Abstract
Background
Vγ9Vδ2 T lymphocytes are regarded as promising mediators of cancer immunotherapy due to their capacity to eliminate multiple experimental tumors, particularly within those of hematopoietic origin. However, Vγ9Vδ2 T-cell based lymphoma clinical trials have suffered from the lack of biomarkers that can be used as prognostic of therapeutic success.
Design and Methods
We have conducted a comprehensive study of gene expression in acute lymphoblastic leukemias and non-Hodgkin’s lymphomas, aimed at identifying markers of susceptibility versus resistance to Vγ9Vδ2 T cell-mediated cytotoxicity. We employed cDNA microarrays and quantitative real-time PCR to screen 20 leukemia and lymphoma cell lines, and 23 primary hematopoietic tumor samples. These data were analyzed using state-of-the-art bioinformatics, and gene expression patterns were correlated with susceptibility to Vγ9Vδ2 T cell mediated cytolysis in vitro.
Results
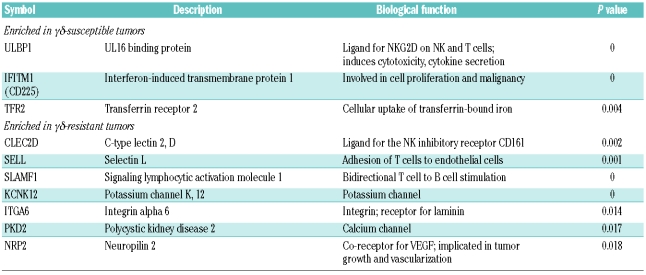
We identified a panel of 10 genes encoding cell surface proteins that were statistically differentially expressed between “γδ-susceptible” and “γδ-resistant” hematopoietic tumors. Within this panel, 3 genes (ULBP1, TFR2 and IFITM1) were associated with increased susceptibility to Vγ9Vδ2 T-cell cytotoxicity, whereas the other 7 (CLEC2D, NRP2, SELL, PKD2, KCNK12, ITGA6 and SLAMF1) were enriched in resistant tumors. Furthermore, some of these candidates displayed a striking variance of expression among primary follicular lymphomas and T-cell acute lymphoblastic leukemias.
Conclusions
Our results suggest that hematopoietic tumors display a highly variable repertoire of surface proteins that can impact on Vγ9Vδ2 cell-mediated immunotargeting. The prognostic value of the proposed markers can now be evaluated in upcoming Vγ9Vδ2 T cell-based lymphoma/leukemia clinical trials.
Keywords: biomarkers, Vγ9Vδ2 T-lymphocytes, hematopoietic tumors, lymphoma cell lines
Introduction
γδ T lymphocytes display potent innate anti-tumor activity in both humans1 and mice.2,3 For example, mice genetically devoid of γδ T cells displayed increased susceptibility to skin tumor development induced experimentally by carcinogens,2,3 and to transgenic adenocarcinoma of the mouse prostate model (TRAMP).4 More importantly, murine γδ T cells were shown to prevent (through perforin-mediated cytotoxicity) the development of spontaneous B-cell lymphomas.5
The major γδ T-cell subset in human peripheral blood, Vγ9Vδ2 T lymphocytes, exert potent cytotoxicity towards tumor cell lines upon activation with small non-peptidic prenyl pyrophosphate intermediates of isoprenoid biosynthesis.6 We and others have shown that, among such “phosphoantigens”, 4-hydroxy-3-methyl-but-2-enylpyrophosphate (HMB-PP), a metabolite found in Eubacteria and Protozoa, is a very potent agonist of the Vγ9Vδ2 T-cell receptor (TCR) that promotes cytotoxicity and the secretion of anti-tumor cytokines such as interferon-γ (IFN-γ) and tumor necrosis factor α (TNF-α).6,7
Phosphoantigen-activated Vγ9Vδ2 T cells can kill various solid tumor cell lines,1 and a particularly large number of hematopoietic cell-derived tumors,7–9 as well as freshly isolated tumor cells from patients with follicular B-cell lymphoma or chronic lymphocytic leukemia (CLL).10
The well-established anti-tumor activity of Vγ9Vδ2 T cells has been recently explored in clinical trials for solid/epithelial11–13 or liquid/hematopoietic tumors (14–16), which were collectively promising even though they showed limited success. The lack of response to therapy of some patients was attributed to deficient expansion of effector Vγ9Vδ2 T cells.11,13,14 However, a large proportion of patients exhibiting significant and sustained in vivo activation and proliferation of Vγ9Vδ2 T cells also failed to respond to treatment. Thus, in both prostate carcinoma11 and non-Hodgkin’s lymphoma,14 objective responses (partial remissions) were observed in just 33% of the patients who activated/expanded their Vγ9Vδ2 T cells. These data emphasize the need for tumor biomarkers with prognostic value for γδ peripheral blood lymphocyte (γδ-PBL)-mediated immunotherapy.
Here we have conducted a comprehensive genome-wide expression study aimed at identifying lymphoma/leukemia markers of susceptibility or resistance to γδ-PBL cytotoxicity. We set up an experimental system consisting of lymphoma/leukemia cell lines with various degrees of susceptibility to γδ-PBL-mediated lysis, and performed comparative cDNA microarray analyses to characterize their gene expression profiles. These were validated through bioinformatics and quantitative real-time PCR (RT-qPCR), allowing us to define a panel of 10 candidate biomarkers whose expression displayed very marked variability among non-Hodgkin’s lymphoma and acute lymphoblastic leukemia patients.
Design and Methods
In vitro cultures of human γδ-PBL and tumor cell lines
Peripheral blood was collected from healthy volunteers and peripheral blood mononuclear cells (PBMCs) were isolated as previously described.7 γδ-PBL were expanded from isolated PBMCs for 12 days in RPMI 1640 complete media7 supplemented with 100 U/mL of rhIL-2 (Roche Applied Science) and 1 nM HMB-PP (4-hydroxy-3-methyl-but-2-enylpyrophosphate) (Sup-RPMI). The percentage of Vγ9+ T cells in peripheral blood increased from 3–14% at day 0 to 90–98% at day 12 (Online Supplementary Figure S1). All tumor cell lines were cultured in complete 10% RPMI-1640 as previously described.7
Leukemia and lymphoma primary samples
Pediatric B- or T-cell acute lymphoblastic leukemia cells containing high (> 80%) leukemia involvement were obtained from the peripheral blood and/or the bone marrow of patients at presentation after informed consent and institutional review board approval (Instituto Português de Oncologia, Lisbon, Portugal) had been obtained. Fresh leukemia samples were enriched by density centrifugation over Ficoll-Paque and then washed twice in 10% RPMI-1640 medium supplemented with 2 mM L-glutamine (Sup-RPMI). For lymphoma biopsies, lymph nodes were surgically removed, immediately frozen in liquid nitrogen and kept at −80°C until further use (Department of Pathology, Hospital de Santa Maria, CHLN, Lisbon, Portugal). Upon diagnosis, we selected lymph nodes from lymphoma cases and reactive lymph nodes for our studies.
In vitro killing assays
For cytotoxicity assays, tumor cells (cell lines or primary samples) were stained with DDAO-SE (Molecular Probes, Invitrogen) and incubated at a ratio of 1:10 with γδ T cells in Sup-RPMI. Typically, 3×105 HMB-PP-activated γδ-PBL (>90% Vγ9+) were co-incubated with 3×104 tumor cells (pre-labeled with 1 μM DDAO-SE) for 3–4h, then stained with Annexin V-FITC (BD Biosciences) and analyzed by flow cytometry.
RNA isolation, RT-qPCR and Affymetrix Microarrays
Total RNA from tumor cell lines was extracted using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). RNA from leukemia cells and samples was extracted with TRIzol Reagent (Invitrogen) and purified with RNeasy Mini Kit according to the manufacturer’s instructions. Concentration and purity was determined by spectrophotometry and integrity was confirmed using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano Assay (Agilent Technologies, Palo Alto, CA, USA). Total RNA was reverse-transcribed into cDNA as previously described.7 qPCR was performed on Rotor-Gene 6000 (Corbett) using SYBR Green detection system (PE Applied Biosystems). Glucoronidase beta (GUSB) and proteasome subunit beta type 6 (PSMB6) were used as endogenous controls in relative quantification using the standard curve method. Primers were designed using the Roche Design Centre (for sequences see Online Supplementary Table S1).
For genome-wide analyses, RNA from two independent cultures of each cell line (DAUDI, RAJI, RCH-ACV and 697) was processed for use on Affymetrix (Santa Clara, CA, USA) GeneChip HuGene 1.0 ST Arrays, according to the manufacturer’s Whole Transcript Sense Target Labeling Assay.
Microarray data analysis
All the microarray data analysis was performed with R and several packages available from CRAN17 and Bioconductor.18 The raw data (CEL files) were normalized and summarized with the Robust MultiArray Average method from the “affy” package.19 Unsupervised clustering analysis of the gene expression profiles for entire probe set data was assessed through hierarchical clustering (Euclidean distance and complete agglomeration method) and principal component analysis (prcomp function which calls a singular value decomposition method for non-symmetric matrices) as implemented in the statistical computing package.17 Differentially expressed genes for each comparison were selected using linear models and empirical Bayes methods20 as implemented in the Limma package,21 verifying the P values corresponding to moderated F-statistics, and selecting as differentially expressed genes those that had adjusted P values adjusted using the Benjamini and Hochberg method22 lower than 0.05.
The enrichment of biological functions and pathways was analyzed using Ingenuity Pathway Analysis software (Ingenuity Systems, Mountain View, CA, USA) and all genes present in the Affymetrix Human Gene 1.0 ST as control.
Results
Highly variable susceptibility of acute leukemias and non-Hodgkin’s lymphomas to γδ-PBL cytotoxicity
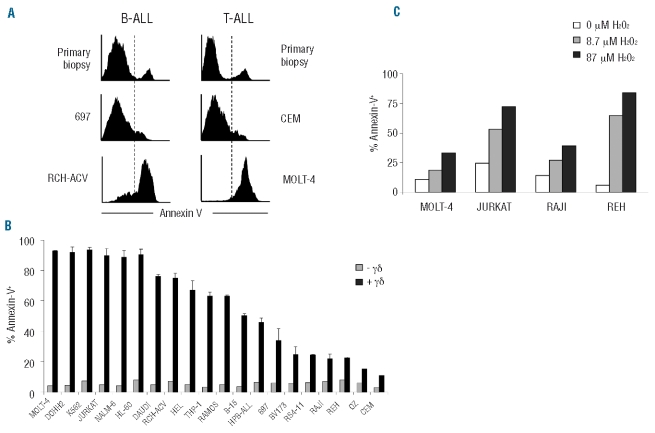
In our laboratory we have studied a collection of 23 samples of acute lymphoblastic leukemias and non-Hodgkin’s lymphomas, and a panel of 20 tumor cell lines of hematopoietic origin. The latter included acute lymphoblastic leukemia (ALL) (JURKAT, MOLT4, RCH-ACV, 697, CEM, TOM-1, RS4-11, B15, REH, Bv173) and acute myelogenous leukemia (AML) (HL-60, HEL, THP-1) cell lines; and non-Hodgkin Burkitt’s (DAUDI, RAJI, RAMOS), follicular (DOHH2) and lymphoblastic (Oz) lymphoma cell lines (for detailed description of these cell lines see Online Supplementary Table S2). Although the capacity of peripheral blood γδ T cells to target multiple tumor cell lines of hematopoietic origin is well documented,7–9 we observed that a substantial fraction of cell lines (Figure 1A and B) and patient samples (Figure 1A and data not shown) were strikingly resistant to γδ-PBL (obtained from healthy donors) pre-activated (as illustrated by high CD69 levels) with HMB-PP, the most potent natural Vγ9Vδ2 T-cell activator known to date (6, 7) (Online Supplementary Figure S1). For example, the BALL cell lines Bv173, REH and 697 (Figure 1 A and B), and six primary samples obtained from B-ALL patients (Figure 1A and data not shown) remained mostly alive (Annexin V-) in co-cultures with fully-activated (100% CD69+; data not shown) γδ-PBL. Similar data were obtained with primary T-ALL samples and the cell line CEM (Figure 1A). This resistance to γδ-PBL cytotoxicity contrasted sharply with the extensive killing observed for the B-ALL line RCH-ACV and the T-ALL line MOLT-4 (Figure 1A), among various other hematopoietic tumors (Figure 1B).
Figure 1.
Differential susceptibility of leukemia and lymphoma cells to γδ-PBL cytotoxicity. (A) Annexin V staining for apoptotic tumor cells after 4 h of co-incubation with HMB-PP-activated γδ-PBL. Tumors were B-ALL (left panels) or T-ALL (right panels) cells, either primary samples or the indicated cell lines. (B) Summary of killing assays (as in A) with 20 leukemia or lymphoma cell lines (described in Online Supplementary Table S2). Error bars correspond to triplicate assays. (C) Effect of increasing concentrations of H2O2 on leukemia/lymphoma cell apoptosis (% Annexin V+).
For systematic analysis of our killing assay data, we considered tumor samples with over 70% lysis as susceptible to γδ-PBL-mediated lysis (“γδ-susceptible”), and those under 30% lysed as “γδ-resistant”. Importantly, susceptibility was independent of the γδ-PBL donor, as the pattern of susceptible/resistant lines was equivalent for 3 independent healthy donors (Online Supplementary Figure S2A). Moreover, the differences in susceptibility to γδ T cells were maintained when tumor cell lines were incubated with γδ T cells activated for a shorter time (12h) (Online Supplementary Figure S2B), further supporting the segregation between susceptible and resistant cell lines. As primary samples to reproduce and expand experiments aimed at dissecting the molecular mechanisms of tumor susceptibility to γδ-PBL cytotoxicity are difficult to obtain, we focused on our well-established panel of cell lines for the initial candidate searches and later extended our findings to patient samples.
We first considered that tumor resistance to γδ-PBL cytotoxicity could stem from intrinsic anti-apoptotic mechanisms developed by some leukemia/lymphoma cell lines. However, when we tested the effect of a pro-apoptotic stimulus (H2O2) we observed no association between resistance to apoptosis and to γδ-PBL cytotoxicity. Namely, the cell lines Jurkat (γδ-susceptible) and REH (γδ-resistant) were more sensitive to non-saturating concentrations of H2O2 than the cell lines MOLT-4 (γδ-susceptible) and RAJI (γδ-resistant) (Figure 1C). This suggests that susceptibility to γδ-PBL cytotoxicity is not related to the response to other death stimuli and probably involves a specific protein expression program (involved in tumor/γδ-PBL interactions) that we set out to characterize.
Genome-wide comparisons between γδ-susceptible and γδ-resistant hematopoietic tumors
The observed differences in susceptibility γδ-PBL cytotoxicity among hematopoietic tumors emphasize the importance of defining gene signatures that may predict the effectiveness of γδ T-cell based immunotherapies in the clinic. We performed a genome-wide analysis aimed at comparing the mRNA expression profiles of γδ-susceptible and γδ-resistant tumors. We employed cDNA microarrays to examine two pairs of hematopoietic tumor cell lines sharing the same cytogenetic alterations and cellular phenotypes (Online Supplementary Table S2): the Burkitt’s lymphomas DAUDI (susceptible) and RAJI (resistant), and the B-ALL lines RCH-ACV (susceptible) and 697 (resistant).
First, samples were grouped according to the similarity of gene expression patterns using unsupervised clustering analysis (no group specification a priori). Based on the entire probe set data, two main groups could be defined which corresponded to the original cell type (Figure 2A): pre-B (697 and RCH-ACV) and mature B cells (DAUDI and RAJI). We next applied principal component analysis (PCA), which identifies new variables, to the principal components, which are linear combinations of the original variables (gene expression levels) and represent the largest variation found between samples.23 Although the original cell type was the major source of variation between all samples (53.3% of total variation), PCA showed that component 3 was responsible for the segregation (16.4% of total variation) according to the susceptibility to γδ-PBL cytotoxicity (Figure 2B): susceptible (DAUDI and RCH-ACV) versus resistant (RAJI and 697).
Figure 2.
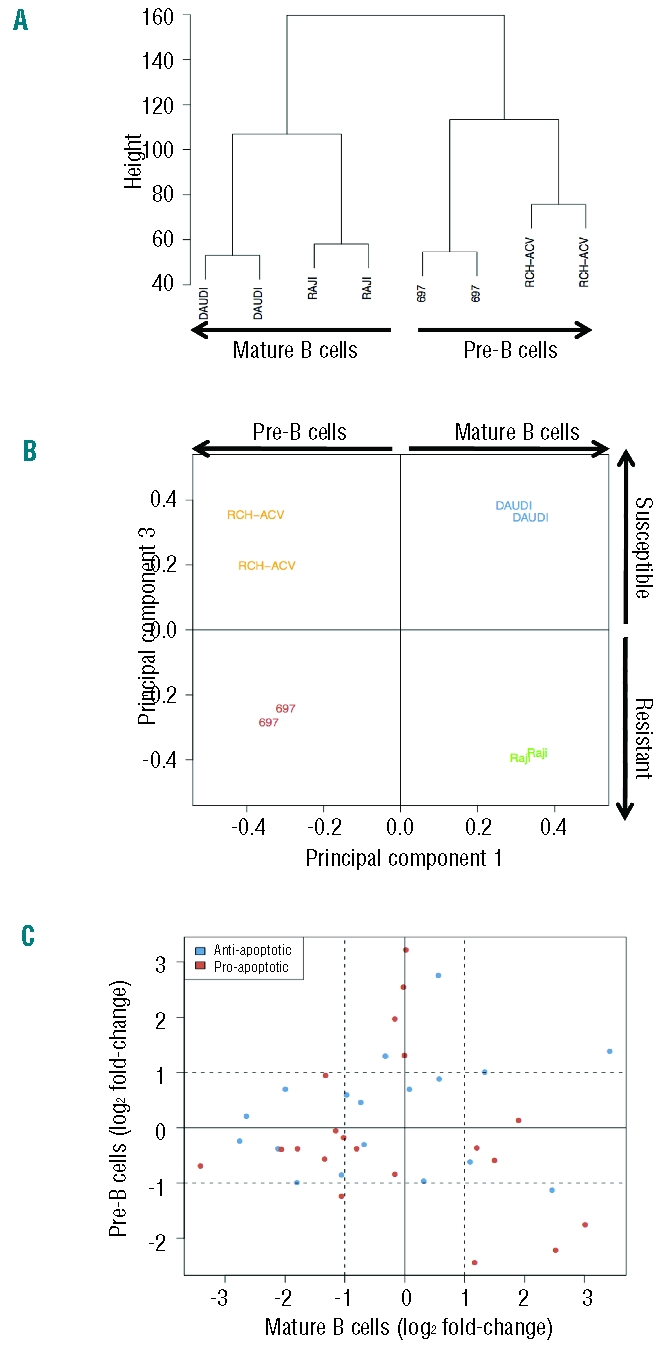
Comparison of gene expression in tumor cell lines susceptible or resistant to γδ-PBL cytotoxicity. Bioinformatics analyses of cDNA microarray comparisons between the Burkitt’s lymphomas DAUDI and RAJI; and the B-ALL lines RCH-ACV and 697. (A) Unsupervised hierarchical clustering analysis. Samples with similar gene expression patterns are grouped together and connected with branches, producing a clustering tree (or dendrogram) on which the branch length inversely reflects the degree of similarity between samples. (B) Principal Component Analysis. The samples are plotted according to the first and third principal components (corresponding to the largest variation found between samples). (C) Variations in expression levels of anti- or pro-apoptotic genes in susceptible versus resistant tumor cell lines. Dashed lines indicate 2 fold-changes (in logarithmic scale) in the expression ratio susceptible/resistant.
To identify gene expression variations associated with susceptibility to γδ-PBL cytotoxicity, and to suppress the variations due to the transformed cell type (pre-B or mature B cells), we first compared tumors with identical origin, i.e. DAUDI versus RAJI, and RCH-ACV versus 697 (Online Supplementary Tables S3 and S4). We then used Bayesian linear models20 and selected the common genes between both analyses: 340 genes (155 up- and 185 down-regulated in γδ-susceptible tumors) presented similar gene expression variations and were considered for further analysis (Online Supplementary Table S5). Bioinformatics analysis revealed an enrichment for functions related to cell-to-cell signaling and interaction, hematologic system development and function, immune cell trafficking (P value < 0.05; Online Supplementary Table S6). Some of the top pathways affected were interferon signaling, crosstalk between dendritic cells and natural killer cells, and molecular mechanisms of cancer (P value < 0.05; Online Supplementary Table S7).
The gene expression variations observed also suggested that, consistent with our previous experimental data (Figure 1C), the segregation between susceptible and resistant tumors is not associated with expression of anti- or pro-apoptotic genes (Figure 2C and Online Supplementary Table S8). Thus, up-/down-regulation of pro-/anti-apoptotic genes did not correlate with susceptibility to γδ-PBL cytotoxicity. Moreover, apoptotic related functions and pathways were not enriched in the panel of 340 genes (Online Supplementary Table S7). Based on these results, we favored the hypothesis that susceptibility or resistance to γδ-PBL cytotoxicity is conferred by signals presented at the tumor/γδ-PBL interface, i.e. on the surface of leukemia/lymphoma cells.
A set of cell surface proteins segregates between γδ-susceptible and γδ-resistant leukemia/lymphoma cell lines
T cells recognize their targets through cell surface antigens. We, therefore, focused our analysis of the panel of 340 genes on those encoding plasma membrane proteins (with extracellular domains), using a fold change threshold of 2 (log FC >1). These consisted of 8 genes up-regulated and 19 genes down-regulated in γδ-susceptible tumors when compared to resistant tumors (Online Supplementary Table S9). The mRNA expression levels of the 27 candidates were assessed by RT-qPCR (in independent samples) to validate the microarray results. Upon statistical analysis of the data, 22 out of the 27 genes were confirmed as differentially expressed in the two pairs of cell lines used for microarray comparisons: of these, 6 genes were up-regulated and 16 genes were down-regulated in γδ-susceptible tumors (Figure 3A). In order to have more stringent selection criteria, we extended our expression studies to a broader panel of cell lines, including 6 susceptible and 4 resistant cell lines (Online Supplementary Figure S3). This showed 10 genes with significant expression variation between susceptible and resistant tumors (P value < 0.05, Mann-Whitney test) (Figure 3B). Thus, our final panel of candidate markers of susceptibility to γδ-PBL cytotoxicity consisted of 3 genes enriched in γδ-suceptible tumors (ULBP1, TFR2 and IFITM1), and 7 genes enriched in γδ-resistant leukemias/lymphomas (CLEC2D, NRP2, SELL, PKD2, KCNK12, ITGA6 and SLAMF1) (Table 1).
Figure 3.
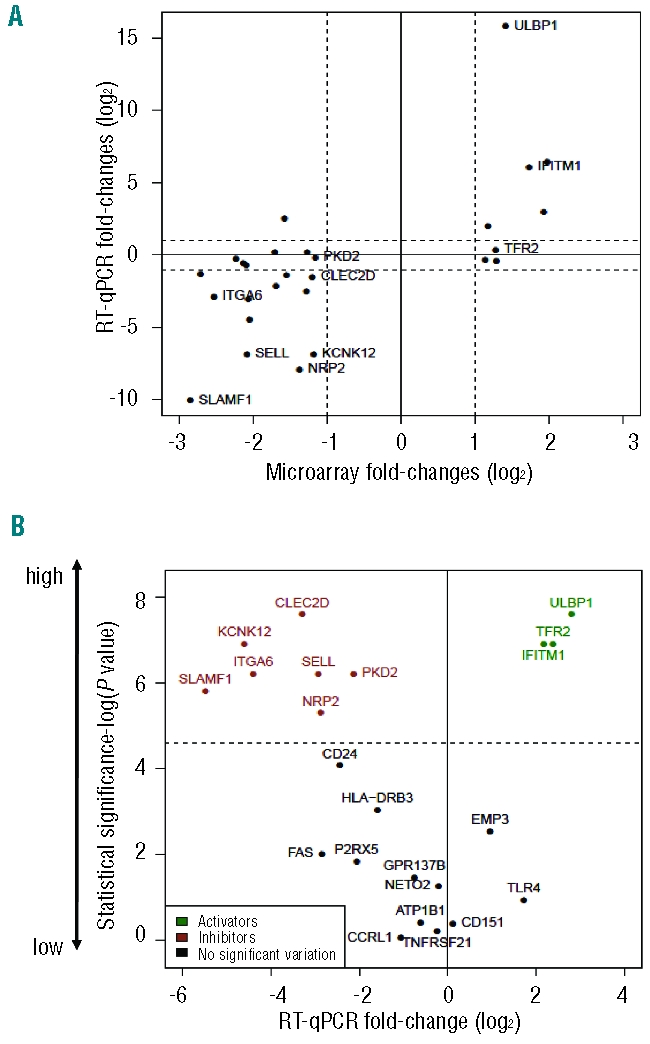
Variations in expression of genes encoding cell surface proteins that segregate between γδ-susceptible and γδ-resistant leukemia/lymphoma cell lines. (A) RT-qPCR validation of microarray results for the comparisons of Figure 2. The mRNA expression levels were normalized to GUSB and PSMB6 for each cell line. Plotted are the averages of relative expression levels in DAUDI versus RAJI (DAUDI/RAJI) and RCH-ACV versus 697 (RCH-ACV/697). Dashed lines indicate 2 fold-change values (in logarithmic scale). (B) Statistical analysis of RT-qPCR results (detailed in Figure 4) in 6 susceptible and 4 resistant cell lines. Statistical significance was assessed by Mann-Whitney test (−log P value). Dashed line represents the statistical threshold P=0.01.
Table 1.
Panel of cell surface proteins associated with the susceptibility or resistance of lymphomas/leukemias to γδ T-cell cytotoxicity. The statistical difference between the average gene expression in the 6 susceptible versus the 4 resistant tumors of Figure 4 was assessed by Mann-Whitney test (P<0.05).
Heterogeneity of expression of candidate markers in primary leukemia and lymphoma samples
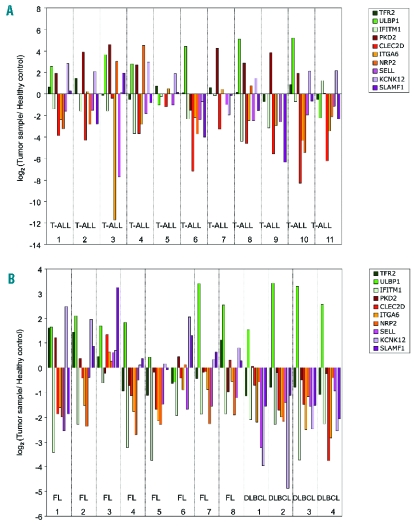
We next determined the expression levels of each candidate marker in primary samples obtained from T-cell acute lymphoblastic leukemia (T-ALL) and non-Hodgkin’s lymphoma (NHL) patients. Within the latter group, we sampled patients with common indolent (follicular) or aggressive (diffuse large B-cell - DLBCL) lymphomas. Gene expression levels in samples were compared with healthy PBMCs (for ALL) and reactive follicles (for NHL), taken as references (0 on log scale) in Figure 4A and B. Hence, a positive or negative (log scale) variation indicates higher or lower expression in tumors than in the control samples, respectively. Overall, the tumors exhibited very variable gene expression profiles. For example, among susceptibility-associated genes, ULBP1 was over-expressed in a large number of primary samples, while TFR2 was only enriched in three FL samples (FL 1, FL 2 and FL 8), and IFITM1 was strongly depleted in various tumors (Figure 4A and B). On the other hand, all resistance-associated genes were over-expressed in FL sample 3, in contrast to the majority of primary samples analyzed. Moreover, there was no essential difference in some markers, such as ITGA6 or SELL, between the various patients (Figure 4A and B). Collectively, these data revealed a striking heterogeneity in the expression of particular candidate genes in primary tumors. When compared to our results with tumor cell lines (Online Supplementary Figure S3), these clinical data possibly reflect distinct selective pressures on the expression of the genes that compose the candidate panel, the consequence of which should now be evaluated in clinical trials.
Figure 4.
Quantification of mRNA expression levels of γδ-susceptibility markers in acute lymphoblastic leukemia and non-Hodgkin’s lymphoma patients. (A) RT-qPCR analysis of mRNA expression in 11 T-cell acute lymphoblastic leukemia (T-ALL) samples, normalized to housekeeping genes (GUSB and PSMB6) and to reference PBMCs from healthy individuals. Values were converted to logarithmic scale. (B) RT-qPCR analysis of mRNA expression in 8 follicular lymphoma (FL) and 4 diffuse large B cell lymphoma (DLBCL) samples, normalized to housekeeping genes (GUSB and PSMB6) and to a reference sample - reactive follicles – obtained through the same procedure. Values were converted to logarithmic scale.
Discussion
The success of immunotherapy to tackle tumors, in particular those that prevail after chemo-or radiotherapy, critically depends on two factors: the specific activation of effector anti-tumor lymphocytes and the molecular recognition of tumor cells by activated lymphocytes. Concerning γδ T cells, research over the last 15 years has identified very potent and specific phosphoantigens, most notably HMB-PP,6,7 that seem to fulfill the first requirement. There have been suggestions that phospho-antigens themselves,6,24,25 or an F1-ATPase-related structure complexed with delipidated apolipoprotein A-I,26 or the non-classical MHC protein ULBP427 could be responsible for tumor cell recognition by Vγ9Vδ2 PBL. However, despite this, the issue is still highly controversial. This naturally impacts on our ability to design effective therapeutic protocols based on γδ-PBL immunotargeting of tumors. Thus, only 33% of patients with prostate carcinoma11 or non-Hodgkin’s lymphoma14 showed objective responses despite large activation and expansion of their Vγ9Vδ2 T cells in vivo. These considerations stress the importance of identifying tumor molecular signatures that may predict the response to activated γδ-PBL.
In this study, we set out to identify cell surface proteins involved in interactions between leukemia/lymphoma cells and γδ-PBL. Taking in vitro tumor cytolysis as functional readout, we screened a panel of 20 leukemia and lymphoma cell lines that faithfully reproduced the susceptibility/resistance of primary tumors (Figure 1A). The use of cell lines permitted experimental reproducibility and hence statistical robustness for the gene expression undertaken. Upon the identification of candidate markers, we analyzed their expression in 23 samples derived from T-ALL and NHL (FL and DLBCL) patients.
The choice of cDNA microarrays as screening tools was based on a multiplicity of previous studies that demonstrated how powerful and reliable they are in defining cancer molecular signatures.28 Our analyses led to the identification of a large panel of genes differentially expressed between “γδ-susceptible” and “γδ-resistant” tumors. Importantly, we verified that there was no correlation between intrinsic anti-apoptotic properties and resistance to γδ-PBL cytotoxicity, both in terms of gene expression and response to a death stimulus. Thus, susceptibility or resistance to γδ-mediated lysis is more likely to be related to tumor recognition and immune evasion strategies, the molecular basis of which remains to be clarified. Of note, MHC class Ia expression did not consistently segregate between γδ-susceptible and γδ-resistant tumor cell lines (Online Supplementary Figure S4). For example, among susceptible lines, DAUDI and MOLT-4 expressed very low or undetectable levels, whereas JURKAT and RCH-ACV displayed high levels of surface MHC class I (Online Supplementary Figure S4). These data exclude a mechanism of “missing self” as the basis for γδ T-cell recognition of hematopoietic tumors.
Building upon stringent biological and statistical selection criteria, we narrowed our microarray data down to 10 genes encoding cell surface proteins (with extracellular domains), whose expression segregated with susceptibility versus resistance to γδ-PBL cytotoxicity. We believe it is important to make this gene profile available to the biomedical community. Thus, we propose the expression of each candidate gene to be evaluated during upcoming γδ T-cell based clinical trials. The genes with highest predictive value will constitute novel leukemia/lymphoma biomarkers, for which standardized quantification essays should be developed. This will provide clinicians with a key tool for the indication and monitoring of γδ T-cell based immunotherapies.
Furthermore, within the panel of 10 candidate markers, some are likely to play non-redundant roles in leukemia/lymphoma cell recognition by γδ-PBL. Thus, proteins that are enriched in γδ-susceptible tumors may provide activation signals, whereas markers of resistance may convey inhibitory signals to γδ-PBL. Provocatively, 7 of the candidates are known to intervene in immune responses: 4 of them (ULBP1, IFITM1, CLEC2D and SLAMF1) provide stimulatory (or inhibitory) signals through receptors expressed on lymphocytes, while 3 (NRP2, SELL and ITGA6) control lymphocyte adhesion. ULBP1 is a ligand for the NKG2D receptor expressed on all cytotoxic lymphocyte lineages, including 100% of Vγ9Vδ2 T cells, which has been clearly implicated in anti-tumor responses.29–32 IFITM1 was shown to modulate NK cell responses and its expression correlated with improved survival of gastric cancer patients.33 By contrast, the expression of CLEC2D, a ligand for the inhibitory receptor CD161, inhibits NK cell responses and was associated with increased malignancy grade of glioblastoma.34 NRP2 is another protein that can favor cancer progression by acting as a coreceptor for vascular endothelial growth factor (VEGF) and stimulating tumor growth (35). We will now proceed with individual knock-down (RNA interference) experiments in a functional (tumor killing) bioassay to dissect the role of each of the candidates in γδ-PBL targeting of leukemias and lymphomas. Given that some of these molecules can also provide costimulatory or inhibitory signals to NK cells, we also plan to address their role in NK cell targeting of hematopoietic malignancies.
In summary, this report establishes a panel of 10 putative markers of leukemia/lymphoma susceptibility to γδ-PBL cytotoxicity. The expression data collected from primary samples showed a striking heterogeneity for particular candidate genes, most notably ULBP1, whereas other genes, such as IFITM1, ITGA6 or SELL, essentially did not vary among patients. It is, therefore, predictable that different components of the proposed panel will behave in very distinct ways when associated to therapeutic outcome in clinical trials. It will also be interesting to evaluate to what extent immunoselection may have conditioned the expression of these markers in tumors evolving in a dynamic interaction with γδ T lymphocytes. This will significantly add to our understanding of anti-tumor immunity and to our capacity to modulate it for cancer immunotherapy.
Acknowledgments
the authors thank Serviço de Anatomia Patológica and Serviço de Hematologia of HSM-CHLN, and Serviço de Pediatria and Serviço de Hematologia of IPOLFG, for provision of tumor samples; and Dr. José Cabeçadas for helpful discussions.
Footnotes
Funding: this work was supported by the European Molecular Biology Organization (YIP Installation Grant to BS-S), Fundação Calouste Gulbenkian (SDH Oncologia 2008 - Projecto 99293) and Fundação para a Ciência e Tecnologia/FCT (PTDC/BIA-BCM/71663/2006).
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
BSS was the principal investigator and takes primary responsibility for the paper. AQG, DVC and TL performed the laboratory work for this study. ARG performed the bioinformatics analysis of the data. CF, JFL, JTB and MGS provided clinical samples and suggestions. AQG, DVC and BSS wrote the manuscript.
The authors reported no potential conflicts of interest.
References
- 1.Kabelitz D, Wesch D, He W. Perspectives of gammadelta T cells in tumor immunology. Cancer Res. 2007;67(1):5–8. doi: 10.1158/0008-5472.CAN-06-3069. [DOI] [PubMed] [Google Scholar]
- 2.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, et al. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294 (5542):605–9. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Yang W, Pan M, Scully E, Girardi M, Augenlicht LH, et al. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J Exp Med. 2003;198(3):433–42. doi: 10.1084/jem.20030584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, Eltoum IE, Guo B, Beck BH, Cloud GA, Lopez RD. Protective immunosurveillance and therapeutic antitumor activity of gammadelta T cells demonstrated in a mouse model of prostate cancer. J Immunol. 2008;180(9):6044–53. doi: 10.4049/jimmunol.180.9.6044. [DOI] [PubMed] [Google Scholar]
- 5.Street SE, Hayakawa Y, Zhan Y, Lew AM, MacGregor D, Jamieson AM, et al. Innate immune surveillance of spontaneous B cell lymphomas by natural killer cells and gammadelta T cells. J Exp Med. 2004;199(6):879–84. doi: 10.1084/jem.20031981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita CT, Jin C, Sarikonda G, Wang H. Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vgamma2Vdelta2 T cells: discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol Rev. 2007;215:59–76. doi: 10.1111/j.1600-065X.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 7.Correia DV, d’Orey F, Cardoso BA, Lanca T, Grosso AR, deBarros A, et al. Highly active microbial phosphoantigen induces rapid yet sustained MEK/Erk- and PI-3K/Akt-mediated signal transduction in anti-tumor human gammadelta T-cells. PLoS One. 2009;4(5):e5657. doi: 10.1371/journal.pone.0005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sicard H, Al Saati T, Delsol G, Fournie JJ. Synthetic phosphoantigens enhance human Vgamma9Vdelta2 T lymphocytes killing of non-Hodgkin’s B lymphoma. Mol Med. 2001;7(10):711–22. [PMC free article] [PubMed] [Google Scholar]
- 9.Kunzmann V, Wilhelm M. Anti-lymphoma effect of gammadelta T cells. Leuk Lymphoma. 2005;46(5):671–80. doi: 10.1080/10428190500051893. [DOI] [PubMed] [Google Scholar]
- 10.Tokuyama H, Hagi T, Mattarollo SR, Morley J, Wang Q, Fai-So H, et al. V gamma 9 V delta 2 T cell cytotoxicity against tumor cells is enhanced by monoclonal antibody drugs--rituximab and trastuzumab. Int J Cancer. 2008;122(11):2526–34. doi: 10.1002/ijc.23365. [DOI] [PubMed] [Google Scholar]
- 11.Dieli F, Vermijlen D, Fulfaro F, Caccamo N, Meraviglia S, Cicero G, et al. Targeting human {gamma}delta} T cells with zoledronate and interleukin-2 for immunotherapy of hormone-refractory prostate cancer. Cancer Res. 2007;67(15):7450–7. doi: 10.1158/0008-5472.CAN-07-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi H, Tanaka Y, Yagi J, Osaka Y, Nakazawa H, Uchiyama T, et al. Safety profile and anti-tumor effects of adoptive immunotherapy using gamma-delta T cells against advanced renal cell carcinoma: a pilot study. Cancer Immunol Immunother. 2007;56(4):469–76. doi: 10.1007/s00262-006-0199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennouna J, Bompas E, Neidhardt EM, Rolland F, Philip I, Galea C, et al. Phase-I study of Innacell gammadelta, an autologous cell-therapy product highly enriched in gamma9delta2 T lymphocytes, in combination with IL-2, in patients with metastatic renal cell carcinoma. Cancer Immunol Immunother. 2008;57(11):1599–609. doi: 10.1007/s00262-008-0491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilhelm M, Kunzmann V, Eckstein S, Reimer P, Weissinger F, Ruediger T, et al. Gammadelta T cells for immune therapy of patients with lymphoid malignancies. Blood. 2003;102(1):200–6. doi: 10.1182/blood-2002-12-3665. [DOI] [PubMed] [Google Scholar]
- 15.Gertner-Dardenne J, Bonnafous C, Bezombes C, Capietto AH, Scaglione V, Ingoure S, et al. Bromohydrin pyrophosphate enhances antibody-dependent cell-mediated cytotoxicity induced by therapeutic antibodies. Blood. 2009;113(20):4875–84. doi: 10.1182/blood-2008-08-172296. [DOI] [PubMed] [Google Scholar]
- 16.Abe Y, Muto M, Nieda M, Nakagawa Y, Nicol A, Kaneko T, et al. Clinical and immunological evaluation of zoledronate-activated Vgamma9gammadelta T-cell-based immunotherapy for patients with multiple myeloma. Exp Hematol. 2009;37(8):956–68. doi: 10.1016/j.exphem.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Team RDC Computing RFfS. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2009. [Google Scholar]
- 18.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 21.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry WH R, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 22.Benjamini YH, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 23.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26(3):303–4. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Herold MJ, Kimmel B, Muller I, Rincon-Orozco B, Kunzmann V, et al. Reduced expression of the mevalonate pathway enzyme farnesyl pyrophosphate synthase unveils recognition of tumor cells by Vgamma9Vdelta2 T cells. J Immunol. 2009;182(12):8118–24. doi: 10.4049/jimmunol.0900101. [DOI] [PubMed] [Google Scholar]
- 25.Vantourout P, Mookerjee-Basu J, Rolland C, Pont F, Martin H, Davrinche C, et al. Specific requirements for Vgamma9Vdelta2 T cell stimulation by a natural adenylated phosphoantigen. J Immunol. 2009;183(6):3848–57. doi: 10.4049/jimmunol.0901085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scotet E, Martinez LO, Grant E, Barbaras R, Jeno P, Guiraud M, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity. 2005;22(1):71–80. doi: 10.1016/j.immuni.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Kong Y, Cao W, Xi X, Ma C, Cui L, He W. The NKG2D ligand ULBP4 binds to TCRgamma9/delta2 and induces cytotoxicity to tumor cells through both TCRgammadelta and NKG2D. Blood. 2009;114(2):310–7. doi: 10.1182/blood-2008-12-196287. [DOI] [PubMed] [Google Scholar]
- 28.Sole X, Bonifaci N, Lopez-Bigas N, Berenguer A, Hernandez P, Reina O, et al. Biological convergence of cancer signatures. PLoS One. 2009;4(2):e4544. doi: 10.1371/journal.pone.0004544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3(10):781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 30.Gomes AQ, Correia DV, Silva-Santos B. Non-classical major histocompatibility complex proteins as determinants of tumour immunosurveillance. EMBO Rep. 2007;8(11):1024–30. doi: 10.1038/sj.embor.7401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28(4):571–80. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strid J, Roberts SJ, Filler RB, Lewis JM, Kwong BY, Schpero W, et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat Immunol. 2008;9(2):146–54. doi: 10.1038/ni1556. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Lee JH, Kim KY, Song HK, Kim JK, Yoon SR, et al. The interferon-inducible 9–27 gene modulates the susceptibility to natural killer cells and the invasiveness of gastric cancer cells. Cancer Lett. 2005;221(2):191–200. doi: 10.1016/j.canlet.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Roth P, Mittelbronn M, Wick W, Meyermann R, Tatagiba M, Weller M. Malignant glioma cells counteract antitumor immune responses through expression of lectin-like transcript-1. Cancer Res. 2007;67(8):3540–4. doi: 10.1158/0008-5472.CAN-06-4783. [DOI] [PubMed] [Google Scholar]
- 35.Pellet-Many C, Frankel P, Jia H, Zachary I. Neuropilins: structure, function and role in disease. Biochem J. 2008;411(2):211–26. doi: 10.1042/BJ20071639. [DOI] [PubMed] [Google Scholar]