (Figure 5A). Similar results were obtained with median expression (p=0.028) or average expression (p=0.017) as cut-off values for NF1 expression. Importantly, NF1 retains prognostic significance in combination with any of the four clinically used prognostic markers: MYCN amplification, deletion of chromosomal band 1p36 (LOH 1p36), age (<18 months) and stage in a multivariate analysis (Table S2).
Figure 5. NF1 expression predicts outcome of neuroblastoma and the combined expression status of NF1 and ZNF423 is a powerful prognostic marker.
(A) Kaplan-Meier analysis of the AMC cohort (n=88) documenting increased progression-free survival (PFS) of neuroblastoma patients with tumors that have high NF1 expression (NF1 high) versus patients with tumors that have low NF1 expression (NF1 low), using the NF1 cut-off value determined as described in the text. See also supplemental Table S2.
(B) Kaplan-Meier analysis of PFS for the AMC cohort classified by the combined expression status of NF1 and ZNF423. The NF1 cut-off value was the same as above and ZNF423 cut-off value was as described previously (Huang et al., 2009).
(C) Kaplan-Meier analysis of PFS for a second independent set of 102 patients diagnosed with metastatic neuroblastomas lacking MYCN amplification from CHLA (Asgharzadeh et al., 2006). These patients were classified using the same NF1 cut-off value determined from the AMC cohort.
(D) Kaplan-Meier analysis of PFS for the CHLA cohort classified by the combined expression status of NF1 and ZNF423. The cut-off values for NF1 and ZNF423 were the same as above.
(E and F) Kaplan-Meier analysis of PFS for the Oberthuer cohort (validation set, n=126) classified by the expression status of NF1 (E) and the combined expression status of NF1 and ZNF423 (F). The cut-off values for NF1 and ZNF423 were determined in the Oberthuer training set (n=125).
We then validated the prognostic value of NF1 expression in an independent cohort of 102 neuroblastoma patients with metastatic tumors lacking MYCN amplification from the Childrens Hospital Los Angeles (CHLA), for which progression-free survival data are known (Asgharzadeh et al., 2006). Since the same microarray platform was used, the NF1 expression from both datasets was normalized and the cut-off value used in AMC series was applied to the validation cohort. Again, high level of NF1 expression correlated with longer progression-free survival and in return low NF1 expression was associated with poor outcome (Figure 5C). When we combined the expression status of NF1 and ZNF423, patients classified as NF1low/ZNF423low had the poorest disease outcome, whereas the NF1high/ZNF423high groups had the best outcome (Figure 5B, D). High expression of either NF1 or ZNF423 alone was correlated with an intermediate progression free survival. As the CHLA validation cohort was restricted to a subset of high-risk tumors without MYCN amplification, we analyzed a third independent data set that covers all stages including MYCN amplified tumors (Oberthuer et al., 2006). These gene expression data were generated on a different microarray platform. Therefore, we randomly divided the entire cohort into a training (n=125) and validation set (n=126) and cut-off values for NF1 and ZNF423 were determined as described above in the training set and then applied to the validation set. Again, low expression of NF1 and ZNF423 correlated with poor outcome (Figure 5E, and data not shown). When we combined NF1 and ZNF423 expression, patients classified as NF1low/ZNF423low had the poorest disease outcome, whereas the NF1high/ZNF423high group had the best outcome (Figure 5F). Prognostic significance of NF1 and the combined NF1/ZNF423 expression status was independent of MYCN amplification in a multivariate analysis (Table S2). Differences in progression-free survival were consistent and statistically significant in all three patient cohorts. Moreover, the ZNF423 low group was significantly enriched for NF1 low tumors (Wilcoxon test, p<0.001 in all three cohorts). This is in line with the functional interaction between NF1 and ZNF423 that we observed in cell lines. In summary, the combined expression status of the two functionally connected genes, NF1 and ZNF423, is a powerful prognostic marker in neuroblastoma.
NF1 mutations in neuroblastoma
Most of the patients with neurofibromatosis type 1 harbor mutations of the NF1 gene, ranging from single point mutations to larger deletions (Messiaen et al., 2000; Wimmer et al., 2006; Zatkova et al., 2004). NF1 mutations and deletions were also found in 23% of glioblastomas, a highly aggressive adulthood brain tumor (TCGA_Network, 2008). A role for NF1 in the pathogenesis of neuroblastoma has been discussed based on cell line observations, but never studied systematically in a large cohort of primary neuroblastomas (The et al., 1993).
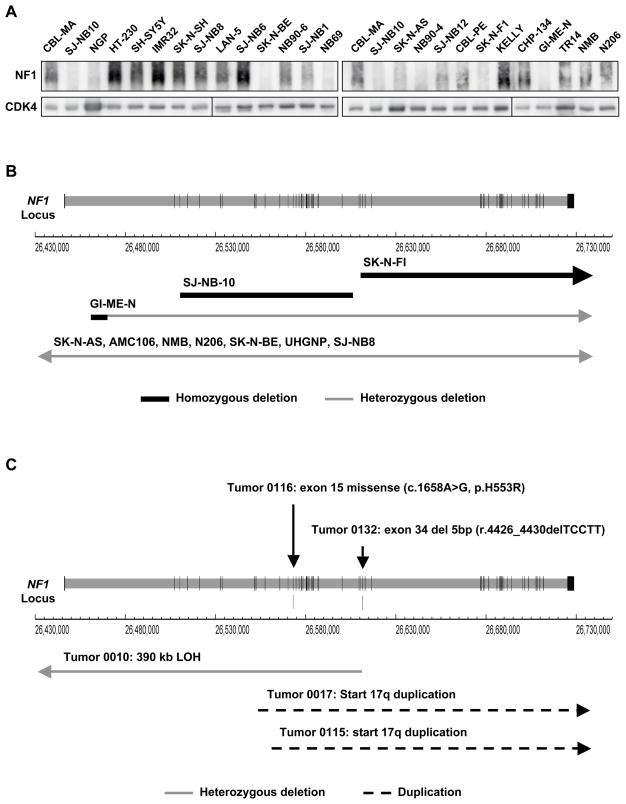
In a panel of 25 neuroblastoma cell lines and we found loss of NF1 protein expression in approximately one third (Figure 6A). Absence of NF1 protein was significantly associated with resistance to RA and also lower expression of ZNF423 (Figure S3). An overview of genomic defects of NF1 was obtained by a comprehensive SNP analysis (Illumina 657K platform) in 20 neuroblastoma cell lines (Figure 6B). We detected homozygous micro-deletions in SJ-NB10 (NB90-9) and SK-N-FI cells, deleting part of NF1 or NF1 and a neighboring gene (Figure S4A, B). The cell line GI-ME-N showed a 3.6 Mb hemizygous deletion encompassing NF1, while the second NF1 allele had a micro-deletion. AMC106 and SK-N-AS cells showed hemizygous deletions of the NF1 region of 3.7 and 5.4 Mb. Five more cell lines (NMB, N206, SK-N-BE, UHGNP and SJ-NB8) showed loss of heterozygosity encompassing the NF1 region. Only 10/20 cell lines had two apparently normal NF1 alleles.
Figure 6. NF1 mutations in neuroblastoma.
(A) NF1 protein levels in a panel of 25 neuroblastoma cell lines as determined by Western blotting. See also supplemental figure S3.
(B) Schematic representation of genomic aberrations in the NF1 gene in neuroblastoma cell lines. Vertical lines in the NF1 locus correspond to exons and horizontal arrows depict regions of hemizygous or homozygous deletions. See also supplemental figure S4.
(C) Schematic representation of genomic aberrations in the NF1 gene in primary neuroblastomas. Vertical arrows indicate the position of the identified NF1 mutations. Horizontal arrows depict regions of hemizygous or homozygous deletion and dashed lines indicate the regions affected by the duplication of 17q. See also supplemental Figure S4.
These results urged us to screen for mutations of NF1 in the AMC patient cohort that covers a representative spectrum of neuroblastoma and ganglioneuroma cases including different disease stages, ages and MYCN amplifications. Sequencing of the full coding region in cDNA identified two mutations, which were both confirmed by sequencing of the genomic exons (Figure 6C). One neuroblastoma carried a 5 base pair deletion in exon 34 inducing a frame shift. A ganglioneuroma was detected with a missense mutation in exon 15, which is also known from neurofibromatosis type 1 patients (Griffiths et al., 2007). Re-analysis of the clinical records revealed that the patient in our series was indeed diagnosed with neurofibromatosis type 1. Sequencing of the exonic DNA of this tumor revealed that besides the mutated allele, also a normal NF1 allele was present. However, at the cDNA level, only expression of the mutant allele was observed, suggesting epigenetic silencing of the normal NF1 allele (Figure S4C). Finally, we performed a SNP analysis of 50 neuroblastoma patients from the AMC series. This identified one tumor with a 390 kb heterozygous deletion covering NF1, as well as two tumors with translocations involving chromosome 17 with breakpoints in NF1, resulting in gain of distal chromosome 17q starting in NF1 (Figure 6C). We conclude that NF1 aberrations are detected in primary neuroblastomas, but at lower frequency than in neuroblastoma cell lines.
Restoration of RA responsiveness
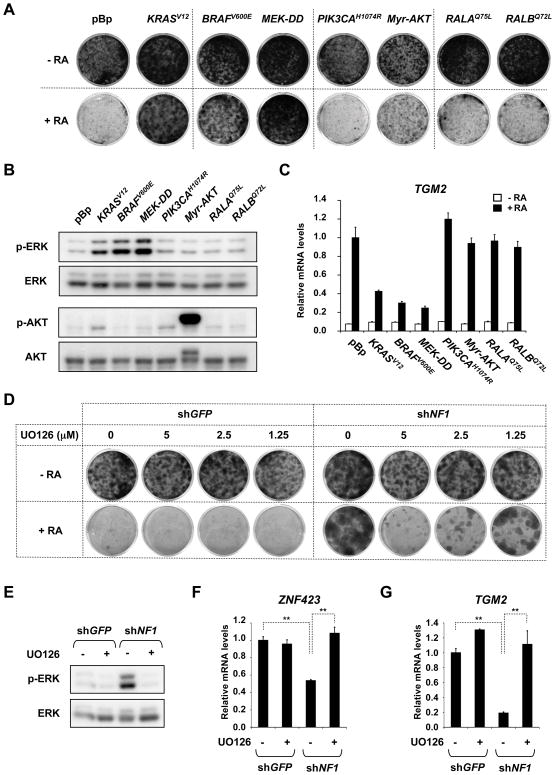
RA maintenance therapy has a modest, but significant, benefit for high-risk neuroblastoma patients. Our results suggest that tumors with activated RAS signaling e.g. by NF1 loss/mutation and low expression of ZNF423 would respond poorly to RA. We reasoned that inhibitors of downstream components of RAS could potentially restore sensitivity to RA in these tumors. To address this concept, we first dissected the contribution of the major RAS downstream effector pathways by expressing respective active alleles. BRAFV600E and MEK-DD conferred resistance to RA, in contrast to PIK3CAH1047R, RALAQ75L and RALBQ72L (Figure 7A). Noteworthy, the activation of AKT by PIK3CAH1047R was comparable to KRASV12 (Figure 7B). Even expression of highly active myristylated AKT failed to potently rescue from RA mediated growth arrest. Further, only KRASV12, BRAFV600E and MEK-DD blocked RA target gene activation (Figure 7C). Therefore, the RAF-MEK cascade is the predominant pathway responsible for resistance to RA by NF1 loss in neuroblastoma cells.
Figure 7. Restoration of RA responsiveness.
(A) Constitutively active RAF (BRAFV600E) and MEK (MEK-DD, MEK1S218D,S222D) recapitulate resistance to RA caused by KRASV12. SH-SY5Y cells expressing pBabe vector control or the indicated active alleles of RAS effector pathways were cultured for 12 and 20 days in the absence or presence of 1μM RA and then fixed, stained and photographed.
(B) Level of phosphorylated ERK and AKT in the SH-SY5Y cells described in figure 7A.
(C) Activation of the RA target gene TGM2 in SH-SY5Y cells described in figure 7A. Cells were grown for 14 days in the absence or presence of 1μM RA.
(D) MEK inhibition restores RA sensitivity in NF1 knockdown cells. SH-SY5Y cells expressing shGFP control or shNF1 were grown in the absence or presence of 1 μM RA for 13 and 18 days. Cells were additionally treated with or without the MEK inhibitor U0126 at various concentrations. Cells were then fixed, stained and photographed.
(E to G) Inhibition of MEK signaling and re-expression of ZNF423 and TGM2 in NF1 knockdown cells treated with MEK inhibitor U0126. SH-SY5Y cells expressing shGFP control or shNF1 grown in the presence of 1 μM RA for 9 days and then additionally treated with or without U0126 for 2 days. Levels of phosphorylated ERK were detected by Western blotting (E). ZNF423 and TGM2 mRNA expression was determined by qRT-PCR. Error bars denote SD. **p<0.001 (t-test for comparison of three biological independent experiments). See also supplemental figure S5.
Next, we added increasing concentrations of the MEK inhibitor U0126 with or without RA to SH-SY5Y cells expressing either shGFP or shNF1 (Figure 7D). Addition of U0126 restored sensitivity to RA in NF1 knockdown cells that were otherwise resistant. Application of U0126 affected cell growth only moderately in the absence of RA. Importantly, MEK inhibition partially restored ZNF423 expression and subsequently RA target gene expression in NF1 knockdown cells and also other NF1low neuroblastoma cell lines (Figure 7E–G and S5). Thus, the combination of RA and a MEK inhibitor might be a valid strategy for the maintenance therapy of high-risk neuroblastoma, in particular for tumors with low or mutated NF1.
Discussion
We identify here an unexpected role for the tumor suppressor NF1 in RA signaling through an unbiased large-scale RNAi screen in neuroblastoma cells. The NF1 protein is known to antagonize activation of RAS proteins, but it has also been implicated in other pathways such as cAMP/PKA signaling (Tong et al., 2002). In this study, we provide several lines of evidence that NF1 controls the response to RA in neuroblastoma through the RAS-MEK signaling cascade. We identify here a mechanistic connection between NF1-RAS-MEK and RA signaling by showing that NF1-RAS-MEK cascade suppresses expression of the RAR/RXR co-activator ZNF423 (Figure 4A, B and 7B). We have shown recently that ZNF423 is critically required for the response to RA in neuroblastoma (Huang et al., 2009). Restoration of ZNF423 expression in NF1low neuroblastomas partially restored their responsiveness to RA, underscoring the relevance of the regulation of ZNF423 by NF1. Furthermore, we find that inhibition of the MEK kinase, a downstream effector of RAS signaling, with a small molecule inhibitor restored ZNF423 expression in NF1 knockdown cells and restored both RA target gene expression and sensitivity to RA. These experiments place ZNF423 downstream of the NF1-RAS-MEK signaling cascade and provide a rationale for the combination of RA with MEK inhibitors in the clinic to induce differentiation, apoptosis and growth arrest in NF1low tumors, a phenotype seen in over half of primary neuroblastomas.
Other studies have also suggested a role for MAPK signaling in regulation of responses to RA in different cellular systems (Antonyak et al., 2003; Bruck et al., 2009; Delaune et al., 2008; Gianni et al., 2006; Kosa et al., 1995; Quinlan et al., 2008; Verheijen et al., 1999). These data indicate that the effects of MAPK signaling on RA responses can be heterogeneous. This may be explained by cell type specific regulatory mechanisms. Indeed, RARs can use a multitude of co-activators, some of which are cell type specific, including ZNF423. Moreover, there is evidence that short-term responses to MAPK signaling are quite distinct from the long-term effects we studied here. For example, p38MAPK activates RAR signaling by phosphorylation of RARα by MSK1, a p38MAPK activated kinase, but also inhibited RAR signaling later by phosphorylation and degradation of the RARα coactivator SRC3 (Gianni et al., 2006). However, a role for the p38MAPK pathway in RA-mediated long-term growth arrest in neuroblastoma has not been established.
Since RA plays a central role in neuronal differentiation and given that Rarα-knockout mice exhibit neural crest defects (McCaffery et al., 2003), we reasoned that NF1 expression could potentially impact the clinical course of neuroblastoma and even act as a tumor suppressor in human neuroblastoma. In three independent gene expression data sets of 316 neuroblastoma patients in total, we found that low expression of NF1 (NF1low), in particular in combination with ZNF423low, correlated with a very poor outcome (Figure 5). Tumors having both NF1low and ZNF423low constitute a clinically significant subgroup. In contrast, high expression of both was associated with the best prognosis. NF1low tumors were significantly enriched in the ZNF423low subgroup, suggesting that hyperactive RAS-MEK signaling also contributes to suppression of ZNF423 in primary neuroblastomas. Proteasomal degradation of NF1, as observed in glioblastoma, could also account for active RAS signaling and suppression of ZNF423 (McGillicuddy et al., 2009). As RAS mutations occur rarely if ever in primary neuroblastoma, other negative or positive regulators of RAS signaling could be involved as well. In addition, control of ZNF423 level by independent signaling pathways could antagonize or synergize with RAS signaling.
Germ line mutations in NF1 are linked to neurofibromatosis type 1 and somatic mutations and deletions are frequent in glioblastoma (Messiaen et al., 2000; TCGA_Network, 2008). The data demonstrating the poor prognosis of patients with NF1low expression sparked our interest in asking if NF1 could also be lost by mutation in neuroblastoma. We found two sequence mutations and three genomic aberrations (microdeletion, 17q duplication) in the NF1 gene out of 83 analyzed tumors. Based on a few incidental case reports, a predisposition of neurofibromatosis type 1 patients for neuroblastoma has been proposed (Martinsson et al., 1997; Origone et al., 2003). Indeed, the identified H553R missense mutation in NF1 proved to be a neurofibromatosis type 1 associated germ line mutation. The germ line status is unknown for the other patient having a five base pair deletion in NF1. Remarkably, this patient was diagnosed as stage 2 disease, which in general has an excellent prognosis. Nevertheless, this patient was the only one of the 23 stage 1/2 neuroblastoma patients in the AMC cohort that died of disease.
The mutation rate of NF1 in neuroblastoma was lower than reported for glioblastoma. It is reasonable to expect fewer mutations in an embryonal tumor like neuroblastoma than in a highly genomic unstable and largely p53 deficient adulthood malignancy. In addition, low- and intermediate-risk neuroblastomas (~50% of the AMC cohort) have a very good disease outcome and since low expression of NF1 correlated with poor outcome, we speculate that NF1 mutations are more prevalent in high-risk disease. However, larger cohorts will be required to investigate this hypothesis.
In conclusion, we found genomic aberrations of the NF1 gene in 6% (5/83) of primary neuroblastomas, supporting a direct role for NF1 in the pathogenesis of the disease, at least in a subset of cases. Noteworthy, activating mutations in the protein-tyrosine phosphatase PTPN11 (SHP2), an enhancer of RAS signaling, have been previously described in 3.4% (3/89) of neuroblastomas (Bentires-Alj et al., 2004). Dominant mutations in PTPN11 cause the developmental disorder Noonan syndrome and possibly predispose to neuroblastoma (Cotton and Williams, 1995; Tartaglia et al., 2001). Together with our functional, mutational and survival studies on NF1, it further substantiates a role for aberrant RAS signaling in the development of neuroblastoma, even though RAS mutations are rarely, if ever, found in this type of cancer (Moley et al., 1991).
Our data predict that low levels of ZNF423 and NF1 will be associated with poor response to RA-based therapies in the clinic. To begin to address this, we asked if we could observe a correlation between ZNF423 levels and RA responsiveness in a panel of 26 neuroblastoma cell lines. We reasoned that loss of NF1 causes suppression of ZNF423 expression (Figure 4A, B) and hence that ZNF423 levels would also report NF1 status in neuroblastoma cell lines. Indeed, we found that high expression of ZNF423 significantly correlated with sensitivity to RA (Figure 4E). The range of ZNF423 expression in the cell line panel corresponds well to the 3 to 4-fold repression of ZNF423 caused by loss of NF1.
RA is used as a maintenance therapy with some success for high-risk neuroblastoma and levels of NF1 and/or ZNF423 may well account for at least a fraction of non-responders in the clinic. Our finding that MEK inhibitors can restore responsiveness to RA in vitro in NF1low cells suggests a potential strategy to improve the therapeutic efficacy of retinoids in the treatment of neuroblastoma. As highly selective MEK inhibitors are currently evaluated in clinical trials, their combination with RA could be an attractive therapeutic approach.
Most, if not all, established predictive biomarkers for targeted therapies affect the target itself or key downstream components of the signaling pathway targeted by the drug. For example, KRAS mutations confer resistance to EGFR targeted therapy in colon cancer and PIK3CA mutations or lack of PTEN expression correlate with poor response to trastuzumab in HER2-positive breast cancer (Berns et al., 2007; Karapetis et al., 2008). Our results reveal a new paradigm for predictive biomarkers by demonstrating that aberrant activity of lateral signaling pathways can determine the response to a targeted therapeutic without mutations in any of the core components of the pathway itself. Our data highlight the importance of mapping crosstalk between signaling pathways to predict responses to targeted therapies in cancer. Unbiased genetic screens are powerful tools to identify these genetic interactions and the present case even suggests a therapeutic strategy to restore drug response in resistant patients.
EXPERIMENTAL PROCEDURES
shRNA Barcode Screen
The NKI shRNA library and the barcode screen are as described (Berns et al., 2004; Brummelkamp et al., 2006). Additional details can be found at www.screeninc.nki.nl.
Cell Proliferation Assays
Single cell suspensions were seeded into 6-well plates (1–2 ×104 cells/well) and cultured both in the absence and presence of RA. At the endpoints of colony formation assays, cells were fixed, stained with crystal violet and photographed. All knockdown and overexpression experiments were done by retroviral infection. For the experiments to measure RA responsiveness in the panel of 26 neuroblastoma cell lines, each line was subjected to long-term colony formation assays in triplicates in the absence or presence of 100nM RA. Treated and untreated dishes of each cell line were harvested at the same time. Crystal violet stain was then extracted using 10% acetic acid and quantified at OD 590nm. The growth curves were performed according to the standard 3T3 protocol. All relevant assays were performed independently at least three times.
Patient Samples
For all studies presented, the expression data were obtained using Affymetrix micro-array analyses on the untreated primary tumor samples at the time of diagnosis. The patient samples for both cohorts and the definition of an event (Progression-free survival) for both cohorts are as described (Asgharzadeh et al., 2006; Huang et al., 2009). Written informed consent was obtained from patients’ parents or guardians in accordance with institutional review board policies and procedures for research dealing with tumor specimen and clinical information. The institutional review board at Childrens Hospital Los Angeles (CHLA) and the medical-ethics committee of the Academic Medical Center (AMC) in Amsterdam approved the study.
Statistical Analysis
To determine the optimal value to set as a cut-off for NF1 expression in the AMC cohort, kaplan scanning was used as described for the cut-off for ZNF423 (Huang et al., 2009). The cut-off value was validated using a second independent set of 102 CHLA patients (Asgharzadeh et al., 2006). See supplemental experimental procedures for more details. Progression-free survival was measured for all outcome analysis presented in this study using the same NF1 or/and ZNF423 cut-off values.
Supplementary Material
Acknowledgments
We thank M. Hauptmann for statistical advice, the NKI microarray facility for supporting gene expression studies and C. Sun for assistance. This work was supported by The Netherlands Genomics Initiative (NGI), grants from the US National Institutes of Health R01-CA60104 (RCS) and K12-CA60104 (SA), and a grant from the Dutch Cancer Society (KWF).
References
- Antonyak MA, McNeill CJ, Wakshlag JJ, Boehm JE, Cerione RA. Activation of the Ras-ERK pathway inhibits retinoic acid-induced stimulation of tissue transglutaminase expression in NIH3T3 cells. J Biol Chem. 2003;278:15859–15866. doi: 10.1074/jbc.M300037200. [DOI] [PubMed] [Google Scholar]
- Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, Matthay K, Buckley J, Ortega A, Seeger RC. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, Brodeur GM, Cohn SL, Matthay KK, Maris JM. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Bentires-Alj M, Paez JG, David FS, Keilhack H, Halmos B, Naoki K, Maris JM, Richardson A, Bardelli A, Sugarbaker DJ, Richards WG, Du J, Girard L, Minna JD, Loh ML, Fisher DE, Velculescu VE, Vogelstein B, Meyerson M, Sellers WR, Neel BG. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, Beijersbergen RL, Mills GB, van de Vijver MJ, Bernards R. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Bruck N, Vitoux D, Ferry C, Duong V, Bauer A, de The H, Rochette-Egly C. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 2009;28:34–47. doi: 10.1038/emboj.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- Bunone G, Borrello MG, Picetti R, Bongarzone I, Peverali FA, de Franciscis V, Della Valle G, Pierotti MA. Induction of RET proto-oncogene expression in neuroblastoma cells precedes neuronal differentiation and is not mediated by protein synthesis. Experimental cell research. 1995;217:92–99. doi: 10.1006/excr.1995.1067. [DOI] [PubMed] [Google Scholar]
- Caron H, van Sluis P, de Kraker J, Bokkerink J, Egeler M, Laureys G, Slater R, Westerveld A, Voute PA, Versteeg R. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–230. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- Cotton JL, Williams RG. Noonan syndrome and neuroblastoma. Arch Pediatr Adolesc Med. 1995;149:1280–1281. doi: 10.1001/archpedi.1995.02170240098019. [DOI] [PubMed] [Google Scholar]
- Delaune A, Corbiere C, Benjelloun FD, Legrand E, Vannier JP, Ripoll C, Vasse M. Promyelocytic leukemia-nuclear body formation is an early event leading to retinoic acid-induced differentiation of neuroblastoma cells. J Neurochem. 2008;104:89–99. doi: 10.1111/j.1471-4159.2007.05019.x. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Gianni M, Parrella E, Raska I, Jr, Gaillard E, Nigro EA, Gaudon C, Garattini E, Rochette-Egly C. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 2006;25:739–751. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Thompson P, Frayling I, Upadhyaya M. Molecular diagnosis of neurofibromatosis type 1: 2 years experience. Fam Cancer. 2007;6:21–34. doi: 10.1007/s10689-006-9001-3. [DOI] [PubMed] [Google Scholar]
- Huang S, Laoukili J, Epping MT, Koster J, Holzel M, Westerman BA, Nijkamp W, Hata A, Asgharzadeh S, Seeger RC, Versteeg R, Beijersbergen RL, Bernards R. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328–340. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Kosa K, Jones CS, De Luca LM. The H-ras oncogene interferes with retinoic acid signaling and metabolism in NIH3T3 cells. Cancer Res. 1995;55:4850–4854. [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O’Connell P, Cawthon RM, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Martinsson T, Sjoberg RM, Hedborg F, Kogner P. Homozygous deletion of the neurofibromatosis-1 gene in the tumor of a patient with neuroblastoma. Cancer Genet Cytogenet. 1997;95:183–189. doi: 10.1016/s0165-4608(96)00259-2. [DOI] [PubMed] [Google Scholar]
- Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, Gerbing RB, London WB, Villablanca JG. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children’s oncology group study. J Clin Oncol. 2009;27:1007–1013. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT, Brodeur GM, Gerbing RB, Reynolds CP. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- McCaffery PJ, Adams J, Maden M, Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. The European journal of neuroscience. 2003;18:457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, Cloughesy TF, Mischel PS, Parret A, Seiler J, Moldenhauer G, Scheffzek K, Stemmer-Rachamimov AO, Sawyers CL, Brennan C, Messiaen L, Mellinghoff IK, Cichowski K. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen LM, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, Speleman F, Paepe AD. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Moley JF, Brother MB, Wells SA, Spengler BA, Biedler JL, Brodeur GM. Low frequency of ras gene mutations in neuroblastomas, pheochromocytomas, and medullary thyroid cancers. Cancer Res. 1991;51:1596–1599. [PubMed] [Google Scholar]
- Mosse YP, Wood A, Maris JM. Inhibition of ALK signaling for cancer therapy. Clin Cancer Res. 2009;15:5609–5614. doi: 10.1158/1078-0432.CCR-08-2762. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, Ernestus K, Konig R, Haas S, Eils R, Schwab M, Brors B, Westermann F, Fischer M. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- Origone P, Defferrari R, Mazzocco K, Lo Cunsolo C, De Bernardi B, Tonini GP. Homozygous inactivation of NF1 gene in a patient with familial NF1 and disseminated neuroblastoma. Am J Med Genet A. 2003;118A:309–313. doi: 10.1002/ajmg.a.10167. [DOI] [PubMed] [Google Scholar]
- Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol Cell Biol. 2008;28:2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette-Egly C, Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs) Nucl Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- Sidell N, Altman A, Haussler MR, Seeger RC. Effects of retinoic acid (RA) on the growth and phenotypic expression of several human neuroblastoma cell lines. Experimental cell research. 1983;148:21–30. doi: 10.1016/0014-4827(83)90184-2. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- TCGA_Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The I, Murthy AE, Hannigan GE, Jacoby LB, Menon AG, Gusella JF, Bernards A. Neurofibromatosis type 1 gene mutations in neuroblastoma. Nat Genet. 1993;3:62–66. doi: 10.1038/ng0193-62. [DOI] [PubMed] [Google Scholar]
- Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- van Limpt V, Schramm A, van Lakeman A, Sluis P, Chan A, van Noesel M, Baas F, Caron H, Eggert A, Versteeg R. The Phox2B homeobox gene is mutated in sporadic neuroblastomas. Oncogene. 2004;23:9280–9288. doi: 10.1038/sj.onc.1208157. [DOI] [PubMed] [Google Scholar]
- Verheijen MH, Wolthuis RM, Bos JL, Defize LH. The Ras/Erk pathway induces primitive endoderm but prevents parietal endoderm differentiation of F9 embryonal carcinoma cells. J Biol Chem. 1999;274:1487–1494. doi: 10.1074/jbc.274.3.1487. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Yao S, Claes K, Kehrer-Sawatzki H, Tinschert S, De Raedt T, Legius E, Callens T, Beiglbock H, Maertens O, Messiaen L. Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer. 2006;45:265–276. doi: 10.1002/gcc.20289. [DOI] [PubMed] [Google Scholar]
- Zatkova A, Messiaen L, Vandenbroucke I, Wieser R, Fonatsch C, Krainer AR, Wimmer K. Disruption of exonic splicing enhancer elements is the principal cause of exon skipping associated with seven nonsense or missense alleles of NF1. Hum Mutat. 2004;24:491–501. doi: 10.1002/humu.20103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.