Abstract
A major pathway of beta-alanine synthesis in insects is through the alpha-decarboxylation of aspartate, but the enzyme involved in the decarboxylation of aspartate has not been clearly defined in mosquitoes and characterized in any insect species. In this study, we expressed two putative mosquito glutamate decarboxylase-like enzymes of mosquitoes and critically analyzed their substrate specificity and biochemical properties. Our results provide clear biochemical evidence establishing that one of them is an aspartate decarboxylase and the other is a glutamate decarboxylase. The mosquito aspartate decarboxylase functions exclusively on the production of beta-alanine with no activity with glutamate. Likewise the mosquito glutamate decarboxylase is highly specific to glutamate with essentially no activity with aspartate. Although insect aspartate decarboxylase shares high sequence identity with glutamate decarboxylase, we are able to closely predict aspartate decarboxylase from glutamate decarboxylase based on the difference of their active site residues.
Keywords: Aspartate decarboxylase, Glutamate decarboxylase, Mosquito, Beta-alanine, Cuticle tanning
Introduction
β–alanine plays several critical roles in insect physiology. N-β–alanyl dopamine (NBAD) is formed from β–alanine and dopamine [1, 2]. It has been well known that NBAD is required for the process of cuticle sclerotization, therefore β–alanine is also required [3]. Recently it has been apparent that β–alanine also plays a pivotal role in synthesis of carcinine (N-β–alanyl histamine), a mechanism that is necessary for normal eye function [4]. It has also been shown that higher levels of sclerotization, stimulated by raising the levels of β–alanine, increase mating success and provide an adaptive advantage in populations where UV radiation provides selective pressure [5]. Conversely, Drosophila mutants that are unable to produce normal levels of β–alanine show markedly increased levels of melanization [6] as insufficient β–alanine to completely conjugate dopamine to NBAD results in excess dopamine being shuttled into a melanization pathway, ultimately forming 5,6-dihydroxyindole melanin, a reportedly cytotoxic chemical process [7]. β–alanine also appears to play a role in pupae at eclosion [3] in what has been speculated to be an ecdysone stimulated mechanism [2]. Furthermore, β–alanine can be conjugated with histidine to form the dipeptide carnosine, which has been shown to improve the longetivity of male Drosophila [8]. Thus, it would seem reasonable to conclude that abnormally low levels of β–alanine will incur a fitness cost, while increased levels may provide longevity through, as yet, unestablished mechanisms.
In vivo β–alanine can be produced from aspartate through the decarboxylation of its α-carboxyl group by decarboxylase-mediated reactions. In Drosophila, mutation of a specific gene leads to a black phenotype (a more intense black cuticle than wild type), and therefore the corresponding gene has been named black. Two Drosophila glutamate decarboxylase-like cDNAs, termed Drosophila glutamate decarboxylase-1 (GAD1) and glutamate decarboxylase-2 (GAD2) respectively, have been cloned [9, 10]. Subsequently, it was confirmed that the Drosophila GAD2 corresponds to Drosophila black [2]. While the phenotype of the Drosophila black mutant suggests that Drosophila GAD2 (or black) has aspartate decarboxylase (ADC) activity [2], its glutamate decarboxylase (GDC) function is, as yet, unconfirmed. Recently, a decarboxylase from Tribolium castaneum was identified. The mutant of this gene has the same phenotype as the Drosophila black mutant. The decarboxylase has been proposed as an ADC [11].
Glutamate decarboxylation produces γ-amino butyric acid (GABA), an amino acid that functions as an inhibitory neurotransmitter and has been linked with spatial orientation memory in Drosophila [12]. Interestingly, a number of novel putative GABA/glutamate receptors have been reported in neuronal lamina of the Drosophila visual system [13]. Because glutamate and aspartate are structurally similar, it is reasonable to speculate that decarboxylases capable of binding glutamate and mediating its decarboxylation might be able to do the same for aspartate. Although researchers, using the black mutant, were able to establish that Drosophila GAD2 has a higher specificity for aspartate than its GAD1 [2], to date, the relationship between Drosophila GAD2 and GABA in vivo is somewhat murky as researchers have been unable to definitively differentiate between glutamate decarboxylase activity from Drosophila GAD2 and GAD1 [9].
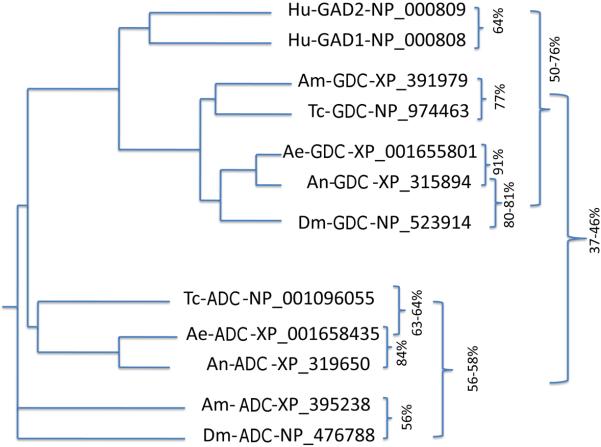
A blast search revealed that the counterparts of Drosophila GAD1 and GAD2 are present in all insect species whose genomes are currently available (Fig. 1), suggesting that the β–alanine and GABA pathways may be indispensable in insects. However, the ambiguity in substrate specificity and overall biochemical properties of the putative insect GAD1 and GAD2 enzymes have become a major barrier to comprehending the physiology and biochemistry of insect β–alanine and GABA synthesis and regulation. The biological significance of β–alanine and GABA in insects apparently warrants an extensive effort to unambiguously establish the biochemical characteristics of the decarboxylases involved in their production. To understand the biochemical properties of insect GAD1- and GAD2-like enzymes, it is necessary to obtain pure forms for each of them so that their substrate specificities and biochemical behaviors can be clearly established. In this study, we expressed the Anopheles gambiae XP_315894 and Aedes aegypti XP_001655801 sequences, the equivalent of Drosophila GAD1 and also the An. gambiae XP_319650 and Ae. aegypti XM_001658385 sequences, the counterpart of Drosophila GAD2. Of these, only An. gambiae XP_315894 and Ae. aegypti XM_001658385 sequences were soluble and active. In this report, we provide data that demonstrate that the An. gambiae XP_315894 sequence (Drosophila GAD1 equivalent, thereafter, abbreviated as AnGDC) is a highly specific glutamate decarboxylase (GDC) and that Ae. aegypti XM_001658385 (Drosophila GAD2 equivalent, thereafter, abbreviated as AeADC) is a rigidly defined ADC. Our results provide clear evidence establishing that a specific ADC and GDC are present in mosquitoes. Because the mosquito ADC and GDC equivalents are present in all currently available insect genomes (see Fig. 1), it is reasonable to propose that each of these insects also has one specific ADC and GDC with similarly categorically rigid roles.
Fig. 1.
Presence of Drosophila GAD1 and GAD2 equivalents in other insects. The Drosophila XP_476788 sequence was named GAD2 in its database. Based on results from this study, we reclassified some of the proposed GDC sequence to ADC based on high specificity of the mosquito XP_001658435 sequence to aspartate. Ae, Ae. aegypti; An, An. gambiae; Am, Apis mellifera; Dm, Drosophila malanogaster; Hu, Homo sapiens; Tc, Tribolium castaneum.
MATERIALS AND METHODS
Chemicals
All chemicals used in this paper were from Sigma-Aldrich unless specified otherwise.
cDNA synthesis
Total mRNAs were isolated from Ae. aegypti or An. gambiae larvae and earlier pupae using Trizol reagent (Gibco-BRL) according to the manufacturer's instructions. First-strand larval and pupal cDNA were synthesized using poly-T primers and subsequently used for amplification of the putative GDC/ADC sequences of AeADC and AnGDC sequences.
cDNA amplification
A pair of forward (CATATGCCAGCCAACGGAATGTTC) and reverse primer (GAATTCACAAATCTGCAGCAAGTCGTT) were designed for the amplification of the AeADC coding sequence, and another forward (CATATGACGTCGATAAACGTGGACA) and reverse primer (GAATTCACAAATCCTGACCAAGACGATC) were designed for the amplification of the AnGDC coding sequence. The underlined nucleotides in the forward primers and reverse primers correspond to an NdeI and an EcoRI endonuclease restriction site, respectively. The primers were synthesized and used for amplification of their cDNA from combined larval and pupal cDNA pools. The amplified cDNA fragments were cloned into a TA cloning vector and then subcloned into an ImpactTM-CN plasmid (New England Biolabs) for expression of their fusion protein containing a chitin-binding domain. The recombinant plasmids were propagated in Escherichia coli and the cDNA inserts were verified to be in frame by DNA sequencing.
Recombinant protein expression
Recombinant ImpactTM-CN plasmids containing AnGDC or AeADC coding sequence were transformed into E. coli cells and selected on bacterial agar plate containing ampicillin. Transformed cells were cultured at 37 °C. After induction with 0.2 mM isopropyl-1-thio-β-D-galactopyranoside, the cells were cultured at 15 °C for 24 hr. The cells were harvested by centrifugation (3000 g for 20 min at 4 °C) for subsequent isolation of the AnGDC or AeADC recombinant proteins.
Recombinant protein purification
Proteins in transformed E. coli cells were solubilized by sonication and supernatants containing recombinant AnGDC or AeADC protein were chromatographed on a column packed with chitin beads. Recombinant proteins were subsequently hydrolyzed under reducing conditions. The affinity purification resulted in the isolation of recombinant AnGDC or AeADC protein at 40–60% purity. Further purification of their recombinant proteins was achieved by Mono-Q, hydroxylapatite and gel-filtration chromatographies. Protein concentration was determined by the Bradford method (Bio-Rad protein assay kit) using bovine serum albumin as a standard. Purity of the recombinant AnGDC or AeADC recombinant protein was assessed by SDS-PAGE.
GDC and ADC activity screening
The AnGDC or AeADC recombinant protein was screened for GDC and ADC activities. Briefly, a reaction mixture of 100 μl containing varying amounts of purified recombinant protein and 30 mM concentrations of glutamate or aspartate was prepared in 0.2 M phosphate buffer (pH 7.0) containing 0.1 mM pyridoxal-5'-phosphate (PLP). The reaction mixture was incubated for 2 min at 25 °C and then was derivatized by o-phthaldialdehyde thiol (OPT) agent as described in a previous method [14]. Determination of β-alanine or GABA in the reaction mixtures was based on detection of OPT-β- alanine or GABA derivatives by reverse-phase HPLC with electrochemical detection.
Kinetic analysis
Once the GDC and/or ADC activity of a given recombinant protein was verified, the affinity and catalytic efficiency of the recombinant protein to aspartate and/or glutamate were determined by incubating the protein in the presence of varying concentrations of aspartate or glutamate and the kinetic parameters were derived by Lineweaver-Burk plot.
RESULTS
Protein expression and purification
Using combined larval and pupal cDNA pools, AnGDC and AeADC coding sequences were amplified from An. gambiae and Ae. aegypti, respectively, suggesting that both proteins are expressed in mosquito larvae and pupae. After Mono-Q, hydroxylapatite and gel filtration chromatographies of AnGDC and AeADC recombinant proteins, each displayed as a single band on SDS polyacrylamide gel (Fig. 2A and Fig. 2B). During gel filtration chromatography, AnGDC and AeADC recombinant proteins behaved approximately as a 110 KD protein, suggesting that they are present as a dimer (not shown). Spectral analysis of the AnGDC recombinant protein revealed the presence of two absorbance peaks in the visible region with λmax of 339 nm and 418 nm (Fig. 2A), respectively. The presence of recombinant protein peaks at both 339 nm and 418 nm indicates that the PLP cofactor is linked to the recombinant protein through its conserved lysine residue to form an internal aldimine that is present in either its protonated or unprotonated form. The spectrum of AeADC recombinant protein in the visible region (Fig. 2B) was similar to that observed for the AnGDC recombinant protein.
Fig. 2.
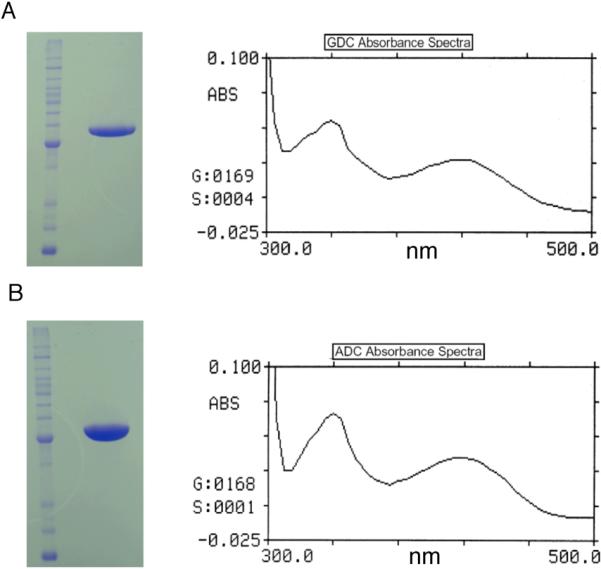
Analysis of purified mosquito GDC and ADC by SDS-PAGE and spectrometry. A: Purified AnGDC on SDS polyacrylamide gel and spectrum of AnGDC in 50 mM phosphate buffer (pH 7.0). B: Purified AeADC on SDS-polyacrylamide gel and spectrum of ADC in 50 mM phosphate buffer (pH 7.0). The presence of absorption peak with a λmax at 339 and 418 nm indicates the association of PLP cofactor with AnGDC and AeADC.
Substrate specificity
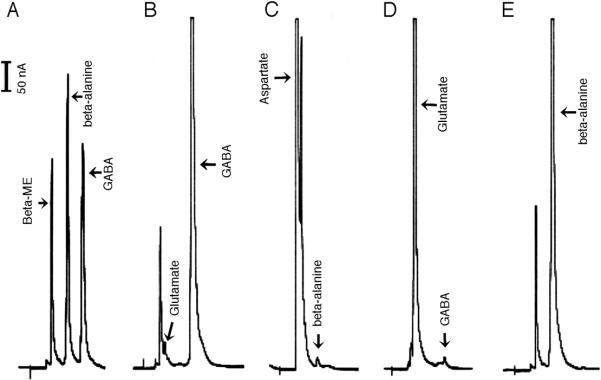
Incubation of the AnGDC recombinant protein with glutamate resulted in a rapid accumulation of GABA in the reaction. In 1.0 ml of a reaction mixture containing 5.0 mM glutamate and 50 μg AnGDC recombinant protein, more than 90% of the substrate was converted to GABA during a 20 min incubation period (Fig. 3B). When aspartate was used as a substrate in the reaction mixture, only a trace amount of β–alanine was produced in the reaction mixture (Fig. 3C). These data suggest that the AnGDC recombinant protein is specific for glutamate, and is consequently a highly specific GDC.
Fig. 3.
Substrate specificity of mosquito GDC and ADC. Reaction mixtures (1.0 ml each) containing 50 μg of AnGDC or AeADC and 5 mM glutamate or aspartate were incubated at 25 °C. At 10 min after incubation, the reaction mixture was mixed with equal volume of OPT derivatizing agent and then analyzed by HPLC with electrochemical detection. Chromatogram A shows the retention time of β-alanine (4.9 min) and GABA (6.8 min) standard under the applied HPLC conditions. Chromatograms B & C illustrate the relative amounts of GABA and β-alanine, produced in the AnGDC reaction mixture containing glutamate and aspartate respectively. Chromatograms D & E shows the relative amounts of β-alanine and GABA, produced in AeADC reaction mixtures containing aspartate and glutamate respectively.
Incubation of the AeADC recombinant protein with glutamate did not result in the production of GABA during a 20 min incubation period (Fig. 3D). In contrast, when aspartate was used as a substrate, more than 95% of the substrate was converted to β–alanine during a 20 min incubation period (Fig. 3E). These data provide the basis for suggesting that the AeADC recombinant protein is a highly specific ADC. An excess of GDC or ADC was used in the reaction mixtures to unambiguously determine the substrate specificity of these proteins.
Kinetic properties of mosquito GDC and ADC
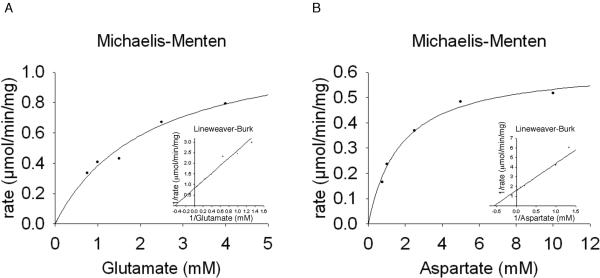
At the applied conditions, the Km of AnGDC to glutamate was 2.2 mM with a calculated Vmax of 1.2 μmol min−1 mg−1. The mosquito ADC displayed a Km of 1.8 mM to aspartate with a Vmax of 0.63 μmol min−1 mg−1(Fig. 4). Although the Km values of the two enzymes are relatively high, they apparently can, quite efficiently, catalyze the production of GABA and β-alanine due to the relatively abundance of these two amino acids in animals and also the irreversible nature of their reactions (Fig. 3), which in-turn raises an interesting question regarding the regulation of both GDC- and ADC-catalyzed reactions in vivo. The followings are the enzyme-catalyzed reactions:
Fig. 4.
Michaelis-Menten plots and Lineweaver-Burk plots derived from reactions with AnGDC (A) and AeADC (B) utilizing varying amounts of glutamate and aspartate as substrates respectively. Reactions were stopped after two minutes and derivatized with OPT reagent to determine product formation.
DISCUSSION
In mammals there are two GDC sequences, termed GAD1 and GAD2 (or GAD67 and GAD65), respectively [15, 16]. GDC knockout studies in mice demonstrate that both GAD1 and GAD2 are essential. For example, Gad1−/− mice have substantially reduced GABA levels and die at birth of severe cleft palate [17]. Gad2−/− mice have normal basal levels of GABA and appear normal at birth, but develop fatal seizures and anxiety phenotypes [18]. The physiological importance of GABA has attracted considerable attention to the proteins that are responsible for GABA production, leading to the cloning and functional verification of a number of mammalian GDC sequences. The availability of the verified mammalian GDC sequences facilitates the prediction of GDC from other species once their corresponding sequences are cloned. For example, both the proposed Drosophila GAD1 and GAD2 share high sequence identity with the mammalian GAD1 and GAD2 and their initial assignment as a GDC [10, 19] was based primarily on their similarity with mammalian GDC sequences. Likewise, both the mosquito GDC and ADC share a high sequence identity (>43%) with mammalian GDC sequences, which explains their assignment as GDCs in the mosquito genomic database. Our data about the high degree of specificity of mosquito GDC and ADC raises a critical question regarding other sequences that were previously classified as GDC sequences based solely on sequence similarity with mammalian GDCs. Also, the question of the structural basis of this substrate specificity is raised by the mutually exclusive substrate specificity of mosquito GDC for glutamate and ADC for aspartate in spite of their high sequence identity.
Glutamate and aspartate have similar structural and chemical properties (Fig. 5). It has generally been considered that GDC, responsible for catalyzing the production of GABA from glutamate, also has aspartate activity. Although this presumed aspartate activity of GDC is mentioned occasionally in the introduction or discussion in some GDC-related reports, it has never been critically analyzed or clearly discussed. In this study, we provide data that demonstrate that the true mosquito GDC is highly specific with essentially no activity to aspartate. Likewise the mosquito ADC functions exclusively on the production of β-alanine with no activity to glutamate. Although high substrate specificity may be unique to insect GDC and ADC, perhaps due in part to the specific physiological requirements during their development, it would be prudent to substantiate the putative aspartate activity of GDC proteins in other species.
In insects β-alanine is involved in several biochemical processes/events which do not seem to occur in other species. For example, β-alanine serves as a precursor for the production of N-β-alanyldopamine, a cross-linking precursor in insect cuticular sclerotization. Periodically, insects need to shed off their old cuticle and produce a new one. The new cuticle, once formed, must be hardened rapidly for protection by sclerotization and melanization. Therefore, cuticle sclerotization is essential for the growth and survival of insects. Histamine serves as a neurotransmitter in photoreceptor response in insects, but upon being released from synaptic vesicles in photoreceptor terminals and transmitted its nerve impulses, histamine needs to be inactivated in the synaptic cleft and β-alanine has been determined as the compound used by insects to inactivate photoreceptive histamine through the formation of β-alanyl-histamine by ebony-mediated reactions [4, 20, 21]. A recent report suggests that β-alanine is also indirectly involved in the regulation of circadian rhythms through its conjugation with dopamine in central nervous system [22]. To fulfill these functions, the availability of β-alanine apparently is a prerequisite. These physiological requirements may explain why a highly specific ADC has been evolved in mosquitoes.
AnGDC shares 50–55% sequence identity to mammalian GDC sequences. The 3-dimenssional structures of both human GDC1 and GAD2 have been available [23]. Comparison of the AnGDC sequence with human GDC sequences determined that their residues involved in substrate and PLP binding are conserved [23]. The great similarity of the AnGDC primary sequence and the conservation of its ligand binding residues (Fig. 6) at its predicted active site with those of human GAD1 explain the activity of mosquito enzyme towards glutamate. We attempted to express four decarboxylases, putative GDC proteins from An. gambiae and Ae. aegypti, putative ADC proteins from An. gambiae and Ae. aegypti, but we only got soluble and active recombinant proteins of putative An. gambiae GDC and putative Ae. aegypti ADC. The use of putative GDC and ADC proteins from different mosquito species for comparative studies is less than ideal. However, each of the two decarboxylases from the two mosquito species share a high degree of amino acid sequence identity with their counterparts; putative ADC proteins from An. gambiae and Ae. aegypti are 84% identical, and their putative GDC proteins are 91% identical. Based on human GAD1-ligand complex structure [23] and amino acid sequence alignment, we identified the active residues of the four proteins for substrate binding. GDCs from both mosquito species share exactly same substrate binding residues with human GAD1 (QLS--F--L--R), and ADCs from the two mosquito species share the same substrate binding sites (QLF--Y--L--R), but not with human GAD1. It is generally accepted that substrate specificity is mainly determined by the binding residues in the enzyme active site; therefore the substrate specificity from an ADC from one mosquito species could represent that of other mosquito ADCs and same with GDCs.
The AeADC sequence shares 43% and 45% sequence identity with human GAD1 and GAD2, respectively. Obviously this level of sequence identity is sufficient to classify the AeADC sequence a GDC beyond reasonable doubt. Moreover, the corresponding PLP binding residues, identified in human GAD1 (Q190, H91, E373, K396 and Y434 and R567) [23] are conserved in the AeADC sequence. However, the 3-dimentional structural analysis and substrate binding residue identification may help understand the substrate specificity of these two decarboxylases. AeADC has two different substrate binding residues from human GAD1, S vs F, and F vs Y. Phenylalanine residue in AeADC is bigger than serine residue, the counterpart residue in human GAD1, occupies more space. Therefore, the substrate binding site of AeADC might be too small to bind glutamate. However, to fully understand the structural basis of its substrate specificity, we will need enzyme-ligand complex structures and enzyme mutation studies.
ACKNOWLEDGEMENT
This work was supported by NIH grant AI 19769.
Abbreviations
- ADC
aspartate decarboxylase
- DAD
glutamate decarboxylase
- GABA
γ-amino butyric acid
- NBAD
N-β–alanyl dopamine
- PLP
pyridoxal-5'-phosphate
- OPT
o-phthaldialdehyde thiol
REFERENCES
- 1.Wright TR. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv Genet. 1987;24:127–222. [PubMed] [Google Scholar]
- 2.Phillips AM, Smart R, Strauss R, Brembs B, Kelly LE. The Drosophila black enigma: the molecular and behavioural characterization of the black1 mutant allele. Gene. 2005;351:131–142. doi: 10.1016/j.gene.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Hodgetts RB. Biochemical characterization of mutants affecting the metabolism of -alanine in Drosophila. J Insect Physiol. 1972;18:937–947. doi: 10.1016/0022-1910(72)90031-5. [DOI] [PubMed] [Google Scholar]
- 4.Gavin BA, Arruda SE, Dolph PJ. The role of carcinine in signaling at the Drosophila photoreceptor synapse. PLoS Genet. 2007;3:e206. doi: 10.1371/journal.pgen.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs ME. Beta-alanine and adaptation in Drosophila. J Insect Physiol. 1974;20:859–866. doi: 10.1016/0022-1910(74)90175-9. [DOI] [PubMed] [Google Scholar]
- 6.Hodgetts R, Choi A. Beta alanine and cuticle maturation in Drosophila. Nature. 1974;252:710–711. doi: 10.1038/252710a0. [DOI] [PubMed] [Google Scholar]
- 7.Urabe K, Aroca P, Tsukamoto K, Mascagna D, Palumbo A, Prota G, Hearing VJ. The inherent cytotoxicity of melanin precursors: a revision. Biochim Biophys Acta. 1994;1221:272–278. doi: 10.1016/0167-4889(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 8.Yuneva AO, Kramarenko GG, Vetreshchak TV, Gallant S, Boldyrev AA. Effect of carnosine on Drosophila melanogaster lifespan. Bull Exp Biol Med. 2002;133:559–561. doi: 10.1023/a:1020273506970. [DOI] [PubMed] [Google Scholar]
- 9.Chude O, Roberts E, Wu JY. Partial purification of Drosophila glutamate decarboxylase. J Neurochem. 1979;32:1409–1415. doi: 10.1111/j.1471-4159.1979.tb11078.x. [DOI] [PubMed] [Google Scholar]
- 10.Phillips AM, Salkoff LB, Kelly LE. A neural gene from Drosophila melanogaster with homology to vertebrate and invertebrate glutamate decarboxylases. J Neurochem. 1993;61:1291–1301. doi: 10.1111/j.1471-4159.1993.tb13621.x. [DOI] [PubMed] [Google Scholar]
- 11.Arakane Y, Lomakin J, Beeman RW, Muthukrishnan S, Gehrke SH, Kanost MR, Kramer KJ. Molecular and functional analyses of amino acid decarboxylases involved in cuticle tanning in Tribolium castaneum. J Biol Chem. 2009;284:16584–16594. doi: 10.1074/jbc.M901629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuser K, Triphan T, Mronz M, Poeck B, Strauss R. Analysis of a spatial orientation memory in Drosophila. Nature. 2008;453:1244–1247. doi: 10.1038/nature07003. [DOI] [PubMed] [Google Scholar]
- 13.Kolodziejczyk A, Sun X, Meinertzhagen IA, Nassel DR. Glutamate, GABA and acetylcholine signaling components in the lamina of the Drosophila visual system. PLoS One. 2008;3:e2110. doi: 10.1371/journal.pone.0002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Q, Fang J, Li J. Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: identity with aspartate aminotransferase. Biochem J. 2001;360:617–623. doi: 10.1042/0264-6021:3600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bu DF, Erlander MG, Hitz BC, Tillakaratne NJ, Kaufman DL, Wagner-McPherson CB, Evans GA, Tobin AJ. Two human glutamate decarboxylases, 65-kDa GAD and 67-kDa GAD, are each encoded by a single gene. Proc Natl Acad Sci U S A. 1992;89:2115–2119. doi: 10.1073/pnas.89.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang WM, Reed-Fourquet L, Wu E, Wu JY. Molecular cloning and amino acid sequence of brain L-glutamate decarboxylase. Proc Natl Acad Sci U S A. 1990;87:8491–8495. doi: 10.1073/pnas.87.21.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, Kuzume H, Sanbo M, Yagi T, Obata K. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci U S A. 1997;94:6496–6499. doi: 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asada H, Kawamura Y, Maruyama K, Kume H, Ding R, Ji FY, Kanbara N, Kuzume H, Sanbo M, Yagi T, Obata K. Mice lacking the 65 kDa isoform of glutamic acid decarboxylase (GAD65) maintain normal levels of GAD67 and GABA in their brains but are susceptible to seizures. Biochem Biophys Res Commun. 1996;229:891–895. doi: 10.1006/bbrc.1996.1898. [DOI] [PubMed] [Google Scholar]
- 19.Jackson FR, Newby LM, Kulkarni SJ. Drosophila GABAergic systems: sequence and expression of glutamic acid decarboxylase. J Neurochem. 1990;54:1068–1078. doi: 10.1111/j.1471-4159.1990.tb02359.x. [DOI] [PubMed] [Google Scholar]
- 20.Stuart AE, Borycz J, Meinertzhagen IA. The dynamics of signaling at the histaminergic photoreceptor synapse of arthropods. Prog Neurobiol. 2007;82:202–227. doi: 10.1016/j.pneurobio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou SR, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenalti G, Law RH, Buckle AM, Langendorf C, Tuck K, Rosado CJ, Faux NG, Mahmood K, Hampe CS, Banga JP, Wilce M, Schmidberger J, Rossjohn J, El-Kabbani O, Pike RN, Smith AI, Mackay IR, Rowley MJ, Whisstock JC. GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop. Nat Struct Mol Biol. 2007;14:280–286. doi: 10.1038/nsmb1228. [DOI] [PubMed] [Google Scholar]