Abstract
This paper proposes a novel and robust approach to the groupwise point-sets registration problem in the presence of large amounts of noise and outliers. Each of the point sets is represented by a mixture of Gaussians and the point-sets registration is treated as a problem of aligning the multiple mixtures. We develop a novel divergence measure which is defined between any arbitrary number of probability distributions based on L2 distance, and we call this new divergence measure ”Generalized L2-divergence”. We derive a closed-form expression for the Generalized-L2 divergence between multiple Gaussian mixtures, which in turn leads to a computationally effcient registration algorithm. This new algorithm has an intuitive interpretation, is simple to implement and exhibits inherent statistical robustness. Experimental results indicate that our algorithm achieves very good performance in terms of both robustness and accuracy.
1 Introduction
The need for groupwise shape matching occurs in diverse sub-domains of engineering and science e.g., computer vision, medical imaging, sports science, archaeology, and others. In model-based image segmentation for example, constructing an atlas typically requires us to bring the pre-segmented shapes into alignment. Shape features are frequently used in image retrieval as well, and need a shape alignment algorithm. And in cardiac applications, if the shapes of the heart chamber are extracted, the septal wall motion tracking problem requires us to solve for shape correspondences in the cardiac cycle.
However, matching multiple shapes can be a daunting task due to the lack of the correspondence information across the shapes. Typically correspondences can be estimated once the point-sets are properly aligned with appropriate spatial transformations. If the objects under consideration are deformable, the adequate transformation would obviously be a non-rigid spatial mapping. Solving for nonrigid deformations between point-sets with unknown correspondence is a challenging problem. A second problem we face in multiple shape matching context is the robustness issue. Some of the raw features present in one image may not be present in the other. Finally, it is desirable for a registration to be unbiased, i.e., if one arbitrarily chooses any one of the given data sets as a reference, the estimated transformation would be biased toward this chosen reference and it would be desirable to avoid such a bias.
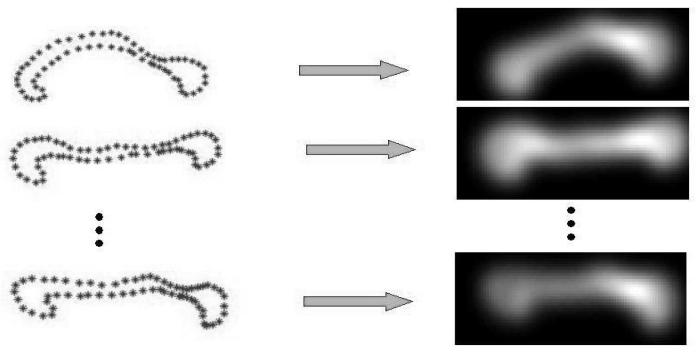
One possible solution for such situations to apply probabilistic shape matching techniques. A recent approach models each point-set by a probability density function, and then quantify the distance between these probability densities using an information-theoretic measure[1,2,3]. Figure 1 illustrates this idea, wherein the right column of the figure depicts the density functions corresponding to the point-sets (representing the boundary and shape) drawn from corpus callosum shapes shown in the left column. Using this approach, the correspondence issue is avoided since we now draw comparisons between density functions instead of point features. The robustness problem is also alleviated since we are matching holistic density functions making it robust to the point-sets of different sizes and the presence of missing features/data. Furthermore, if an unbiased information theoretic measure is chosen to quantify the multiple densities representing the shapes, the matching results can potentially be unbiased to any of the given point-sets [3].
Fig. 1.
Illustration of corpus callosum shapes (point-sets) represented as density functions.
In this paper, we develop a new non-rigid registration method for multiple point-sets. It is based on a novel information theoretic matching criterion called Generalized L2 (GL2) divergence, which is used measure the similarity between the probability densities representing the shapes. Both the Jensen-Shannon (JS) divergence [2] and our Generalized L2-divergence are shown to belong the so-called Generalized Linear Divergence family. In this paper, we use Generalized L2-divergence for achieving non-rigid registration between multiple shapes represented using the points features. We show that the Generalized L2 divergence can be expressed in a closed-form expression for registering mixtures of Gaussians. We also derive the analytic gradient of this match measure in order to achieve effcient and accurate non-rigid registration. The GL2 measure is then minimized over a class of smooth non-rigid transformations expressed in a thin-plate spline basis. The key strengths of our proposed nonrigid registration scheme are: (1) The cost function and its derivative can be expressed in closed form for the point-sets represented as Gaussian Mixtures, and they are computationally less expensive than the rival approaches; (2) The Generalized L2 divergence is inherently more robust than the rival Jensen-Shannon divergences; (3) it can accommodate point-sets to be registered of varying size.
2 Previous Work
Several articles have been reported on point-set alignment in the recent literature that utilizing the information-theoretic measures. For instance, in Wang et al. [4], the relative entropy measure (Kullback-Leibler distance) is used to find a similarity transformation between two point-sets. Their approach only solves the pairwise rigid matching problem, which is a lot easier than the non-rigid matching problem that we tackle in this paper. Jian et al. [1] introduced a novel and robust algorithm for rigidly and non-rigidly registering pairs of data sets using the L2 distance between mixtures of Gaussians representing the point-set data. They derived a closed form expression for the L2 distance between the mixtures of Gaussians, and their algorithm is very fast in comparison to existing methods on point-set registration and the results shown are quantitatively satisfactory. However, they do not actually fit a mixture density to each point-set, choosing instead to allow each point in each set be a cluster center. Consequently, their method is actually more similar to the image matching method of [5] discussed below, but with the advantage of not having to evaluate a cost function involving spatial integrals numerically, since a closed form expression is derived for the same. Roy et al. [6] used a similar approach as in [1], except that they fit a density function to the data via maximum likelihood before the registration step. Both of the methods however have not been extended to the problem of unbiased simultaneous matching of multiple point-sets being addressed in this paper. These methods are similar to our work since we also model each of the point-sets by a kernel density function and then quantify the (dis)similarity between them using an information-theoretic measure. This is followed by an optimization of a (dis)similarity function over a space of coordinate transformations yielding the desired transformation. The difference lies in the fact that GL2-divergence used in our work can be shown to be more general than the information-theoretic measures used in [1,4], and can easily cope with multiple point-sets. Recently, in [5], Glaunes et al. represent points as delta functions and match them using the dual norm in a reproducing kernel Hilbert space. The main problem with this technique is that it needs the numerical evaluation of a 3D spatial integral. In contrast, we compute the GL2-divergence using an empirical framework where the computations converge in the limit to the true values. We will show that our method when applied to matching point-sets, achieves very good performance in terms of both robustness and accuracy. Finally, a related work by Twining et al. [7] has used minimum description length (MDL) for groupwise image registration where contiguity constraints between pixels could be utilized.
Perhaps the methods that are closest in spirit to our approach are the recent work in [2,3]. They minimize the Jensen-Shannon divergence and the CDF-based Jensen-Shannon divergence respectively between the feature point-sets with respect to non-rigid deformation. The divergence measures are then estimated using the law of large numbers, which is computationally expensive and takes large amount of storage (memory). In contrast, the proposed method in this paper is much simpler, and thus less time-consuming than the previously reported methods. Furthermore, the JS-divergence and CDF-JS cannot be computed in closed form for a mixture model. In sharp contrast, our distance between the densities is expressed in closed form, making it computationally attractive. More importantly, Generalized L2 divergence is a special case to a family of divergence measures, which is called Generalized Linear Divergence, and JS-divergence is one of its special case (when we choose D to be KL-divergence in the expression of the Generalized Linear divergence).
3 Generalized L2 Divergence: A New Divergence Measure between Distributions
In this section, we define our new information theoretic measure and present some of its' property/theorems. To motivate the derivation of generalized L2 divergence, we observe that an earlier divergence measure called Jensen-Shannon (JS) divergence had addressed the groupwise point-sets registration problem [2], where each point-set Xi is modeled as probability density functions.
The JS-divergence between probability density functions pi can be written as
| (1) |
where π = {π1, π2, …, πn|πi > 0, Σ πi = 1 are the weights of the probability densities pi and H(pi) is the Shannon entropy.
The JS-divergence and popular Kullback-Leibler (KL) divergence are related by the following equation
| (2) |
where p is the convex combination of the n probability densities, p = Σπipi.
If we extend the Jensen-Shannon divergence by replacing the KL-divergence with a general distance measure between two densities, we get a generalized divergence measure between multiple distributions, which we call Generalized Linear (GL) divergence,
| (3) |
In particular, if we use the L2 distance to quantify the distance between two density function in Eqn. 3 because of its proven robustness property (Jian & Vemuri [1]), we get
| (4) |
For example, when n = 2, the Generalized L2-divergence become L2 distance between two density functions, .
3.1 Properties of the Generalized Linear Divergence
Definition 1: Recall the definition of the Density Power Divergence between two PDFs p and q,
when α = 1, then P1(p, q) = L2(p, q), and when α = 0, then P0(p, q) = DKL(p, q).
Definition 2: If we choose the pairwise distance measure D to be the Density Power Divergence in definition of the Generalized Linear divergence in Eqn. (3), and we get the Generalized Power Divergence between multiple PDFs,
| (5) |
where p is the convex combination of the n probability densities, p = Σπipi, and Pα(·) is the density power divergence.
Theorem 1: When α → 0, generalized Power Divergence will converge to Jensen-Shannon divergence; When α → 1, generalized Density Divergence converges to GL2-divergence, i.e.
| (6) |
Proof: The theorem can be derived easily from the property of Density Power Divergence. We will omit it for brevity.
For general 0 < α < 1, the class of generalized power divergences provides a smooth bridge between the JS divergence and the GL2 divergence. Furthermore, this single parameter α controls the trade-off between robustness and asymptotic efficiency of the parameter estimators which are the minimizers of this family of divergences. The fact that the L2E is inherently superior to MLE in terms of robustness can be well explained by viewing the minimum density power divergence estimators as a particular case of M-estimators [8]. For in-depth discussion on this issue, we refer the reader to [9].
Proposition 1: For a convex symmetric distance function D, the generalized divergence is a convex function of p1, p2 …, pn; conversely, if D is a concave function, the generalized divergence is a concave function of p1, p2, …, pn.
4 Multiple Point-Sets Registration with Generalized-L2 Divergence
We now present the framework of simultaneous non-rigid shape matching of multiple point-sets using the Generalized L2-divergence. The main idea is to measure the similarity between multiple finite point-sets by considering their continuous approximations. In this context, one can relate a point-set to a probability density function. Considering the point set as a collection of Dirac Delta functions, it is natural to think of Gaussian Mixture Model as representation of a point-set, which is defined as a convex combination of Gaussian component densities G(x − μi, Σi), where μi is the mean vector and Σi is the covariance matrix. The probability density function is explicitly given as where
| (7) |
and wi is the weights associate with the components. In a simplified setting, the number of components is the number of the points in the set. And for each component, the mean vector is given by the location of each point. For a dense point cloud, a mixture model-based clustering or grouping may be performed as preprocessing procedure.
Let the N point-sets to be registered be denoted by {Xa, a ∈ {1, …, N}}. Each point-set Xa consists of points , where ka is the total number of points contained in the ath point-set. Each point set is represented by a probability density function Pa. Let the atlas or the mean shape be denoted by Z. Each point set is related to the atlas through a nonrigid transformation fi parameterized by a set of transformation parameters μi. We now pose the simultaneous registration of multiple point sets as the problem of determining the transformation parameters μi such that the Generalized-L2 (GL2) divergence between the probability density functions of the transformed points is minimized. The groupwise registration problem can then be formulated as the problem of minimizing
| (8) |
where Lfi is a regularization term to control the nature of deformation. Having introduced the cost function, now the task is to design an efficient way to estimate the empirical GL2-divergence from the Gaussian mixtures and derive the analytic gradient of the estimated divergence in order to achieve the optimal solution efficiently.
4.1 Estimating Empirical Generalized L2-Divergence with GMMs
Using the Gaussian Mixture model, the density function for the ath point-set can be expressed as . Assume is the total number of points contained in the N point-sets, the pooled points in all point-sets can be expressed as .
For the convex combination ΣπiPi, if we choose , we have the following,
| (9) |
where τ : {1, …, M} → {1, …, N} is a mapping function that maps the index of a point to the index of the point-set. Therefore the linear combination of the GMMs can be expressed as a single Gaussian Mixture centered on the pooled point-sets. Consequently, we have the L2 distance between P = ΣπiPi ans Pi
| (10) |
Use the fact that
| (11) |
the L2 distance can be expressed as
| (12) |
Let us introduce a Gaussian kernel matrix G with , and define an indicator vectors Ia (of length M) for ath point-set, i.e. Ia(i) = 1 if ui is from the ath point-set (therefore τ(i) = a). IM is a vector of length M whose elements are all ones. Eqn. (12) can be rewritten as
| (13) |
Therefore, the final closed-form Generalized L2-divergence becomes
| (14) |
Based on Eqn. (14), we can derive the gradient of the GL2-divergence with respect to the transformation parameters μa, which can be expressed as
| (15) |
The details of the derivation are omitted here due to space limitations. Once we have the analytical gradient, the cost function optimization of Eqn. (8) is achieved very efficiently using a quasi-Newton method.
The Gaussian Kernel in matrix G can be replaced by other kernels (e.g. radial-basis-function, cauchy kernel etc.) leading to a “Generalized L2 divergence family”. Using Gaussian mixtures keeps the complexity O(L2) and the estimation computationally simple. Since GL2 is a generalization of the popular L2 measure, our method is equivalent to the algorithms presented in Jian et al.[1] and Roy et al. [6] when applied to align pairwise pointsets.
5 Experiment Results
We now present experimental results on the application of our algorithm to both synthetic and real data sets. First, to demonstrate the robustness and accuracy of our algorithm, we show the alignment results by applying the GL2 divergence to the matching of pairs of point-sets. Then, we will present the groupwise registration results achieved for the shapes from cardiac echocardiographic videos as well as human brain MRI. For the non-rigid registration experiments, we choose the thin-plate spline (TPS) to represent the deformation, which is similar to [2].
5.1 Pair-Wise Point-Sets Alignment
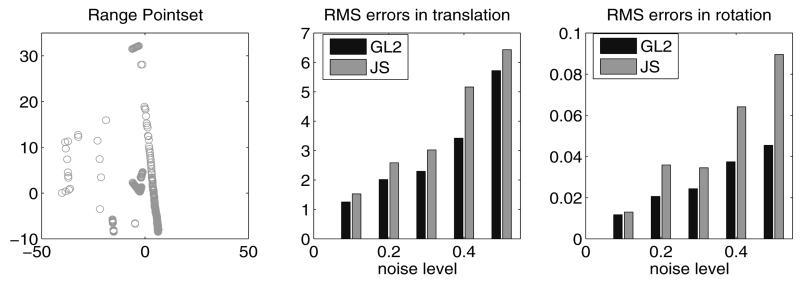
Next, we validate our method based on comparing the recovered motion parameters against the synthetical transformation parameters. We begin with a 2D range data set X of a road (Figure 2) consisting of 277 points (which was also used in Jian & Vemuri's experiment [1]). 30 randomly generated rigid transformations were applied to transform the range pointsets to obtain a transformed pointset Y, and we then remove 20% of the points in Y to get a reduced set and this is done so that the two mixture densities have a large discrepancy in the number of centroids. Different level of noise is then added to perturb reduced set. We then match X to the reduced Y using both GL2 and Jensen-Shannon algorithms. From the error plots in Figure 2, we observe that our method exhibits stronger resistance to noise than the JS method. Furthermore, the average running time for all synthetic examples are 2.151s of our methods compared with 3.738s with the Jensen-Shannon algorithm, and both algorithms are tested on the same laptop with a 1.66 GHZ processor.
Fig. 2.
Left: 2D range road pointset; Middle & Right: Errors in estimated rigid transform vs. noise strength
5.2 Multipoint Registration of Cardiac Echo Videos
Our next experiments were conducted over cardiac echo videos. The data sets depicted over 500 heart beat cycles chosen from over 50 patients with a number of cardiac diseases including cardiomyopathy, hypokinesia, mitral regurgitation, etc. For each disease class, we collected videos depicting similar views (long axis, short axis, four chamber views). An Active Shape Model (ASM) was used to characterize each such view, feature points corresponding to identifiable landmarks on heart wall boundaries were automatically extracted and tracked again as described in [10] to obtain a 3D point set.
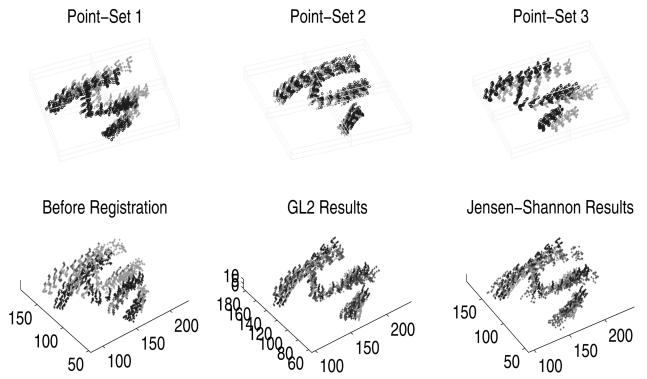
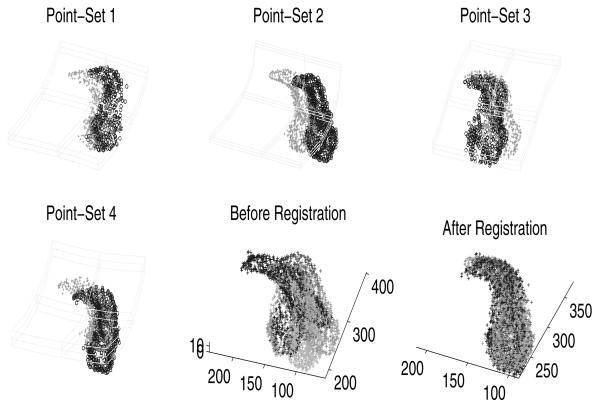
For a set of Parasternal Long Axis (PLA) views, the points from all frames of a single cycle are then stacked together for five patients to form a 3D pointset, and the time axis is normalized. The point-sets are different in size, with the number of points 1026, 988, 988 in each point-set respectively. As shown in Figure 3, the recovered deformation between each point-set (‘o’) and the mean shape (‘+’) are superimposed on the first row in Figure 3. The point-sets before and after registration results are shown in second and third image of the second row of Figure 3. The registration results generated using GL2 is shown in the lower-right for comparison, from which we can observe that the results generated using our algorithm exhibits more similarity than the JS approach. This example clearly demonstrates that our joint matching and atlas construction algorithm can simultaneously align multiple shapes (modeled by sample point-sets) and compute a meaningful mean shape.
Fig. 3.
Experimental results on three 3D echocardiogram point-sets (see text for details).
The advantage of GL2 over JS in registering point sets also exists within each disease categories. In Table 1, we show the remaining point variance after registration of videos from a number of diseases: regional wall motion abnormality (RWMA), mitral regurgitation (MR), and myocardial infarction (MI). For all diseases, the GL2 variance is lower than the corresponding JS variance, showing that GL2 performs a superior registration of the point sets. As in Figure 3, the pointsets are drawn from ASM tracking of wall boundaries in PLA views of the heart.
Table 1.
Within different cardiac diseases, registration variance is lower with our GL2 approach compared with JS, showing we achieve a better overall registration
| Cardiac disease | |||
|---|---|---|---|
| RWMA | MR | MI | |
| GL2 variance | 0.1051 | 0.1194 | 0.1103 |
| JS variance | 0.1212 | 0.1384 | 0.1222 |
5.3 Nonrigid Registration of Point-Sets from Human Brain MRI
In this we show examples of our algorithm for 2D corpus callosum atlas estimation and describe a 3D implementation on real hippocampal data sets. The structure we are interested in this experiment is the corpus callosum as it appears in MR brain images. Constructing an atlas for the corpus callosum and subsequently analyzing the individual shape variation from “normal” anatomy has been regarded as potentially valuable for the study of brain diseases such as agenesis of the corpus callosum(ACC), and fetal alcohol syndrome(FAS).
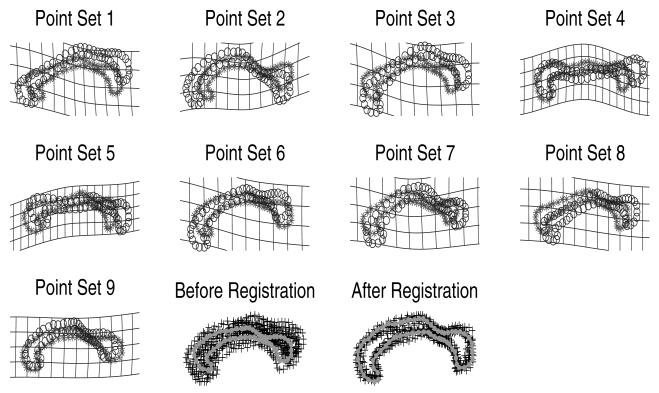
We manually extracted points on the outer contour of the corpus callosum from nine normal subjects, (as shown Figure 4, indicated by ‘o’). The recovered deformation between each point-set and the mean shape are superimposed on the first two rows in Figure 4. The resulting atlas (mean point-set) is shown in third row of Figure 4, and is superimposed over all the point-sets. As we described earlier, all these results are computed simultaneously and automatically.
Fig. 4.
Experiment results on nine 2D corpus callosum point-sets (see text for details)
Next, we present results on 3D hippocampal point-sets. Four 3D point-sets were extracted from epilepsy patients with right anterior temporal lobe foci identified with EEG. An interactive segmentation tool was used to segment the hippocampus from the 3D brain MRI scans of 4 subjects. The point-sets differ in shape, with the number of points 412, 763, 573, 644 in each point-set respectively. In the first four images of Figure 5, the first row and the left image in second row shows the deformation of each point-set to the atlas (represented as cluster centers), superimposed with initial point set (show in ‘o’) and deformed point-set (shown in ‘+’). In second row of the Figure 5, we also show the scatter plot of original point-sets along with all the point-sets after the non-rigid warping. An examination of the two scatter plots clearly shows the efficacy of our recovered non-rigid warping. Note that validation of an atlas shape for real data sets is a difficult problem and is a problem to be addressed in future.
Fig. 5.
Registration of four 3D hippocampal point sets (see text for details)
6 Conclusions
In this paper, we presented a new and robust algorithm that utilizes a novel information theoretic measure, namely Generalized L2-divergence, to simultaneously register multiple unlabeled point-sets. We have shown that it is possible to obtain a closed form solution to the non-rigid registration problem leading to computational efficiency in registration. While we used Gaussian kernels to represent the probability density of point sets, the formalism holds for other kernels as well. Experiments were depicted with both 2D and 3D point sets from medical domain. Future work will focus on generalizing the non-rigid deformations to diffeomorphic mappings.
Footnotes
This research was in part funded by the NIH grant RO1-NS046812.
References
- 1.Jian B, Vemuri B. A robust algorithm for point set registration using mixture of gaussians. ICCV 2005. 2005:1246–1251. doi: 10.1109/ICCV.2005.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Vemuri BC, Rangarajan A, Eisenschenk SJ. Simultaneous nonrigid registration of multiple point sets and atlas construction. IEEE Trans. Pattern Anal. Mach. Intell. 2008;30(11):2011–2022. doi: 10.1109/TPAMI.2007.70829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Vemuri BC, Rangarajan A. Groupwise point pattern registration using a novel CDF-based Jensen-Shannon divergence. CVPR. 2006:1283–1288. doi: 10.1109/CVPR.2006.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Woods K, McClain M. Information-theoretic matching of two point sets. IEEE Transactions on Image Processing. 2002;11(8):868–872. doi: 10.1109/TIP.2002.801120. [DOI] [PubMed] [Google Scholar]
- 5.Glaunes J, Trouvé A, Younes L. Diffeomorphic matching of distributions: A new approach for unlabelled point-sets and sub-manifolds matching. CVPR. 2004;2004(2):712–718. [Google Scholar]
- 6.Roy A, Gopinath A, Rangarajan A. Deformable density matching for 3d non-rigid registration of shapes. In: Ayache N, Ourselin S, Maeder A, editors. MICCAI 2007, Part I. LNCS. Vol. 4791. Springer; Heidelberg: 2007. pp. 942–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twining CJ, Cootes TF, Marsland S, Petrovic VS, Schestowitz R, Taylor CJ. A unified information-theoretic approach to groupwise non-rigid registration and model building. In: Christensen GE, Sonka M, editors. IPMI. 2005. LNCS. Vol. 3565. Springer; Heidelberg: 2005. pp. 1–14. [DOI] [PubMed] [Google Scholar]
- 8.Huber P. Robust Statistics. John Wiley & Sons; Chichester: 1981. [Google Scholar]
- 9.Scott D. Parametric statistical modeling by minimum integrated square Error. Technometrics. 2001;43(3):274–285. [Google Scholar]
- 10.Syeda-Mahmood T, Wang F, Beymer D, London M, Reddy R. Characterizing spatio-temporal patterns for disease discrimination in cardiac echo videos. In: Ayache N, Ourselin S, Maeder A, editors. MICCAI 2007, Part I. LNCS. Vol. 4791. Springer; Heidelberg: 2007. pp. 261–269. [DOI] [PubMed] [Google Scholar]