SUMMARY
We report an interaction between poxA, encoding a paralog of lysyl tRNA-synthetase, and the closely linked yjeK gene, encoding a putative 2,3-β-lysine aminomutase, that is critical for virulence and stress resistance in Salmonella enterica. Salmonella poxA and yjeK mutants share extensive phenotypic pleiotropy including attenuated virulence in mice, an increased ability to respire under nutrient limiting conditions, hypersusceptibility to a variety of diverse growth inhibitors and altered expression of multiple proteins including several encoded on the SPI-1 pathogenicity island. PoxA mediates post-translational modification of bacterial elongation factor P (EF-P), analogous to the modification of the eukaryotic EF-P homologue, eIF5A, with hypusine. The modification of EF-P is a mechanism of regulation whereby PoxA acts as an aminoacyl-tRNA synthetase that attaches an amino acid to a protein resembling tRNA rather than to a tRNA.
INTRODUCTION
Salmonella enterica has the ability to withstand many of the varied effector mechanisms employed by the host immune system during infection (Prost et al., 2007). Salmonella mutants that are deficient for replication in macrophages are avirulent (Baumler et al., 1994; Fields et al., 1986). An important aspect of Salmonella virulence is the ability to reside and multiply in host cells within a specialized compartment known as a Salmonella-containing vacuole (SCV, (Bakowski et al., 2008)). Salmonella adjusts many aspects of its physiology during the transition from life as an extracellular pathogen to one that resides within a host cell. Type-III and flagellar secretion systems important for the initial process of intestinal infection are rapidly turned off while a separate SPI-2 encoded type-III secretion system is induced to prevent fusion of the nascent Salmonella-containing endosome with membrane compartments of the endosomal/lysosomal pathway (Brumell and Grinstein, 2004; Steele-Mortimer, 2008).
Intracellular bacterial pathogens must also make metabolic adaptations to utilize the array of nutrients available within the host cell, particularly since the host cell can sequester certain metabolites including tryptophan, magnesium, manganese and iron (Appelberg, 2006; Schaible and Kaufmann, 2005; Thompson et al., 2006). Salmonella strains deficient in glycolysis or in the biosynthesis of chorismate, nucleotides, or valine and isoleucine are attenuated in infection models (Fields et al., 1986; Hoiseth and Stocker, 1981; Leung and Finlay, 1991). Substrates utilized by intracellular pathogens also vary during the course of infection, suggesting that adaptive changes in metabolism must occur over time. Isocitrate lyase, an enzyme required for growth on C2 carbon sources such as acetate and acetyl-CoA derived from fatty acids, is dispensable during acute Salmonella enterica Sv. Typhimurium (S. Typhimurium) infection but is required for growth of the bacteria during long-term chronic infection (Fang et al., 2005). Furthermore, phagocyte-derived reactive oxygen and nitrogen species can interfere with bacterial growth by damaging iron- and thiol-containing enzymes necessary for redox balance and biosynthesis of important metabolites including branched-chain amino acids (Ren et al., 2008; Richardson et al., 2006; Richardson et al., 2008). Staphylococcus aureus employs an NO•-inducible lactate dehydrogenase to regenerate oxidizing equivalents (NAD+) from the buildup of NADH caused by the disruption of pyruvate-formate lyase and pyruvate dehydrogenase by NO• (Richardson et al., 2008).
During a screen for Salmonella mutants resistant to S-nitrosoglutathione, we uncovered mutations in two poorly characterized genes poxA (yjeA or STM4344) and yjeK (STM4333), putatively involved in tRNA and lysine biosynthesis, respectively. In a number of phylogenetically distant bacterial species, the poxA and yjeK genes are linked to each other and to a third gene, efp, encoding the bacterial elongation factor P (EF-P) involved in protein synthesis (Bailly and de Crecy-Lagard, 2010). Here we report that these genes operate in a common pathway critical for virulence and resistance to several classes of antibiotics. 2D-gel analysis revealed that the poxA and yjeK mutant strains display nearly identical phenotypes and changes in protein expression profiles including several proteins involved in metabolism and factors encoded by the Salmonella pathogenicity island, SPI-1. Furthermore we demonstrate by biochemical means that PoxA is the enzyme responsible for the previously observed post-translational modification of EF-P. Our data demonstrate that YjeK, PoxA and EF-P affect the expression of factors that play an essential role in Salmonella virulence and intrinsic resistance to diverse antimicrobial compounds.
RESULTS
Genetic screen uncovers loci involved in susceptibility to GSNO
Nitric oxide (NO) produced by phagocytes through the action of the inducible nitric oxide synthase (iNOS) inhibits bacterial growth through a variety of mechanisms including modification of protein thiols and metal centers, such as heme groups and iron sulfur clusters (Pullan et al., 2007). Cellular glutathione can scavenge nitric oxide to generate the nitrosylated form of the tripeptide S-nitroso-glutathione (GSNO) (Foster et al., 2003). Exogenously added GSNO is cytostatic for Salmonella and import of GSNO into the cytoplasm is essential for this effect (De Groote et al., 1995). To identify cytosolic factors that may be necessary for the cytostatic effect we initiated a screen to obtain Salmonella mutants that could grow in the presence of 500 μM GSNO.
Salmonella enterica Sv. Typhimurium (S. Typhimurium) strain 14028s was mutagenized with a derivative of transposon Tn10 (Rappleye and Roth, 1997) to generate twenty independently derived pools containing a total of approximately 40,000 independent random insertions. Pools from the library of mutant strains were plated on M9 minimal agar containing 500 μM GSNO to select for colonies that displayed enhanced growth compared to the wild-type strain. 152 individual colonies were picked, and the transposon insertions were introduced into a fresh wild-type background by phage P22-mediated transduction. These fresh transductants were screened again for GSNO resistance. Resistant mutants were mapped using inverse PCR (see Experimental Procedures).
Seven independent transposon insertions conferring high levels of GSNO resistance mapped to the poxA (also called yjeA) gene encoding a protein that has considerable homology (32% identitity, 50% similarity) to the C-terminal domain of Escherichia coli LysRS, a class-II lysyl tRNA synthetase (Figure 1A and Table S1). Class II LysRS ligases are generally composed of a C-terminal catalytic domain and an N-terminal anticodon binding domain necessary for the proper selection of the tRNA. The catalytic domains of Class II LysRS enzymes are responsible for the activation of Lys, through a lysyl-adenylate intermediate, and for the subsequent transfer of Lys on the 3'OH terminus of the tRNA. PoxA is significantly shorter than LysRS and lacks an N-terminal anticodon binding domain. Several truncated aaRS paralogs have been characterized, displaying functions that utilize aminoacyl-adenylates to accomplish reactions mechanistically similar to tRNA aminoacylation. For instance, a truncated asparaginyl-tRNA synthetase paralog was implicated in asparagine biosynthesis (Roy et al., 2003), and a truncated glutamyl-tRNA synthetase was found to post-transcriptionally modify tRNA (Blaise et al., 2005; Campanacci et al., 2004; Dubois et al., 2004; Salazar et al., 2004). Other notable functions for aaRS paralogs include roles in the biosynthesis of biotin and histidine (Artymiuk, 1995; Sissler et al., 1999).
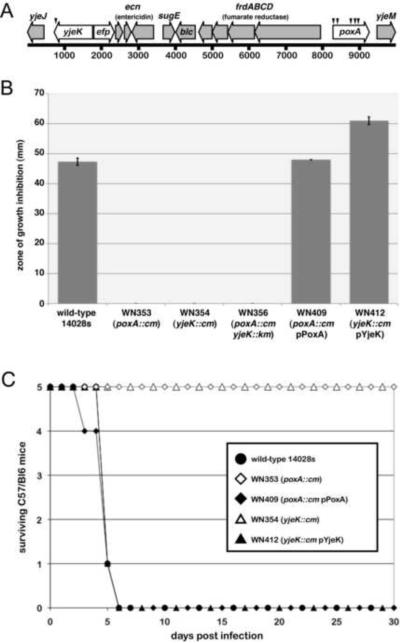
Figure 1. poxA and yjeK mutant S. Typhimurium are attenuated for virulence and resistant to GSNO.
A. The close genetic linkage of poxA, yjeK and efp in S. Typhimurium is shown. Small numbered arrows above the genes in the top row indicate the sites of the independent T-POP insertions as described in Table S1. B. Results from a disk diffusion assay in which 1 cm paper disks saturated with 15 μl of a 500 mM solution of GSNO were placed in the center of bacterial lawns on a 15 cm petri dish containing 25 ml of M9 agar are shown (see Experimental Procedures). Zones of inhibition were measured after 16 h growth and error bars represent standard deviation of three independent experiments. C. Approximately 1200 colony forming units of S. Typhimurium 14028s wild-type (WT), poxA::cm mutant (WN353), yjeK::cm mutant (WN354) and strains complemented in trans with plasmid vectors containing intact copies of poxA or yjeK (WN409 and WN413) were each inoculated intraperitoneally into five 6 week-old female C57/Bl6 mice. Infected animals were monitored for disease and moribund animals euthanized.
The gene encoding poxA was first indentified in a screen for E. coli strains deficient for the ability to convert pyruvate to acetate and CO2 (Chang and Cronan, 1982). A different protein, PoxB (genetically unlinked to poxA), was later identified as the enzyme that catalyzes the conversion of pyruvate to acetate (Chang and Cronan, 1983). The mechanism by which PoxA positively affects the activity of PoxB has not been established. Mutations in poxA have been further characterized in both Salmonella and E. coli. The resulting phenotypes of these strains are highly pleiotropic and, for the most part, independent of PoxB. Salmonella poxA mutants display a mild growth defect, reduced acetolactate synthase and pyruvate oxidase activity, and are sensitive to a variety of antimicrobial agents that are mechanistically and structurally dissimilar to one another (Van Dyk et al., 1987). Salmonella poxA mutants have also been reported to be highly attenuated for virulence (Kaniga et al., 1998).
Another independent transposon insertion that conferred high levels of GSNO resistance mapped to the 3' end of a poorly characterized gene, yjeK, encoding a protein with strong similarity to the family of 2,3-β-lysine aminomutases. These iron-sulfur cluster-containing enzymes catalyze the transfer of the alpha-carbon amino group of lysine to the β-carbon to generate the product β-lysine. Lysine aminomutases display stereospecificity and YjeK belongs to the group that catalyze the conversion of L-lysine to (R)-β-lysine (Behshad et al., 2006). No biological function for this enzyme has been ascribed thus far in either E. coli or Salmonella, although in other bacterial species β-lysine can act as a compatible solute to protect the cell from osmotic stress or as an intermediate for the generation of secondary metabolites (Muller et al., 2005).
To verify that mutations in poxA and yjeK confer increased resistance to GSNO, defined mutations were constructed in each gene using λ red-gam-mediated replacement (Datsenko and Wanner, 2000). The GSNO sensitivity of these newly constructed strains was assessed using a disk diffusion assay, and both strains were markedly more resistant than the Salmonella wild-type strain (Figure 1B). Wild-type levels of sensitivity to GSNO could be restored by reintroducing either poxA or yjeK in trans on plasmids containing the poxA and yjeK open reading frames cloned downstream of the arabinose-inducible PBAD promoter (Guzman et al., 1995). Notably, complete complementation was observed even under conditions known to repress expression from this vector (e.g. glucose-containing medium in the absence of arabinose), suggesting that the PoxA and YjeK proteins do not need to be expressed at high levels for function. Neither poxA nor yjeK mutants displayed increased resistance to other nitric oxide generating compounds including spermine NONOate, nor did they display altered susceptibility to hydrogen peroxide (data not shown), suggesting that the effects observed are specific to GSNO and not due to a generalized change in susceptibility to reactive oxygen or nitrogen species.
The phylogenetic distribution of poxA and yjeK genes among different bacterial species was explored using tools available on the MicrobesOnline website (Alm et al., 2005). This analysis revealed that the two genes are closely linked in several bacterial species within the alpha- and gamma-proteobacteria, often immediately adjacent to one another. In each of these cases the two genes are also linked, sometimes in the same operon, with the gene encoding elongation factor P (EF-P) (Bailly and de Crecy-Lagard, 2010). Genes that cluster together despite relatively large phylogenetic distances often share a common function. Given their genetic proximity and similar phenotypes with regard to GSNO, we explored the possibility that YjeK and PoxA function in the same pathway.
Mutations in poxA and yjeK are attenuated for virulence in mouse models of infection
To determine if poxA and yjeK mutants shared similar virulence phenotypes, each mutant was assayed for its ability to cause disease in mice following intraperitoneal inoculation. Five C57/BL6 mice were infected with approximately 1200 colony forming units of S. Typhimurium 14028s, the isogenic poxA or yjeK mutant strains, or the mutant strains complemented with their corresponding genes in trans on plasmid vectors (Figure 1C). Wild-type Salmonella and complemented strains caused lethality within 5 days of infection whereas both poxA and yjeK mutants were highly attenuated for virulence. The mice infected with these mutants appeared healthy throughout the course of the experiment.
poxA and yjeK mutations phenocopy for a large number of disparate phenotypes including sensitivity to antibiotics
To assess the degree of phenotypic overlap between Salmonella strains carrying mutations in poxA or yjeK, we subjected each of these strains to analysis by “phenotype microarray” (Bochner et al., 2001). Twenty 96-well microplates, with each well containing a different nutrient or growth inhibitor condition, were used to simultaneously assay nearly 1,900 phenotypes. Cellular respiration is measured by the turnover of a tetrazolium redox dye resulting in the formation of a soluble and stable purple compound that accumulates in the well over the incubation period. This color is measured throughout the assay with a robot-controlled camera, and the rate of formation is plotted over time. The area under the curve is subsequently used as a measure of total respiration over the course of the assay.
The results of the phenotype microarray clearly indicate that the phenotypes of cells carrying mutations in either poxA or yjeK are highly pleiotropic and nearly identical for almost all of the 1,900 conditions tested (Figure S1, Table S3). Overall both mutants displayed an increased ability to turnover the tetrazolium dye under a number of nutrient-limiting conditions compared to the wild-type strain. This enhanced respiratory activity was particularly evident for the utilization of amino acids and dipeptides as nitrogen sources (in particular, those containing methionine or branched-chain amino acids) as well as for a number of inorganic and organic sources of phosphate or sulfur (Table S3). One notable exception is the inability of either mutant to use μ-glutamyl-glycine as a nitrogen source. Enhanced metabolism was also observed for poxA and yjeK mutants when using glutamine as a carbon source.
In contrast to their increased respiratory activity in the presence of various nutrient sources, the poxA and yjeK mutants displayed wide-ranging defects in response to a large number of cellular stressors. Both mutants displayed enhanced susceptibility to antibiotics that fall into several distinct pharmacological and structural classes including antimicrobial peptides, detergents, lipophilic chelators, heavy metals, and various inhibitors of cell wall synthesis, protein synthesis, RNA synthesis, and DNA gyrase. The wide spectrum of compounds that dramatically inhibited the growth of these mutants suggests that the defect lies in a general stress response. In total each mutant displayed measurable phenotypes that differed from the wild-type strain under 300 different conditions not including variation between wells that differed only by dose.
poxA and yjeK mutants are epistatic and display increased susceptibility to amino-glycosides and sulfometuron methyl
If poxA and yjeK operate in the same pathway, we predicted that they should display genetic epistasis (i.e., the phenotypes of the poxA/yjeK double mutant should be no more pronounced than that of either single mutant). The concentration of the aminoglycoside gentamicin sufficient to inhibit growth of the poxA, yjeK and double mutant strains was assessed in a liquid growth assay (Figure 2A). Corroborating the results of the phenotype microarray, mutations in either poxA or yjeK led to an increased susceptibility to gentamicin (MIC of 6.3 μg/ml) as compared to 12.5 μg/ml for the wild-type, although the poxA mutant displayed a more pronounced growth suppression in the presence of 6.3 μg/ml gentamicin than the yjeK mutant. The poxA yjeK double mutant was also susceptible to gentamicin, but no more so than a poxA single mutant.
Figure 2. S. Typhimurium poxA and yjeK mutations are epistatic and confer increased sensitivity to gentamicin and sulfometuron methyl.
A. A broth dilution assay was performed to determine the minimal inhibitory concentration of gentamicin for poxA, yjeK and double mutant strains. Mutant and wild-type Salmonella strains were grown in two-fold serial dilutions of gentamicin starting at 25 μg/ml, and growth was detected by a change in turbidity after 16 h. B. Susceptibility of mutant strains to the acetolactate synthase inhibitor, sulfometuron methyl, was assessed by disk diffusion assay, error bars represent standard deviation from three independent experiments. Further evidence of phenotypic overlap is given in Figure S3.
Salmonella poxA mutants have also been reported to be hypersusceptible to sulfometuron methyl, a compound that specifically inhibits acetolactate synthase, the first enzyme in the synthesis of isoleucine and valine from pyruvate (Van Dyk et al., 1987). In disk diffusion assays for growth in the presence of this inhibitor, the poxA yjeK double mutant was more sensitive than wild-type to sulfometuron methyl but not more susceptible than either single mutant (Figure 2B). We noted that our strain was less susceptible than has been previously reported for poxA mutants in the S. Typhimurium LT2 strain background. Differences in alpha-ketobutyrate susceptibility were reported for strains used by Kaniga et al. (Kaniga et al., 1998) and an earlier study by Van Dyk et al. (Van Dyk et al., 1987) using an LT2 strain, suggesting that the LT2 background may have additional determinants that further enhance its susceptibility to inhibitors of isoleucine/valine synthesis.
PoxA post-translationally modifies elongation factor P
To understand the role of PoxA and YjeK, we examined the possibility that they operate through the third closely linked gene, EF-P. The crystal structure of the EF-P protein indicates that it, like many other factors involved in translation, adopts a tertiary structure similar in shape to that of tRNA (Hanawa-Suetsugu et al., 2004). Indeed the EF-P protein has recently been co-crystalized in complex with the ribosome and found to interact with the ribosomal peptidyl-transferase center (Blaha et al., 2009) consistent with a role in translation initiation. Moreover, it has recently been reported that EF-P is post-translationally modified on Lys34 by a moiety with a mass consistent with spermidine or lysine (Aoki et al., 2008). Homologues of EF-P exist in all kingdoms of life (called eIF5A in eukaryotes) and Lys34 is absolutely conserved in all species, although some species encode second paralogous copies of EF-P that lack the conserved lysine (Bailly and de Crecy-Lagard, 2010; Ganoza et al., 2002). In eukaryotes this lysyl residue is modified post-translationally to yield a unique amino acid called hypusine (Park, 2006). Although the enzymes necessary for the hypusine modification are well understood in eukaryotes, no prokaryotic counterparts have been identified.
Given the sequence and structural similarity of PoxA to a class II LysRS, the possibility that PoxA post-translationally modifies EF-P was examined. To test the ability of PoxA to modify EF-P, each protein was purified and added to a reaction containing [14C]-lysine and ATP (Figure 3). EF-P was rapidly labeled in the presence of PoxA, providing direct evidence that PoxA is the enzyme responsible for the observed modification of EF-P purified in vivo. The PoxA-mediated modification of EF-P with lysine results in a 128 Da change in the mass of the protein consistent with the mass previously determined for in vivo modified EF-P (Figure S2; Aoki et al. 2008). PoxA was unable to lysylate mutant EF-P when the conserved Lys34 residue previously shown to carry the modification was substituted with alanine. Like LysRS, PoxA exhibits Lys-dependent ATP-PPi exchange activity consistent with the expected formation of a lysyladenylate intermediate by the class II aaRS-type active site (Figure S3); however PoxA is unable to transfer lysine to tRNALys (data not shown).
Figure 3. Lysylation of EF-P by PoxA in vitro.
[14C]-Lys addition to EF-P catalyzed by PoxA was monitored by SDS-PAGE after various incubation intervals at 37°C. The assay was performed in presence or absence of PoxA and with EF-P wild-type (wt) or the mutant K34A. Proteins were visualized by Coomassie staining (upper panel) and [14C]-Lys-EF-P by phosphorimaging (lower panel). The modification has a mass of 128 Da (Figure S2) and no modification of EF-P by PoxA was observed in the absence of ATP (Figure S3).
Structural determination of the PoxA enzyme
To further explore the mechanism of PoxA the structure of the full-length molecule in complex with AMP was solved through crystallographic methods to a resolution of 1.95 Å (Figures 4 and S4, Table 1, supplementary Experimental Procedures). Molecular replacement, using a model generated automatically from the protein sequence by the Swiss-Model server (http://swissmodel.expasy.org/SWISS-MODEL.html), determined that there were two molecules in the asymmetric unit, and after refinement of the model, a buried surface area of 7580 Å2 was calculated between the two molecules. The structure is deposited in the Protein Data Bank (ID 3G1Z). The interaction of PoxA with its AMP moiety is maintained by a network of stacking interactions and hydrogen bonds with the residues of motif II that are responsible for ATP binding by class II aaRS enzymes (Figure S4).
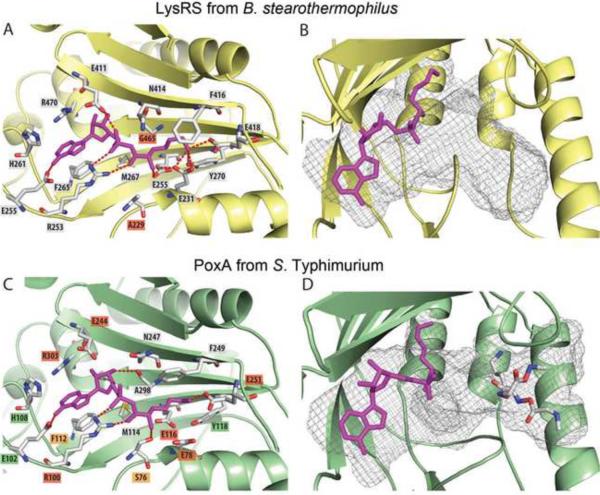
Figure 4. Comparison of PoxA and LysRS active sites.
LysRS of Bacillus stearothermophilus (PDB ID 3E9H, enzyme collored yellow) complexed with L-Lysyl-sulfamoyl adenosine (panels A and B). Docking models of L-Lysyl-adenylate (purple) and of the peptide GKG in the active site of PoxA from S. typhimurium (PDB ID 3G1Z, enzyme colored green) are shown in panels C and D, respectively. The residues constituting the surface of the Lysyl-adenylate binding site are represented and the positions differing in PoxA and LysRS are indicated in red (A). The cavity of the active site is represented by gray mesh (B and D). Salmonella strains expressing PoxA single mutants of the residues forming the active site cavity displayed three categories of phenotype for GSNO resistance and growth on AB2 agar: residues essential for PoxA activity are showed in red, those having no effect in green and those inducing an intermediate phenotype are showed in orange (panel C). Data regarding the specific effects of residue mutations are given in Table S4.
Table 1.
- Data Collection and Refinement Statistics for the PoxA/AMP Complex
| Data collection | |
|---|---|
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å), β (°) | 69.9, 68.9, 73.3, 110.3, |
| Wavelength (Å) | 1.54178 |
| Resolution (Å) | 50–1.85 |
| Rmerge (%)a | 0.146(0.561) |
| I/σI | 20.0(3.8) |
| Completeness (%) | 96.1(97.0) |
| Redundancy | 4.0(4.1) |
| Refinement | |
|---|---|
| Resolution (Å) | 47.5–1.95 |
| No. reflections | 43960(2339) |
| Rwork (%)b | 18.4 |
| Rfree (%)c | 23.9 |
| No. atoms | |
| Protein | 5228 |
| Water | 645 |
| Other (including nucleotide) | 77 |
| B-factors (Å2) | |
| Overall | 25.7 |
| Protein | 24.5 |
| Water | 34.8 |
| Other (including nucleotide) | 28.0 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.4 |
Values in parentheses are for the highest-resolution shell.
Rwork = Σ|Fobs − Fcale|/Σ|Fobs|, where Fobs and Fcalc are the observed and the calculated structure factors, respectively.
Rfree calculated using 5% of total reflections randomly chosen and excluded from the refinement
PoxA exhibits a wider active site cavity than that of the well-characterized LysRS from Bacillus stearothermophilus ((Sakurama et al., 2009) compare Figure 4B and 4D). The residues and the geometry of the backbone forming the cavity of the active site of both enzymes are highly conserved (RMS =1.1 Å) with only residues Gly465 and Ala229 of B. stearothermophilus LysRS substituted in PoxA by Ala298 and Ser76, respectively. We employed molecular modeling techniques to explore how PoxA would be able to accommodate EF-P and the lysyl-adenylate intermediate in its active site (Figure 4). A docking model of PoxA with L-lysyl-adenylate built using Autodock 3.0 revealed that LysRS and PoxA each accommodate the L-lysyl-adenylate in their binding pockets in an extended conformation. The lateral chain and the ε-NH2 group of the lysyl group are located in an acidic pocket found in the active site of both enzymes.
A fragment of EF-P was modeled in the active site of PoxA using the peptide GKG as a mimic of the residue Lys34. Interestingly, the peptide could readily be fitted within a part of the active site that extended the L-lysyl-adenylate pocket. As predicted, the GKG peptide occupies a location that differs from that of a tRNA 3' hydroxyl group in a classII aaRS (Moulinier et al., 2001). The ε-NH2 of the lysyl group of the GKG peptide and the activated α-carbonyl group of the L-lysyl-adenylate are separated by 9 Å. Altogether, these observations indicate that while the L-lysyl-adenylate intermediate binds PoxA and LysRS in a similar way, tRNA and EF-P may bind in different modes.
Identification of residues critical for PoxA activity in vivo
The active site of the PoxA enzyme predicted from our structural analaysis suggested 12 residues that may be essential for function. Alanine mutations were constructed for eleven of these residues (S76, E78, R100, E102, H108, F112, E116, Y118, E244, E251, R303) in the chromosomal poxA gene. The twelfth predicted active site residue, A298, was not mutated in this study. The effect of each Salmonella mutant was assessed for GSNO-resistance and growth on AB2 agar compared to wild-type, which has proven useful as a rapid screen for PoxA activity (Table S4). PoxA mutants were previously described to be defective for growth on AB2 agar, although the underlying reason for this phenotype remains unclear (Kaniga et al., 1998).
Six residues were found to be critical for activity since mutants displayed GSNO resistance and growth on AB2 identical to that of a poxA deletion mutant. Essential residues include the glutamates at position 78, 116 and 244 that form the acidic pocket predicted to interact with the ε-NH2 group of the lysyl side chain. Also required for PoxA activity are residues Arg100, Glu244 and Arg303, that are invariant in class II aaRS enzymes. These positions are crucial for substrate binding and for the formation and stabilization of the aminoacyl-adenylate within the active site (for review see (First, 2005). Arg100, within the conserved motif 2 of class II aaRSs (Eriani et al., 1990) is critical for the positioning of the aa and ATP within the active site by interacting with the α-carboxylate of the amino acid and with the α-phosphate of ATP. These interactions remain intact between the arginine in motif 2 and the corresponding functional groups in the aminoacyl-adenylate after the amino acid activation. The arginine in motif 2 is also critical for catalysis by increasing the electrophilicity of the α-phosphate of ATP, and by stabilizing the transition state of the reaction (Desogus et al., 2000; First, 2005). Arg303 within motif 3 stacks the adenine ring of the aminoacyl-adenylate and binds ATP by forming a salt bridge with its γ-phosphate. Residue Glu244 has been shown, in numerous class II aaRSs, including LysRS, to coordinate Mg2+ bound to ATP and to form hydrogen bonds with the 3'OH group of the ribose of ATP or of the aminoacyl-adenylate (Desogus et al., 2000).
Mutants with alteration of PoxA residues Phe112 and Ser76 displayed intermediate GSNO resistance and could grow on AB2 media indicating only partial loss of activity. The Glu102, His108 and Tyr118 mutants behaved similar to wild-type Salmonella indicating these three residues are dispensable for PoxA activity in vivo (Fig 7). Together the structural, mutational and biochemical analyses indicate that PoxA is in many aspects typical of a class 2 aaRS, in that residues critical for aaRS activity are also critical for PoxA function in vivo.
Analysis of protein expression differences in poxA and yjeK mutants
Given its similarity to a lysyl-tRNA synthetase and ability to modify EF-P, we hypothesized that PoxA acts through EF-P to control gene expression at a post-transcriptional level, perhaps on a specific subset of genes involved in stress adaptation or virulence. To further test this hypothesis, 2D-SDS-PAGE electrophoresis was performed on total bacterial protein to compare the levels of individual proteins in wild-type and isogenic poxA mutant Salmonella. Relative protein expression levels were quantified by DIGE (difference in gel electrophoresis) using data pooled from three different biological replicates with two technical replicates for each (Figure S5).
Eighty-five protein spots were identified that showed altered expression in the poxA mutant strain under the culture conditions employed (selected as those with a p-value < 0.05). Of these, 33 displayed higher expression in the wild-type strain while 52 displayed higher expression in the poxA mutant. 2D-PAGE analysis followed by colloidal Coomassie staining was used to determine that the protein expression profile of a yjeK mutant is nearly identical to that of a poxA mutant (Figure 5). This provides further evidence that YjeK and PoxA operate in the same pathway.
Figure 5. 2D SDS-PAGE of Salmonella poxA and yjeK mutants reveals similar protein expression profiles that differ from wild-type Salmonella.
Colloidal Coomassie-stained polyacrylamide gels after 2D gel electrophoresis on a PAGE gradient 8–16% acrylamide gel are shown. Isoelectric focusing was performed on a non-linear pH gradient ranging from 3–11. Arrows show prominent protein spots that are differ in expression between the wild-type strain (white arrows) and the yjeK/poxA mutants (grey arrows). Data on proteins displaying expression differences between the poxA mutant and wild-type are provided in Table 2 and Figure S5.
The identity of 25 proteins differentially expressed in a poxA mutant could be unambiguously determined using MALDI-TOF (Table 2, Figure S5). The identity of several spots in the dense region of the gel were difficult to unambiguously identify, as these spots contained peptides derived from multiple proteins. Two of the most strongly upregulated proteins included YjhT, a protein of unknown function, and GlnH, a periplasmic glutamine transporter. The most overrepresented class of proteins that were unambiguously identified is associated with the SPI-1 pathogenicity island required for Salmonella invasion of host cells (InvG, PrgH, SicA, SipA, SipC, SipD, SopB). Once Salmonella is internalized by host macrophages, these proteins are downregulated as the Salmonella virulence program switches to express effectors secreted by the type-III secretion system encoded on SPI-2 (Bustamante et al., 2008; Haraga et al., 2008). One of the picked spots identified HilA, a transcription factor that positively-regulates SPI-1 (Ellermeier and Slauch, 2007), as strongly upregulated by poxA, although the presence of other peptides in the sample prevented conclusive identification. The functional consequences of misregulated SPI-1 expression on virulence are discussed further below.
Table 2.
Proteins identified as differentially expressed inpoxA mutants.
| Gene identifier | Gene name | Fold change (wt/poxA)* | p-value | function |
|---|---|---|---|---|
| Protein spots identified with higher expression in poxA mutant than in wild-type 14028s background | ||||
| STM2882 | sipA | −2.06 | 0.02 | SPI-1 secreted effector SipA. Binds actin and induces host cytoskeleton rearrangements to promote membrane ruffling and bacterial internalization. |
| STM2898 | invG | − 2.51 | 0.0087 | Outer membrane secretin precursor InvG. Forms ring-shaped structure in outer membrane as part of TTSS. |
| STM2884 | sipC | − 5.71 | 0.0066 | SPI-1 secreted translocase SipC. Translocates secreted proteins into host cells. |
| STM2874 | prgH | − 1.63 | 0.022 | Needle complex inner membrane protein PrgH. Forms multimeric ring-shaped structures with PrgK as part of TTSS. |
| STM2883 | sipD | − 2.08 | 0.0095 | SPI-1 secreted translocase SipD. Translocates secreted proteins into host cells. |
| STM0830 | glnH | − 6.02 | 0.031 | Glutamine ABC transporter periplasmic protein |
| STM0013 | dnaJ | −2.57 | 0.018 | Chaperone protein DnaJ. Prevents aggregation of stress-denatured proteins in response to hyperosmotic and heat shock |
| STM1130 | yjhT | − 10.9, −6.07 | 0.013 | Kelch-domain containing protein |
| STM2370 | pdxB | −1.67 | 0.04 | Erythronate-4-phosphate dehydrogenase PdxB. Catalyzes the formation of 3-hydroxy-4-phospho-hydroxy-alpha-ketobutyrate from erythronate-4-phosphate. |
| STM0413 | Tsx | − 2.35 | 0.029 | Nucleoside channel Tsx. Receptor of phage T6 and colicin K. |
| STM2886 | sicA | − 3.14 | 0.0087 | SPI-1 secretion chaperone SicA. Partitioning factor for SipB and SipC to prevent their premature association. |
| STM1091 | sopB | − 6.79 | 0.0066 | SPI-1 secreted effector SopB. Hydrolyzes inositol phosphates and phosphoinositides. |
| STM1451 | Gst | −2.97 | 0.0094 | Glutathione S-transferase. Catalyzes the conjugation of glutathione with a wide range of endogenous and xenobiotic alkylating agents. |
| STM0608 | ahpC | −1.88 | 0.0051 | Alkyl hydroperoxide reductase subunit AhpC. Catalyzes the conversion of alkyl hydroperoxides to their corresponding alcohols. |
| STM1133 | STM1133 | −2.48 | 0.013 | Putative dehydrogenase |
| Protein spots identified with higher expression in wild-type than in poxA mutant | ||||
|---|---|---|---|---|
| STM4007 | glnA | 1.68 | 0.0091 | Glutamine synthetase GlnA. Forms glutamine from ammonia and glutamate with the conversion of ATP to ADP. |
| STM1830 | manX | 4.89 | 0.011 | Mannose-specific enzyme IIAB. Imports hexoses (including mannose, glucose, glucosamine, and fructose), releasing the phosphate esters into the cell cytoplasm in preparation for metabolism, primarily via glycolysis. |
| STM4581 | yjjK | 4.58 | 0.0032 | Putative ABC transporter ATP-binding protein |
| STM0215 | Map | 2.6 | 0.0089 | Methionine aminopeptidase Map. Catalyzes the removal of N-terminal amino acids from peptides and arylamides. |
| STM3996 | yihE | 2 | 0.046 | Serine/threonine protein kinase YihE. Catalyzes the phosphorylation of protein substrates at serine and threonine residues. |
| STM3865 | atpD | 3.81 | 0.013 | ATP synthase beta subunit AtpD. Produces ATP from ADP in the presence of a proton gradient across the membrane. The beta chain is a regulatory subunit. |
| STM0831 | Dps | 1.5 | 0.015 | DNA starvation/stationary phase protection protein Dps. Binds DNA nonspecifically to protect DNA from damage. |
| STM4149 | rplK | 1.63 | 0.014 | 50S ribosomal protein L11. Binds directly to 23S ribosomal RNA. |
Proteins with negative fold changes are more highly expressed in the poxA mutant than in the wild-type strain.
Three of the most strongly downregulated proteins in a poxA mutant are ManX, AtpD, and a putative ABC transporter YjjK. ManX is a central component of the IIABCDMan PTS system involved in hexose import that has been shown to be important for virulence (Bowden et al., 2009). AtpD, the β-subunit of the F1F0-ATPase, plays a major role in the energy balance of the cell and is critical for the generation of ATP via respiration (Fillingame and Divall, 1999).
DISCUSSION
We have discovered an interaction between three poorly characterized genes, poxA, yjeK, and efp that is essential for Salmonella virulence and resistance to a variety of unrelated stressors. EF-P was recently found to be post-translationally modified at a highly conserved lysyl-residue, but neither the chemical nature of this modification nor the enzymes responsible were identified (Aoki et al., 2008). Our observations indicate that PoxA can catalyze the ATP-dependent ligation of lysine to EF-P. We propose that YjeK performs further modification by converting the lysine substrate added to EF-P to (R)-β-lysine either before or after its ligation to EF-P (Figure 6). We conclude that PoxA and YjeK operate in a common pathway on the basis of several independent lines of evidence including: (1) genetic proximity in Gram-negative bacterial species that are phylogenetically distant (i.e., alpha- and gamma-proteobacteria); (2) extensive overlap of pleiotropic phenotypes including GSNO-resistance, sensitivity to a large number of antibiotics, loss of virulence, and over three-hundred phenotypes observed on the phenotype microarray; (3) genetic epistasis experiments demonstrating that the phenotypes of yjeK poxA double mutants are no more severe than those displayed by yjeK or poxA single mutants, and; (4) similar effects of each mutation on global protein expression profiles. Despite the general role EF-P is proposed to play in translation, it is clear that mutations in poxA and yjeK affect the expression of a relatively small subset of proteins under the conditions tested. It is likely that the effects of PoxA and YjeK on the expression of some of these proteins is indirect.
Figure 6. Model for PoxA, YjeK and EF-P function.
The eIF5A protein is modified at a conserved lysyl residue by two enzymes to generate the unique amino acid hypusine, which is required for cell growth and viability in eukaryotes. See (Park, 2006) for a review of the hypusine biosynthesis pathway. PoxA catalyzes aminoacylation (with lysine) of the same conserved lysyl residue of EF-P (see Figure 4). YjeK, a 2,3-β-lysine aminomutase, is shown here modifying the Lys-Lys side-group to generate a fully active form of EF-P. Alternatively, YjeK may act on the lysine substrate prior to its ligation to EF-P.
EF-P has homologues in eukaryotes and archaea, identified as eIF5A and aIF5A, respectively (Ganoza et al., 2002). Interestingly the same conserved lysyl residue in eIF5A (Lys50 in humans) is post-translationally modified to generate a unique amino acid (hypusine) in a reaction requiring two enzymes unrelated to PoxA and YjeK, suggesting convergent evolution of this pathway and conservation in all three kingdoms of life (Park, 2006). EF-P has been proposed to position fMet-tRNA for the formation of the first peptide bond during translation initiation (Blaha et al., 2009), and studies in yeast have implicated eIF5A in translation elongation (Saini et al., 2009). Although efp is reported to be essential in E. coli (Aoki et al., 1997), mutations of efp in Agrobacterium tumefacians interestingly lead to a pronounced growth defect, avirulence in plants, and increased susceptibility to detergents (Peng et al., 2001) resembling the phenotypes we have described for poxA and yjeK in Salmonella.
The proteomic analysis points to at least two factors that can account for the virulence and stress-related defects exhibited by poxA and yjeK mutants. First, the misregulated expression of SPI-1 encoded proteins could largely account for the observed attenuation of virulence. The SPI-1 type-III secretion system is necessary for the efficient penetration of the intestinal epithelial barrier and colonization of Peyer's patches (Baumler et al., 1997; Galan and Curtiss, 1989). However, expression of SPI-1 is rapidly terminated upon Salmonella entry into macrophages. Ectopic expression of SPI-1 in macrophages is cytotoxic, rapidly leading to macrophage death and the induction of inflammatory cytokines (Chen et al., 1996; Hersh et al., 1999). A failure to control SPI-1 expression in the macrophage environment would ultimately be detrimental to bacterial growth within the host (Fink and Cookson, 2007; Lara-Tejero et al., 2006). Other examples in which misregulated expression of virulence factors negatively affects Salmonella survival in mammalian hosts include constitutive mutants of the PhoP/PhoQ two-component regulatory system and strains lacking the YdgT virulence gene repressor (Coombes et al., 2005; Miller and Mekalanos, 1990).
Our proteomic analysis along with previous observations regarding PoxB expression also suggest that the PoxA/YjeK-mediated modification of EF-P is essential for the proper expression of proteins necessary for the appropriate utilization of alternative energy sources during nutrient deprivation, as encountered by Salmonella in the intracellular environment. The finding that poxA and yjeK play a role in regulating the use of alternative energy sources is supported by the known function of PoxA as a regulator of pyruvate oxidase activity (Chang and Cronan, 1982). Some PoxA/YjeK-regulated proteins are likely to be important for metabolic adaptation to the host environment. For example ManX(YZ), which displays decreased expression in poxA mutants, has recently been found to be required for Salmonella growth in macrophages (Bowden et al., 2009).
An unexpected result of the phenotype microarray assays is the persistent respiration observed in poxA and yjeK mutants under nutrient-poor conditions in which wild-type cells cease respiration. One interpretation of these findings is that wild-type bacteria enter a state of metabolic stasis during nutrient limitation or stress, whereas poxA and yjeK mutants fail to respond appropriately to these environmental conditions. Several researchers have found that disruption of metabolic pathways caused by errors in the translation of membrane proteins play a critical role in the lethality induced by antibiotics (Girgis et al., 2009; Kohanski et al., 2007; Kohanski et al., 2008; Tamae et al., 2008; van Stelten et al., 2009). A “stress-blind” phenotype in which poxA and yjeK mutants continue to respire under sub-optimal conditions might lead to the formation of toxic oxygen species and help to explain why such mutants are broadly sensitive to a large number of unrelated compounds and cellular stresses.
Finally, defects in poxA were observed in some cases to cause more severe phenotypes than mutations in yjeK, for example, with regard to gentamicin and sulfometuron methyl susceptibility. Similarly, we have observed that poxA and yjeK mutants grow poorly in certain preparations of M9 minimal medium, and the growth defect exhibited by poxA mutants is relatively more severe than that of a yjeK mutant (data not shown). A medium-dependent effect on growth was observed previously for a Salmonella poxA mutant strain that could be exacerbated by the specific formulation of antibiotic number 2 agar manufactured by Difco (Kaniga et al., 1998).
It is presently unknown whether the mRNA species whose translation is directly affected by PoxA-dependent modification of EF-P exhibit common features (e.g., 5' RNA signals or leader peptides). Future studies will address the specific mechanisms responsible for the virulence, metabolic, and stress-resistance defects displayed by PoxA- and YjeK-deficient bacteria as well as further define the role of YjeK and the nature of the EF-P modification.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
Experiments were conducted using wild-type Salmonella enterica serovar Typhimurium 14028s and mutants thereof. The origins and properties of strains used in this study are outlined in Table S2. Null mutations in poxA and yjeK were constructed using the red-gam recombinase as described (Datsenko and Wanner, 2000). Complementing plasmid vectors for poxA and yjeK were generated by cloning open reading frames into the arabinose inducible plasmid pBAD18. Chromosomal poxA mutations were generated by allelic replacement of the poxA T-POP transposons with mutagenized PCR products selected on fusaric acid agar as described (Bochner et al., 1980; Karlinsey, 2007; Maloy and Nunn, 1981). Details on primer sequences used and specific protocols are given in the Supplemental Methods.
Genetic Screen for GSNO Resistant Mutants
Salmonella was mutagenized with the T-POP tetA mini-transposon (Tn10d(del25)) (Rappleye and Roth, 1997) to generate a large set of independently generated pools totaling approximately 40,000 independent transposition events. These pools were plated on M9 agar containing 500 μM GSNO at a cell density of 4 × 103 per plate. Transposon insertions were rapidly mapped using a modification of a previous method by O'Toole and Kolter (O'Toole and Kolter, 1998). Specific details on library construction, mapping and the GSNO resistance screen are given in the Supplemental Methods.
Phenotype microarrays
Phenotype microarray testing was performed under contract by Biolog's PM Services group using a full set of 20 phenotype plates for Salmonella (Hayward, CA). Details are provided in the Supplementary Methods.
Mouse virulence assay
Female 6–8-week-old C56BL/6 (The Jackson Laboratory, Bar Harbor, ME) mice were used for the determination of Salmonella virulence. Salmonella were grown overnight in LB medium and diluted in phosphate-buffered saline (PBS; Difco). Approximately 1200 cfu were administered intraperitoneally and mice monitored twice daily for signs of disease. Moribund mice were euthanized according to the animal care and use regulations of the University of Washington.
Determination of the structure of PoxA
PoxA was cloned into the expression plasmid p15TvLic, and the plasmid was expressed in E. coli BL21(DE3)-RIPL (Stratagene). PoxA was purified using Ni-NTA affinity chromatography, cleaved with recombinant His-tagged TEV protease. Crystallization was performed with protein concentrated to 25 mg/mL at room temperature (21 °C) using sitting-drop vapor diffusion with an optimized sparse matrix crystallization screen (Kimber et al., 2003). The crystal used for the data collection at (see Table S3) was obtained using crystallization liquor containing 1.6 M sodium/potassium dihydrogen phosphate, 0.1 M HEPES, pH 7.5, 0.5 mM ATP and 0.3 mM MgCl2. Crystals were cryoprotected and flash-frozen in liquid nitrogen prior to data collection.
The structure of PoxA was determined by molecular replacement using a model derived by inputting the protein sequence into the SWISS-MODEL server (http://swissmodel.expasy.org/swiss). Diffraction data collected at 100° K on a Rigaku Micromax-007 rotating anode generator equipped with Osmic mirrors, and diffraction data were recorded on an R-Axis IV++ detector and integrated and scaled using HKL2000 (Minor et al., 2006). The molecular replacement program PHASER (McCoy et al., 2005) as part of the CCP4 program suite (1994) was used to determine the initial positions of the individual monomers derived by SWISS-MODEL and subsequently improved through alternate cycles of manual building and water-picking using COOT (Emsley and Cowtan, 2004). All refinement steps were performed using REFMAC (Murshudov et al., 1997) in the CCP4 program suite, with final steps of refinement including TLS parameterization (Winn et al., 2001; Winn et al., 2003). The final model was refined to an Rwork of 18.1% and Rfree of 23.9%. The Ramachandran plot generated by PROCHECK (Laskowski et al., 1993) showed 99.6% of the residues in the most favored and additional allowed regions. Further detail is given in the Supplemental Methods. The structure has been deposited into the Protein Data Bank (www.rcsb.org; PDB ID: 3G1Z).
Lysylation of EF-P by PoxA in vitro
Hexahistidine tagged EF-P and Streptavadin-tagged PoxA were purified as described in the Supplemental Methods. EF-P lysylation was performed at 37°C in a mixture containing 100 mM HEPES –NaOH, pH 7.2, 20 mM MgCl2, 30 mM KCL, 10 mM ATP, 30 μM [14C]-Lys (215 cpm/pmol), 20 μM EF-P, and the reaction was initiated by addition of 20 μM PoxA. At various time intervals, 10 μl aliquots were added to 3 μl of protein loading dye and analyzed by SDS-PAGE. After migration, the gel was stained with Coomassie dye and lysylated EF-P with [14C]-Lys was revealed by phosphorimaging. For MS analysis, EF-P was modified by PoxA in the reaction medium described above containing 15 μM PoxA, 40 μM EF-P and 40 mM Lys. A negative control was simultaneously carried out without addition of Lys. After 4 hours of incubation at 37°C, 10 μl of the reaction mix were analyzed on SDS-PAGE and the proteins were revealed by Coomassie staining. The gel slices containing EF-P were cut and trypsic digest of the protein was analyzed by MS-MS. Structural Docking was performed with Autodock 3.0 (Morris et al., 2008) and active site cavities displayed with PocketPicker (Weisel et al., 2007).
Two-dimensional gel electrophoresis and DIGE
Cytoplasmic proteins from three biological replicates each of both wild type and poxA mutant strains grown to early stationary phase (OD600 = 1.5) were labeled with Cy5 minimal dye (GE Healthcare). Equal amounts of protein from all six samples were pooled to form an internal standard (IS) that was labeled with Cy2 minimal dye (GE Healthcare). IEF was carried out on Immobiline Drystrips (pH 3–11 NL, 24 cm, GE Healthcare) according to manufacturers instructions. Second dimension SDS-PAGE was carried out on 8–16% tris-glycine gels and were scanned using a Typhoon scanner with subsequent analysis carried out with the DeCyder 6.5 software suite (GE Healthcare). Six gels, three biological replicates each of wildtype and poxA, were analyzed using Student's t-test and ANOVA to identify changes in protein abundance. Spots of interest were excised using the Ettan Spot Picker (GE Healthcare) and subjected to an overnight trypsin digest. Following peptide extraction, samples were analyzed by tandem mass spectrometry using the LTQ Ion Trapmass spectrometry at the Advanced Protein Technology Centre (Hospital for Sick Children, Toronto, ON, Canada). Identity of peptides was determined using Mascot (Matrix Science, London, UK; version Mascot) and validated using Scaffold (version Scaffold_2_06_00, Proteome Software Inc., Portland, OR). Details are provided in the Supplemental Methods.
HIGHLIGHTS.
PoxA and YjeK operate together in a pathway necessary for bacterial virulence and drug resistance.
PoxA post-translationally modifies elongation factor P (EF-P) in vitro.
PoxA is a tRNA-synthetase family member that modifies a tRNA-mimic protein, rather than a tRNA.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. Stephen J. Libby, and Anthony Richardson for many helpful discussions. Dr. Eric Alm and the MicrobesOnline website (established and curated by the group of Dr. Adam Arkin at the University of California, Berkeley) were instrumental in uncovering the genetic linkage between poxA, yjeK and efp. We acknowledge Kelly Hughes for strains used for transposon mutagenesis and Dr. Rick Collins, Andrew Keeping and Kathy Lam for extensive advice and support for the 2D-gel electrophoresis and DIGE assays. We thank Dr. Barry Bochner for providing us with excellent support concerning the Biolog Phenotype Microarray. Dr. Hiroyuki Aoki generated the PoxA expression plasmid used in this work. Work on PoxA structure determination was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200700058C. F.C.F. (AI039557, AI050660) and M.I. (GM065183) are supported by grants from the National Institutes of Health. W.W.N. received support from the Damon-Runyon Cancer Research Foundation and an operating grant from the Canadian Institutes of Health Research (MOP-86683).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline Web site for comparative genomics. Genome research. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Dekany K, Adams SL, Ganoza MC. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J Biol Chem. 1997;272:32254–32259. doi: 10.1074/jbc.272.51.32254. [DOI] [PubMed] [Google Scholar]
- Aoki H, Xu J, Emili A, Chosay JG, Golshani A, Ganoza MC. Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 2008;275:671–681. doi: 10.1111/j.1742-4658.2007.06228.x. [DOI] [PubMed] [Google Scholar]
- Appelberg R. Macrophage nutriprive antimicrobial mechanisms. J Leukoc Biol. 2006;79:1117–1128. doi: 10.1189/jlb.0206079. [DOI] [PubMed] [Google Scholar]
- Artymiuk PJ. A sting in the (N-terminal) tail. Nat Struct Biol. 1995;2:1035–1037. doi: 10.1038/nsb1295-1035. [DOI] [PubMed] [Google Scholar]
- Bailly M, de Crecy-Lagard V. Predicting the pathway involved in post-translational modification of Elongation factor P in a subset of bacterial species. Biol Direct. 2010;5:3. doi: 10.1186/1745-6150-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski MA, Braun V, Brumell JH. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler AJ, Tsolis RM, Valentine PJ, Ficht TA, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behshad E, Ruzicka FJ, Mansoorabadi SO, Chen D, Reed GH, Frey PA. Enantiomeric free radicals and enzymatic control of stereochemistry in a radical mechanism: the case of lysine 2,3-aminomutases. Biochemistry. 2006;45:12639–12646. doi: 10.1021/bi061328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise M, Becker HD, Lapointe J, Cambillau C, Giege R, Kern D. Glu-Q-tRNA(Asp) synthetase coded by the yadB gene, a new paralog of aminoacyl-tRNA synthetase that glutamylates tRNA(Asp) anticodon. Biochimie. 2005;87:847–861. doi: 10.1016/j.biochi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome research. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Huang HC, Schieven GL, Ames BN. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden SD, Rowley G, Hinton JC, Thompson A. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar Typhimurium. Infect Immun. 2009;77:3117–3126. doi: 10.1128/IAI.00093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell JH, Grinstein S. Salmonella redirects phagosomal maturation. Curr Opin Microbiol. 2004;7:78–84. doi: 10.1016/j.mib.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Bustamante VH, Martinez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A. 2008;105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V, Dubois DY, Becker HD, Kern D, Spinelli S, Valencia C, Pagot F, Salomoni A, Grisel S, Vincentelli R, et al. The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity. J Mol Biol. 2004;337:273–283. doi: 10.1016/j.jmb.2004.01.027. [DOI] [PubMed] [Google Scholar]
- Chang YY, Cronan JE., Jr. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol. 1982;151:1279–1289. doi: 10.1128/jb.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Cronan JE., Jr. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J Bacteriol. 1983;154:756–762. doi: 10.1128/jb.154.2.756-762.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Kaniga K, Galan JE. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. Negative regulation of Salmonella Pathogenicity Island 2 is required for contextual control of virulence during typhoid. Proc Natl Acad Sci U S A. 2005;102:17460–17465. doi: 10.1073/pnas.0505401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groote MA, Granger D, Xu Y, Campbell G, Prince R, Fang FC. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci U S A. 1995;92:6399–6403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desogus G, Todone F, Brick P, Onesti S. Active site of lysyl-tRNA synthetase: structural studies of the adenylation reaction. Biochemistry. 2000;39:8418–8425. doi: 10.1021/bi0006722. [DOI] [PubMed] [Google Scholar]
- Dubois DY, Blaise M, Becker HD, Campanacci V, Keith G, Giege R, Cambillau C, Lapointe J, Kern D. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp. Proc Natl Acad Sci U S A. 2004;101:7530–7535. doi: 10.1073/pnas.0401634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fang FC, Libby SJ, Castor ME, Fung AM. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect Immun. 2005;73:2547–2549. doi: 10.1128/IAI.73.4.2547-2549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingame RH, Divall S. Proton ATPases in bacteria: comparison to Escherichia coli F1F0 as the prototype. Novartis Found Symp. 1999;221:218–229. doi: 10.1002/9780470515631.ch14. discussion 229–234. [DOI] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 2007;9:2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- First EA. Catalysis of the tRNA Aminoacylation Reaction. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Landes Bioscience; Georgetown, TX: 2005. [Google Scholar]
- Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoza MC, Kiel MC, Aoki H. Evolutionary conservation of reactions in translation. Microbiol Mol Biol Rev. 2002;66:460–485. doi: 10.1128/MMBR.66.3.460-485.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis HS, Hottes AK, Tavazoie S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One. 2009;4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci U S A. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci U S A. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Compton MS, Curtiss R, 3rd, Sundaram P. Molecular and functional characterization of Salmonella enterica serovar Typhimurium poxA gene: effect on attenuation of virulence and protection. Infect Immun. 1998;66:5599–5606. doi: 10.1128/iai.66.12.5599-5606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsey JE. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 2007;421:199–209. doi: 10.1016/S0076-6879(06)21016-4. [DOI] [PubMed] [Google Scholar]
- Kimber MS, Vallee F, Houston S, Necakov A, Skarina T, Evdokimova E, Beasley S, Christendat D, Savchenko A, Arrowsmith CH, et al. Data mining crystallization databases: knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51:562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Sutterwala FS, Ogura Y, Grant EP, Bertin J, Coyle AJ, Flavell RA, Galan JE. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp Med. 2006;203:1407–1412. doi: 10.1084/jem.20060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Leung KY, Finlay BB. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR, Nunn WD. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Miller SI, Mekalanos JJ. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Olson AJ. Using AutoDock for ligand-receptor docking. Current Protocols in Bioinformatics. 2008:8.14.11–18.14.40. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- Moulinier L, Eiler S, Eriani G, Gangloff J, Thierry JC, Gabriel K, McClain WH, Moras D. The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism. EMBO J. 2001;20:5290–5301. doi: 10.1093/emboj/20.18.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller V, Spanheimer R, Santos H. Stress response by solute accumulation in archaea. Curr Opin Microbiol. 2005;8:729–736. doi: 10.1016/j.mib.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT, Banta LM, Charles TC, Nester EW. The chvH locus of Agrobacterium encodes a homologue of an elongation factor involved in protein synthesis. J Bacteriol. 2001;183:36–45. doi: 10.1128/JB.183.1.36-45.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost LR, Sanowar S, Miller SI. Salmonella sensing of anti-microbial mechanisms to promote survival within macrophages. Immunol Rev. 2007;219:55–65. doi: 10.1111/j.1600-065X.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TM, Read RC, Green J, Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Roth JR. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J Bacteriol. 1997;179:5827–5834. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Zhang N, Yang J, Ding H. Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol Microbiol. 2008;70:953–964. doi: 10.1111/j.1365-2958.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc Natl Acad Sci U S A. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurama H, Takita T, Mikami B, Itoh T, Yasukawa K, Inouye K. Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate. J Biochem. 2009;145:555–563. doi: 10.1093/jb/mvp014. [DOI] [PubMed] [Google Scholar]
- Salazar JC, Ambrogelly A, Crain PF, McCloskey JA, Soll D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc Natl Acad Sci U S A. 2004;101:7536–7541. doi: 10.1073/pnas.0401982101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SH. A nutritive view on the host-pathogen interplay. Trends Microbiol. 2005;13:373–380. doi: 10.1016/j.tim.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci U S A. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamae C, Liu A, Kim K, Sitz D, Hong J, Becket E, Bui A, Solaimani P, Tran KP, Yang H, et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol. 2008;190:5981–5988. doi: 10.1128/JB.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Rowley G, Alston M, Danino V, Hinton JC. Salmonella transcriptomics: relating regulons, stimulons and regulatory networks to the process of infection. Curr Opin Microbiol. 2006;9:109–116. doi: 10.1016/j.mib.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Van Dyk TK, Smulski DR, Chang YY. Pleiotropic effects of poxA regulatory mutations of Escherichia coli and Salmonella typhimurium, mutations conferring sulfometuron methyl and alpha-ketobutyrate hypersensitivity. J Bacteriol. 1987;169:4540–4546. doi: 10.1128/jb.169.10.4540-4546.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Stelten J, Silva F, Belin D, Silhavy TJ. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science. 2009;325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel M, Proschak E, Schneider G. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J. 2007;1:7. doi: 10.1186/1752-153X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.