Abstract
Although great effort has been put forth to uncover the complex molecular mechanisms exploited in the pathogenesis of Parkinson’s disease, a satisfactory explanation remains to be discovered. The emergence of several -omics techniques, transcriptomics, proteomics and metabolomics, have been integral in confirming previously identified pathways that are associated with dopaminergic neurodegeneration and subsequently Parkinson’s disease, including mitochondrial and proteasomal function and synaptic neurotransmission. Additionally, these unbiased techniques, particularly in the brain regions uniquely associated with the disease, have greatly enhanced our ability to identify novel pathways, such as axon-guidance, that are potentially involved in Parkinson’s pathogenesis. A comprehensive appraisal of the results obtained by different -omics has also reconfirmed the increase in oxidative stress as a common pathway likely to be critical in Parkinson’s development/progression. It is hoped that further integration of these techniques will yield a more comprehensive understanding of Parkinson’s disease etiology and the biological pathways that mediate neurodegeneration.
Keywords: axon guidance, cerebrospinal fluid, CSF, metabolomics, oxidative stress, Parkinson’s disease, plasma, proteomics, SNpc, substantia nigra pars compacta, transcriptomics
Parkinson’s disease (PD) is the most common debilitating neurodegenerative motor disorder, affecting millions of people in the USA and many more worldwide. The disorder is most easily recognized by its prominent locomotor phenotype, comprised of bradykinesia (slowness of movement), difficulty initiating movement, resting tremor, rigidity and postural instability [1]. These deficits are a direct result of the loss of dopaminergic projections in the putamen and caudate nucleus of the striatum, secondary to the degeneration of dopamine-containing neurons in the substantia nigra pars compacta (SNpc) [2,3]. While these motor deficits are the primary markers used in the clinical diagnosis of PD, there also exists a constellation of non-motor symptoms observed in PD patients, including hyposmia, sleep disturbances, autonomic dysfunction, gastrointestinal dysregulation, anxiety, depression and cognitive decline. Most non-motor symptoms are thought to be a consequence of degeneration of systems other than dopamine, such as serotonergic in the dorsal raphe, noradrenergic in the locus coeruleus and cholinergic neurons in many other brain regions [4], and consequently are not typically treatable with dopamine replacement. In other words, PD is a multisystem disorder rather than a purely dopamine-centric disease.
In addition to the degeneration of specific cell populations, the presence of proteinaceous aggregates, such as Lewy bodies (LBs) and Lewy neurites, is one of the defining pathological features of PD. These inclusions are usually found in the perikarya and cellular processes of dopaminergic neurons of the SNpc and other brain regions [5,6]. While a comprehensive composition of these inclusions is still unknown, recent efforts have demonstrated an eclectic mix of proteins being harbored within these aggregates, including proteins involved in oxidative stress, synaptic vesicle dynamics and protein folding and degradation [7]. Further identification of the protein constituents of LBs and Lewy neurites will aid in elucidating potential molecular pathways involved in the pathogenesis of PD.
While the cause of PD remains unknown, considerable evidence suggests a multifactorial etio-pathogenesis involving exposure to environmental contaminants, as well as genetic susceptibility, leading to the potential dysfunction of several cellular processes, including oxidative/nitrative stress [8–10], excitotoxicity [11,12], inflammation [13], mitochondrial impairment [14,15] and altered proteolysis [16–18]. These processes are believed to form a complex cascade of interrelated events that culminate in neuronal death via apoptosis, a type of programmed cell death [19–22]. While the predominant opinion maintains that the majority of PD cases are idiopathic in nature, research in the last 10 years has uncovered several genetic mutations associated with familial cases of PD [23,24]. The recognition of a possible genetic component of PD has been around for over a century [25–27], but definitive proof has only recently come to light. To date, researchers have identified five definitive genes causing familial PD with a Mendelian pattern of inheritance; specifically, α-synuclein, LRRK2, parkin, DJ-1 and PINK1. However, as fewer than 10% of PD cases can be attributed to purely genetic causes, other factors appear to be involved that function independently or in conjunction with the genetic aspect. Indeed, numerous epidemiological studies have identified exposure to pesticides, as well as other environmental contaminants, as a major risk factor for the development of PD [28–39]. Interestingly, several reports have demonstrated an increased vulnerability to dopaminergic dysregulation when an in vitro or in vivo model of genetic susceptibility is simultaneously exposed to an environmental toxicant, further supporting a multifaceted disease etiology [40].
Given the complexity that surrounds both the etiology of the disease, as well as the pathological cascades that follow, a more comprehensive investigation of PD and the identification of specific disease networks and molecular pathways involved will facilitate our understanding of the disease process and enhance our ability to diagnose and treat this debilitating condition. A recent effort has focused on the use of -omics, particularly transcriptomics, proteomics and metabolomics, in order to elucidate many of the pathogenic aspects of PD. Notably, when we discuss these techniques we are referring to the use of transcriptomics to identify genes and evaluate changes in gene expression, while proteomics is focused on recognizing and measuring respective changes in proteins. Finally, metabolomics is concerned with the identification and quantification of metabolites to provide a signature of the metabolic state at that point in time. In general, these techniques can be applied to various biological media, including tissue, cerebrospinal fluid (CSF), blood and blood constituents and urine, as well as others. These platforms provide a means to generate and analyze a significant amount of data with the intent of facilitating studies to reveal the molecular events that underlie neurodegeneration in PD. Moreover, the integration of gene, protein and metabolic targets would generate a cohesive picture of shared pathways and variations between the healthy and disease state.
Furthermore, the application of these approaches can be extensively utilized in the discovery of biomarkers of PD. Biomarkers are used as indicators of normal biological processes, as well as pathologic processes, and can provide a window into the disease mechanism with the hope of developing specific therapeutic targets of the disease. In addition, from a clinical point of view, the development of biomarkers that allow for the delineation of premotor stages of PD from more advanced pathologic states would greatly enhance the diagnostic power of the clinician and widen the currently narrow therapeutic window available for treatment. In this regard, transcriptomics, proteomics and metabolomics could be used separately or in conjunction to discover biomarkers. Thus, biomarkers are imperative to our understanding of PD pathogenesis and progression.
The intent of this article is to appraise and summarize the current PD research in the context of transcriptomic, proteomic and metabolomic platforms and findings. Given the exponential growth in the utilization of these techniques in PD, as well as other neurodegenerative disorders, we will focus our article on research reports in which human samples (brain tissue, CSF, blood and plasma) have been used. However, a future appraisal of the application of -omics to cellular, as well as animal, models of PD and the translation of these results to human studies would be extremely beneficial to the field. We will first provide a general overview of each -omic technique followed by an examination of PD-related data generated by each platform. Potential caveats and shortcomings of each technique will also be discussed. Finally, we will identify overlapping targets discovered by transcriptomics, proteomics and metabolomics before discussing future directions associated with the use of -omics in PD research.
Transcriptomics
Overview of transcriptomics techniques
Microarray analysis, also known as gene expression profiling, measures the mRNA levels of all known human genes coding for proteins in a given sample simultaneously. A typical microarray is a glass slide with thousands of spots of oligonucleotide (or cDNA) probes attached to it. Each ‘spot’ contains thousands of identical copies of one oligonucleotide corresponding to a specific mRNA target. The goal of a typical microarray experiment is to compare gene expression profiles of two or more samples (for overview see Figure 1). The first step of a microarray experiment is the isolation of high-quality RNA. The sample collection procedure and RNA isolation method have to take into account that RNA is labile and subject to rapid degradation. Stringent quality control is critical to ensure high-quality RNA. The second step generates either fluorescently labeled cDNAs or cRNAs from the RNA samples of interest. The third step involves hybridization of the fluorescently labeled sample to the microarray. Microarray platforms can be divided into one or two color platforms. The former uses a single fluorescent dye (one color) and only one sample is hybridized per array, whereas in the latter case, two samples are cohybridized per array, each labeled with a distinct fluorescent dye (two color).
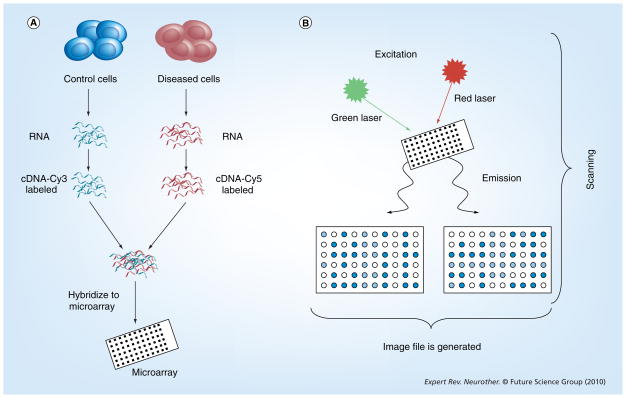
Figure 1. Overview of a microarray experiment.
(A) RNA is isolated from control and diseased cells. cDNA is prepared from the RNA samples and fluorescently labeled with Cy3 or Cy5 dyes. The labeled cDNA samples are cohybridized to the microarray. (B) Following hybridization, washing and drying, fluorescent signals are detected by scanning the microarray with a confocal microscope with appropriate lasers for optimal excitation of the fluorescent dyes and an image file is generated.
Once a set of microarrays has been scanned, the scanner images need to be preprocessed. Preprocessing involves visual inspection of the scanner images, quantification of the fluorescent signal of each spot on the array, quality control and normalization. Normalization is the process of removing the variability within the quantified images that is not due to the biological differences of the samples. A detailed discussion on preprocessing and normalization has been provided by Yang and Speed [41] and Wu and Irizarry [42]. Following preprocessing and normalization a suitable statistical model is chosen to identify differentially expressed genes.
A typical experiment results in hundreds of differentially expressed genes, and it can be challenging to interpret the data. If the objective is biomarker discovery, significance analysis of individual genes is a useful approach, but it does not necessarily provide mechanistic clues. A complementary strategy is to categorize differentially expressed genes into functional groups (e.g., Gene Ontology or GO categories), based on gene annotations, to discover biological themes. Gene Set Analysis methodologies that take information from all genes into account without imposing predefined fold-change and/or p-value cutoffs have been developed. Gene Set Analysis assesses statistical significance of entire sets of genes/pathways and is capable of identifying sets with genes showing modest but concordant changes in expression. From here, targets of interest can be chosen for further validation and evaluation of biological significance.
Findings in transcriptomics
In the last 5–6 years, several gene expression studies have been undertaken in human midbrain tissue in order to better understand the influence of alterations to individual or groups of genes in the pathogenesis of PD. In general, these studies have utilized SNpc tissue collected from post-mortem control and PD patients and employed global microarray platforms to generate extensive lists of genes that exhibit significant changes in expression between the two disease states. In the process we have learned a considerable amount of information concerning molecular pathways that are dysregulated, either as a direct result of the loss of dopaminergic neurons or as an attempt to compensate for this degeneration and maintain nigrostriatal homeostasis.
As pointed out in a recent article by Sutherland et al., when several transcriptomic studies are analyzed for common genes, very few studies demonstrate a consensus as to the individual genes that demonstrate a significant alteration between control and PD [43]. However, when genes are classified into categories based on function, a greater concordance between the data sets is illuminated. Indeed, when examined in concert, the most prominent finding from each of these studies is the altered expression of genes related to dopamine neurotransmission, mitochondrial function, protein degradation and synapse dynamics [43–51]. While not surprising given the almost absolute degeneration of dopaminergic neurons in the SNpc of PD patients, several genes related to dopamine neurotransmission, such as the dopamine transporter, aromatic amino acid decarboxylase and vesicular monoamine transporter 2 (VMAT2), among others, demonstrated substantial reductions in expression [45,48,51]. Although well established, these results, besides confirming the validity of the technology, provide further evidence for the prominent role dopamine and dopaminergic neurodegeneration play in PD.
In addition to alterations in genes associated with dopamine neurotransmission, several studies identified reductions in the expression of genes involved in the functioning of the mitochondria and the proteasome, which have both been shown to be altered in genetic and idiopathic forms of PD [45,46,48–51]. As the mitochondria is considered the major source of ATP utilized by the cell, disruption of its function can have severe repercussions for the normal functioning of the dopamine neuron, through the suppression of energy production and the generation of neurotoxic reactive oxygen species. Indeed, exposure of rats and mice to the mitochondrial complex I inhibitors, rotenone and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), elicits considerable degeneration of dopamine neurons in the SNpc, recapitulating many of the pathological features of PD [52–54]. Furthermore, reductions in complex I expression and function have been recorded in the SNpc of human PD samples [55]. Numerous genes involved in mitochondrial function have also been shown by transcriptional studies to be altered in the SNpc of PD. These genes encompass several aspects of the mitochondria, including complex I–IV, ATP synthase and cytochrome C oxidase [45,46,48–51].
Similar evidence exists for the disruption in function of the ubiquitin proteasome system (UPS). In general, the UPS functions to degrade misfolded or dysfunctional proteins present in the cell, in order to prevent these proteins from further harming the cell [17,56]. Work from the last decade has uncovered proteins involved with the UPS and directly associated with a genetic susceptibility to PD. For example, parkin is an E3 ligase, which helps to couple and target proteins that have been tagged for degradation to the proteasome [57,58]. Reduction in the expression and subsequent function of this protein contributes to the cytoplasmic accumulation of misfolded or damaged proteins in dopamine neurons, precipitating their demise [17]. Moreover, animal studies have shown similar neurodegeneration following exposure to a proteasome inhibitor [59]. These data suggest that alterations to the function of the UPS could have dire consequences for dopamine neurons in the SNpc. In the same line of argument, several transcriptomic studies have demonstrated an alteration in specific genes that are associated with regulating the proper function of the UPS. In particular, SKP1A and HSPA8, which are both involved in modulating the ubiquitination of damaged or misfolded proteins for degradation by the proteasome, were downregulated in PD SNpc [48,49].
Maintaining the integrity of the neuron’s ability to efficiently perform exocytotic and endocytotic functions associated with neurotransmission is imperative to the longevity of the cell. Although the complexity of these events is still being teased out, we do understand that this sequence is a temporally and spatially regulated series of steps that must occur flawlessly in order for synaptic vesicles containing neurotransmitters to be released and for the vesicular constituents to be recovered and reused. The failure of even a single protein in this chain can disrupt the entire sequence. In two independent studies, genes associated with synaptic vesicle dynamics in the SNpc were found to be altered in the SNpc of PD subjects [50,51]. One of these genes, SYT1, which encodes for the vesicle-associated protein synaptotagmin 1, was significantly reduced in the SNpc of PD subjects compared with controls. Synaptotagmin 1 is a membrane-bound protein that resides on the synaptic vesicle membrane and functions to modulate vesicular exocytosis through its involvement in synaptic vesicle docking and subsequent fusion with the presynaptic plasma membrane to facilitate neurotransmitter release. In addition to synaptotagmin 1, reductions in other genes associated with synaptic vesicles, such as syntaxin-binding protein and vesicle-associated membrane protein 1, also known as synaptobrevin 1, were also shown to be decreased in PD, suggesting a decrement in multiple aspects of vesicle handling in PD.
One issue that is constantly at the forefront of array studies that utilize a global expression analysis approach to identify and measure alterations to gene expression in human tissue, especially in the brain, is the heterogeneity of neuronal populations that exist in the region being assayed [60]. For example, although dopamine neurons are the most prominent neuronal population in the SNpc, an abundance of glial cells, as well as other neuronal cell populations, also reside within this region [61]. One technique that has been employed to somewhat circumvent this issue is the use of laser capture microdissection (LCM). When paired with a tool to visualize specific neurons of interest (e.g., dopamine neurons), this technique provides the user with a highly precise method of acquiring single neurons from post-mortem tissue slices. Accordingly, studies have used LCM to isolate dopamine neurons from the SNpc of control and PD subjects, followed by microarray analysis. As these neurons are visualized by the presence of neuromelanin in the cytosol, they were able to accurately capture and analyze a ‘pure’ population of dopamine neurons. A compelling outcome of these studies is the similarity in findings of dysregulated genes comprising pathways associated with synaptic function, mitochondrial energy production and protein degradation, as found in studies that employed a global expression approach to address changes in the transcriptome [45,46,48–51]. In addition, they were able to identify genes involved in programmed cell death, providing supporting evidence for the role of apoptosis in dopaminergic neurodegeneration [62].
As mentioned earlier, although multiple studies have utilized transcriptomics to identify genes that are altered in the SNpc of PD versus control subjects and provided a wealth of information pertaining to the potential pathogenesis of PD, when analyzed more carefully, these datasets provide inconsistent results between studies. The reasons for this disagreement may simply be due to the variability in individual patients or the use of differential microarray platforms and techniques. However, when genes are placed into functional categories and evaluated using Gene Ontology analysis [63], it is found that several genes identified in these studies can be classified into similar pathways. Given the almost absolute loss of dopaminergic neurons in the SNpc of PD patients, it would appear that the substantial reduction in genes in this region could be attributed to dopaminergic neurodegeneration, generating an artificial representation of alterations in gene expression. This would create a predilection towards genes whose expression is reduced simply because dopamine neurons are dying, masking alterations in genes that could reveal molecular pathways and mechanisms truly involved in PD pathogenesis.
Using a carefully devised correction paradigm, a recent report by Sutherland et al. reanalyzed datasets from several transcriptomic-based evaluations of gene expression changes in the SNpc of PD and control subjects [43]. With the application of this analysis technique they were able to reduce the bias towards genes whose differential expression was due to neuronal loss and focus on alterations in cells that were remaining in the SNpc. Following this correction, the dopamine signaling pathway ceases to be an over-represented pathway, which has been demonstrated by multiple studies. However, the ephrin receptor signaling pathway and the axon guidance pathway, which are both involved in nervous system development, emerged as prominent pathways. Interestingly, previous transcriptional studies have also identified the axon guidance pathway in the SNpc of PD subjects compared with controls [64,65] and this pathway appears to be gaining more interest as a potential target in PD pathogenesis [66]. The use of brain tissue can only provide a snapshot of the disease at one single time point and usually at the terminal stage of the disease. However, the use of peripheral tissue sources, such as blood or CSF, allows for a minimally invasive means of sampling a normal and diseased population and provides the researcher with the ability to perform a longitudinal assessment of particular markers of interest in order to give an indication of pathways that may be altered as the disease progresses [67]. Moreover, the use of tissue other than post-mortem brain samples assists in the identification of specific proteins that could provide insight into the modulation or dysfunction of molecular pathways involved in PD disease pathogenesis. In this context, two recent studies by Scherzer et al. have capitalized on the high-throughput capabilities of gene microarrays and the relative availability of blood and blood components from control and PD patients to identify potential biomarkers of PD and to gain a more focused understanding of the role of specific proteins in PD pathogenesis [68,69]. Their initial study of blood taken from control and early stage PD patients identified ST13, a gene that was highly underexpressed in PD samples compared with controls [68]. ST13 is a cofactor for heat shock protein (Hsp)70, a molecular chaperone, and seems to stabilize its chaperone activity. Hsp70 is of particular interest to PD research because of its important function in modulating protein folding, especially of α-synuclein, where it has been demonstrated to attenuate α-synuclein toxicity [70].
Further microarray analysis of blood from PD and control subjects by Scherzer et al. uncovered a significant clue in the transcriptional modulation of α-synuclein [69]. Genetic alteration of α-synuclein, such as point mutations and locus multiplication, can have deleterious effects on the risk of PD. It is suggested that understanding the mechanisms involved in regulating the concentration of α-synuclein may provide a window through which therapeutic interventions aimed at suppressing α-synuclein levels can be developed. Through transcriptional analysis, Scherzer and colleagues were able to identify a transcription factor, GATA-2, that is highly expressed in SNpc and regulates the expression of α-synuclein levels when manipulated in an in vitro model system.
Caveats in transcriptomics
An important caveat that deserves consideration when undertaking a transcriptomic-based evaluation of mRNA expression differences between disease and normal states, is the further validation of the identified mRNA as well as the subsequent protein product [71]. As microarrays have the capabilities to identify several hundred or thousand differentially expressed genes, the validation of these genes and their changes via an independent method is imperative to confirm the findings. The most recognized and reliable method available for this purpose is quantitative real time-PCR. Further validation of gene expression studies can be undertaken in order to evaluate whether alterations in the expression of a gene of interest are translated into changes in levels of its respective protein product. These changes are most easily evaluated through immunoblotting or immunohistochemical techniques utilizing an antibody that specifically recognizes the protein of interest. Also, the biological implications of an increase or decrease in expression of a particular gene can be assayed using in vitro and in vivo model systems that allow for the manipulation of the target gene and the measurement of specified end points. Obviously, this extensive confirmation process is not practically possible for all genes identified that exhibit a change in expression between two disease states; however, they are absolutely necessary in order to reliably identify novel genes and evaluate their biological significance to potential molecular mechanisms and pathways involved with PD pathogenesis. Finally, in transcriptomics, very little, if any, attention has been paid to the non-motor component of PD. This would involve transcriptomic investigation of other neuronal populations, such as serotonergic neurons in the dorsal raphe and noradrenergic neurons in the locus ceruleus, that are also susceptible to degeneration in PD.
Proteomics
Overview of proteomics techniques
Proteomics, a discipline that studies the structure and function of proteins, typically in an unbiased fashion, has rapidly emerged as one of the most powerful technologies, providing the means to study the protein profile of a complex biological system on a large scale [72]. The technology is composed of several integrated technical components, including separation technology, mass spectrometry (MS) and bioinformatics data processing (Figure 2). With the advances in analytical technology, a variety of separation methods have been applied to facilitate the proteomic study of complex biological samples, including liquid chromatography (e.g., strong-cation-exchange, reversed phase, size exclusion), electrophoresis, solid phase extraction and immunoaffinity. Multidimensional separation can be applied to diagonally fractionate a complex sample at either the protein or peptide level to enhance the analytical dynamic range and detection sensitivity. The mass spectrometer is the core component of proteomic technology. High-resolution instruments, such as Fourier transform–ion cyclotron resonance, Orbitrap, quadrupole time-of-flight (TOF) and TOF/TOF, are now available, greatly enhancing the quality of proteomic data. These instruments can be coupled with either electrospray ionization or matrix-assisted laser desorption ionization ion source, providing greater flexibility for different experimental designs. In addition to the widely used collision-induced dissociation method for ion fragmentation, soft collision techniques, such as electron transfer dissociation, have been introduced recently, allowing more sophisticated analysis of post-translational modifications (PTMs), such as phosphorylation and glycosylation. The interpretation of MS data relies on the bioinformatics tools, which may include database development, search algorithms for peptide/protein identification, PTM identification and quantitative analysis. In general, a proteomics data analysis pipeline includes data conversion, database search and verification of peptide/protein identification.
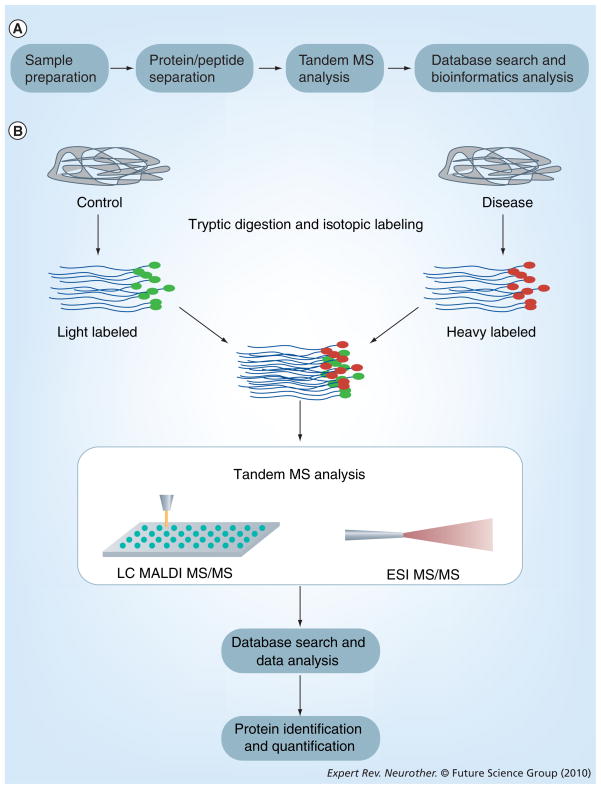
Figure 2. Overview of proteomics workflow and protein quantification.
(A) Samples of brain tissue, cerebrospinal fluid or blood/plasma are collected and prepared for separation by either strong-cation-exchange, reversed phase, or size exclusion chromatography. Samples are then analyzed by tandem MS, followed by identification by database search algorithms and bioinformatics evaluation. (B) For quantitative proteomics, biofluid samples from control and disease patients are obtained and labeled with isotopic tags before being mixed, fractionated and analyzed by tandem MS. The identification and quantification of proteins relies upon protein databases and bioinformatics analysis.
ESI: Electrospray ionization; LC: Liquid chromatography; MALDI: Matrix-assited laser desorption/ionization; MS: Mass spectrometry.
One of the most important areas for clinical proteomics study is the investigation of alterations in protein abundance and PTMs, which may be associated with changes in biological pathways resulting from a disease and its progression. The development of quantitative proteomics, which aims to systematically identify steady or perturbation-induced changes in the protein profile of a biological system, has stimulated great interest in applying the technology to study mechanisms of disease and biomarker discovery. The most widely used approach for quantitative proteomics analysis utilizes stable-isotope labeling to introduce mass tags to distinguish a peptide with the same sequence from different origins (e.g., sample vs control). Therefore, quantification of a peptide can be achieved based on the intensity ratio of heavy and light forms of the peptide. There are a variety of labeling methods, based on different labeling mechanisms, that have been introduced for MS-based quantitative proteomics analysis, including isobaric tag for relative abundance and quantitation (iTRAQ), stable isotope labeling with amino acids in cell culture, isotope-coded affinity tag (ICAT) and 18O labeling. Label-free methods based on spectra count have also been applied for quantitative study. In addition to unbiased quantitative global profiling, very recently, candidate-based quantitative proteomics has been introduced, providing a complementary platform for the targeted detection of candidate peptides/proteins in a complex sample [73]. It is important to note that with all the advancements in technology, a comprehensive proteomics analysis on a clinical sample, such as plasma, serum or tissue, is still a challenging task, requiring careful experimental design and a sophisticated analytical pipeline. The enormous complexity and nonlinear dynamic range in protein abundance in a biological sample is the major hurdle for an in-depth search for low abundant proteins that may be biologically significant. However, with the rapid advances of technology in this field, it is believed that many of the current technical limitations will be transient.
Similar to transcriptomics, proteomics has been widely applied in the study of neurodegenerative disease, providing a high-throughput platform with which to generate extensive amounts of data concerning the identification of proteins and quantification of their relative changes in expression from one disease state, tissue region or cell type to another. Through the use of different body tissues, such as brain tissue, CSF and blood, proteomics has provided a window through which proteins are altered, providing a more in-depth and comprehensive understanding of the potential pathways and mechanisms involved in the pathogenesis of PD. In the following sections we will highlight several recent proteomic results from human subjects, focusing on those findings discovered in brain tissue and CSF.
Human brain tissue
Protein identification
Recent efforts have utilized proteomics in order to characterize the human CNS, especially the cortex and midbrain proteome, with the hopes of gaining a greater insight into the pathophysiology of neurodegenerative disease [74,75]. As discussed previously, the degeneration of dopaminergic neurons in the SNpc of the midbrain are the predominant pathological features that underlie the locomotive phenotype that typifies PD. It is believed that the identification and comprehensive description of the human midbrain will provide a clearer understanding of the course and proteins that initiate and propagate the execution of the dopamine neurons. In keeping with this, we have recently generated an expansive profile of the human midbrain through the coupling of two MS platforms [75]. A comprehensive MS profile of the human midbrain identified over 1200 proteins, many of which have been determined to be associated with PD pathogenesis. These include DJ-1, UCHL-1, MnSOD and glutathione S-transferase omega 1, which have previously identified roles in oxidative stress, mitochondrial function, protein degradation and neuroinflammation.
While a global description of the midbrain proteome has enhanced our overall view of this region and the proteins that may contribute to PD, a more systematic analysis of specific cell populations and constituents of these cells within the midbrain may provide valuable information related to neuronal pathways involved in the disease. This approach, termed subcellular proteomics, relies upon subcellular fractionation techniques to isolate specific subcellular compartments that can then be analyzed for protein composition via a proteomic platform. This technique has recently been applied in an effort to describe the protein components of neuromelanin [76–78]. Neuromelanin is a granular pigment found in most, but not all, catecholaminergic cell populations in the brain, where it is believed to be formed as a byproduct of catecholamine synthesis and breakdown [79]. Although the exact function of neuromelanin remains to be elucidated, it does appear to interact with and sequester multiple compounds found within the cellular cytosol, including iron, lipids, pesticides and neurotoxic compounds, as well as other intracellular components. In this regard, it is believed that neuromelanin retains cytoprotective, but also cytotoxic, roles if these putative sequestration mechanisms are damaged or saturated. It has been speculated that alteration of its function could participate in dopamine degeneration. Pairing subcellular fractionation of neuromelanin granules with a proteomic platform, Tribl et al. have produced an extensive dataset of proteins associated with neuromelanin [76]. These proteins appear to be involved in a diverse set of intracellular functions, including vesicle trafficking, mitochondrial function, endoplasmic reticulum and molecular chaperones, among others. While this study provides valuable insight into the molecular composition of neuromelanin granules, it raises the question of whether association of these proteins with neuromelanin is serving a neuroprotective or neurodegenerative function. More recently, although the significance remains to be elucidated, autoantibodies against neuromelanin have been noted in PD patients, particularly during early stages of the disease [80].
Subcellular isolation techniques have also been employed for the proteomic analysis of the protein composition of LBs in human brain tissue. As mentioned previously, LBs are a major pathologic characteristic of PD, as well as other parkinsonian disorders. Similar to neuromelanin, LBs also reside intracellularly and have been found to be composed of cytosolic components, including VMAT2, α-synuclein and parkin. Thus, further identification of the molecular constituents of LBs could provide a glimpse into their mechanisms of formation and functions in PD. A recent study applied proteomic analysis to biochemically enriched samples of LBs obtained from human tissue [81]. Through this analysis they were able to reliably identify over 40 proteins associated with LBs. Included on this list were proteins involved in molecular chaperone functions, oxidative stress, protein trafficking and proteasomal degradation. As PD pathology progresses, the presence of LBs migrates in a predefined path originating in the brainstem, in the early stages of the disease, until it encompasses the neocortex in more advanced cases of PD [82]. Elucidation of the protein constituents of cortical LBs may shed light on the molecular mechanisms involved in the late stages of the disease. Using LCM to isolate LBs from cortical tissue in patients with dementia with LBs, followed by proteomic analysis, yielded the identification of 156 proteins [83]. One protein in particular, HSC70, was chosen for further validation and was confirmed as a LB-associated protein. HSC70 is important as it has been identified as a molecular chaperone and suggested to participate in multiple cellular processes, including endocytosis, oxidative stress and apoptosis. Interestingly, although this precise protein was not identified by Xia et al., they were able to discover several proteins associated with molecular chaperone functions [81]. It is anticipated that future research will help to clarify the biological implications of many of these proteins in the pathophysiology of PD.
Protein quantification
The identification of proteins in the midbrain, as well as subcellular compartments, has provided an extensive list of proteins that occupy these regions, and can be used as tools or starting points for future discoveries. However, delineating specific proteins whose expression is altered in PD compared with control allows for a more thorough investigation of the biological significance of those proteins in disease pathology. A number of methods have been developed that allow for quantification of protein expression changes in both, in vivo or in vitro systems. The gold standard for this type of investigation is the coupling of 2D gel electrophoresis with MS to identify proteins with relative changes from one group to another. Indeed, a study by Basso et al. utilized these techniques and was able to identify 44 proteins, nine of which demonstrated changes in expression in the SNpc of control and PD subjects [84]. As discussed in the technique portion of the proteomics section, ICAT and iTRAQ labeling of proteins in human tissue has been the most beneficial in identifying and quantitating proteins in control and PD SNpc that exhibit alterations in expression. Subsequently, the application of these techniques has allowed for several advancements in the description of particular proteins involved in PD pathogenesis. Indeed, the application of these techniques in our laboratory has allowed for the demonstration and subsequent validation and investigation of biological importance of several proteins found to be altered in PD SNpc compared with controls.
Analysis of mitochondrial fractions isolated from the SNpc of control and PD patients and labeled with ICAT identified 119 proteins that demonstrated a change in their relative expression in PD compared with control [85]. As one of our premier focuses is to identify and describe novel proteins that have not previously been associated with PD, we chose to further validate and investigate mortalin, which functions as a molecular chaperone in mitochondrial import and energy production, as well as the prevention of oxidative stress. Confirmation of mortalin expression found it significantly reduced in dopamine neurons in the SNpc of PD patients compared with controls and appeared to be specific for the mitochondrial fraction when biochemically isolated, lending evidence to the role of mortalin in mitochondrial function. Biological manipulation of mortalin in an in vitro model of PD found a reduction in cell viability and cellular function following overexpression of mortalin and treatment with the mitochondrial complex I inhibitor, rotenone.
Further evidence for the importance of mortalin in the pathogenesis of PD was uncovered following the evaluation of PD progression in human cortical tissue taken from PD patients exhibiting LB pathology in the brainstem, limbic system and frontal cortex, representing progressively advanced stages of the disease, those with non-motor symptoms in particular [86]. In this study, tissue samples were labeled using iTRAQ reagents and subjected to proteomic analysis. Out of almost 200 proteins exhibiting significant changes in expression from controls, several of them could be considered as candidate proteins that might be important in PD progression. Some of these proteins have been previously linked to neurodegenerative disease and more specifically, PD, such as mortalin, while others were important for CNS function, but as of yet have not been associated with neurodegeneration. Further validation of mortalin in these tissue samples found a somewhat progressive reduction in mortalin expression as the disease advances. Notably, there are at least three subsequent experiments, independently identifying a critical role of mortalin in PD pathogenesis [85–87].
Utilizing a similar approach we isolated synaptosomal fractions from human cortical tissue from relatively early through late stages of PD progression, followed by labeling with iTRAQ and proteomic evaluation. From the proteins confidently identified, 16 that demonstrated changes as PD progressed compared with control were highly represented [88]. From this group we chose to validate and evaluate glutathione S-transferase pi (GSTpi), which is involved in regulating oxidative stress. Interestingly, polymorphisms in the GSTpi gene have been suggested to be risk factors for the development of PD [89]. In our study, GSTpi was found to significantly increase in the synaptosomal fraction as PD progresses from early to advanced stages of the disease. Overexpression of GSTpi demonstrated a neuroprotective function in in vitro models of PD. Interestingly, knockout of GSTpi in mice confers an increase in vulnerability to the dopaminergic neurotoxin, MPTP [90].
An intriguing phenomenon that has been repeatedly observed in PD is the differential vulnerability of the dopamine-containing neurons in the ventral tegmental area (VTA), which is directly apposed to the SNpc in the midbrain. For example, whereas upwards of 90% of dopaminergic neurons are lost in the SNpc, a 40% reduction has been recorded in the VTA [91–95]. A similar pattern of dopaminergic degeneration has been observed in several animal models of PD, including MPTP and rotenone [53,96–100]. This suggests that the dopaminergic neurons of the SNpc possess an endogenous property that renders them substantially more susceptible to the pathogenic mechanism(s) involved in PD. It has been proposed that the differential expression of specific proteins in the SNpc and VTA may explain the distinct pathology observed in these two regions. For instance, the intracellular calcium-binding protein, calbindin D28K is expressed to a greater degree in the VTA compared with the SNpc, and has been shown to colocalize with the dopamine neurons that show resistance to degeneration [101]. In addition, the G-protein inwardly rectifying K+ channel (GIRK) and in particular the GIRK2 isoform, which can regulate dopamine release to the striatum, is specifically expressed in the dopamine neurons of the SNpc [102,103]. Again, using iTRAQ labeling of proteins from SNpc and VTA of control and PD patients paired with shotgun proteomics, we discovered 33 proteins with a greater than 50% change in their relative abundance between these two regions [Caudle et al., Unpublished Data]. Validation and further evaluation of the significance of these proteins to the pathogenesis of PD are currently underway in our lab. Further analysis of these proteins will provide a more in-depth comprehension of the divergent susceptibility to dopaminergic degeneration between the SNpc and VTA in PD.
An area of proteomic research that has been receiving a considerable amount of attention is the identification and quantification of PTMs of proteins and the influence this alteration has on their function and role in disease pathogenesis. In general, the most commonly observed PTMs are phosphorylation, ubiquitination, oxidation and glycosylation of target proteins. The presence of any of these modifications on a protein can elicit both positive and negative effects on its expression, localization and function. Moreover, it is becoming more apparent that modulation of specific proteins by PTMs could have deleterious effects on multiple cellular pathways, leading to disease. For instance, α-synuclein has been shown to be subjected to significant PTMs, usually resulting in an exacerbation of its neurotoxic properties [104].
The use of 2D gel electrophoresis has been extremely powerful in elucidating many of the PTMs associated with specific proteins that may be associated with PD. A series of studies by Choi et al. have characterized the oxidative modification of multiple proteins that have been linked to PD. For example, using human brain tissue, they recorded a significant increase in oxidation of DJ-1, Cu/Zn SOD, as well as UCH-L1 by a combination of carbonyl, cysteine or methionine oxidation, in PD patients compared with controls [105–107]. DJ-1, which functions to mitigate oxidative stress, and its genetic mutations have been previously associated with a genetic susceptibility in PD. It can be speculated that the oxidation of DJ-1 could have significant consequences on its ability to function in attenuating the generation of oxidative stress within the dopamine neuron. Similar to DJ-1, Cu/Zn SOD serves to reduce the amount of intracellular oxidative stress through its dismutation of superoxide, which can readily interact with other radicals to generate highly toxic reactive species. Therefore, modification of its expression and function could interrupt this role and facilitate oxidative damage. Dysregulation of proteasomal function in PD is well documented, with the majority of attention focused on the loss-of-function mutations observed in parkin. Another protein associated with protein degradation and linked to PD is UCH-L1, which removes and recycles the ubiquitin tag from proteins that have been targeted for proteasomal degradation. Recycling of this tag is an important step in sustaining the degradation process. Thus, oxidative modification of UCH-L1 could result in irreversible alteration of its conformation and enzymatic activity, increasing the likelihood of dopaminergic damage.
Although glycosylation is the most prominent PTM, existing as either O-linked (addition of glycan to serine or threonine residues) or N-linked (addition of glycan to asparagine residues), the role of glycosylation and more specifically, aberrant glycosylation of target proteins involved in PD has not been extensively examined [108]. However, it is recognized that a glycosylated form of α-synuclein is ubiquitinated by parkin and targeted for proteasomal degradation [109,110]. As a major function of glycosylation is to assist in the localization of proteins to their appropriate sites of action, alterations of this mechanism could be detrimental to the normal function of the neuron. Recently we have undertaken an unbiased proteomic profiling approach to characterize the glycoproteome in human brain tissue and CSF of controls and patients with Alzheimer’s disease (AD), PD, PD with dementia and dementia with LBs (DLB). Although these studies are still being analyzed and validated and will be published in a forthcoming manuscript, we were able to identify 394 and 283 nonredundant proteins in the human brain and CSF, respectively [111]. When evaluating proteins that were found in both CSF and brain tissue, we found several proteins with known involvement in PD, such as ceruloplasmin and transferrin, which are involved in the regulation of iron and have been reported to be altered in PD [112]. In addition, several overlapping proteins were discovered that are important to general CNS function, including neural cell adhesion molecule and voltage-dependent calcium subunit α2.
Caveats associated with brain tissue proteomics
Like transcriptomics, the use of proteomics has significantly enhanced our understanding of the proteins and pathways involved in PD pathogenesis. Unfortunately, proteomic profiling of human brain tissue is hindered by some of the similar shortcomings, as was seen with transcriptomics. Most prominent is the technology itself, as it is biased towards abundant proteins, that is, signal transduction molecules cannot be identified easily without extensive protein fractionation. The issue can also be confounded by global profiling of the SNpc in control versus PD patients, with the identification of artificial alterations to specific proteins that is congruous with the precipitous loss of dopamine neurons in this region. It can be argued that not all neuronal proteins exhibit a reduction. Indeed, we observe numerous proteins whose relative abundance is increased in the SNpc of PD patients [85]. This is why such rigorous validation and confirmation methods are suggested, followed by evaluation of biological significance, in order to eliminate as much doubt as possible that the changes observed through the proteomic screen are not biased. Extensive validation of candidate proteins revealed by proteomics by an independent method, such as western blotting, is also important because proteins can be identified incorrectly due to a currently incomplete database.
Cerebrospinal fluid
To date, the clinical diagnosis of PD is inadequate, resulting in a significant percentage of misdiagnoses or simply missed diagnosis, with a failure rate of between 50 and 90%, depending on the clinicians’ acumen and experience with such cases. These shortcomings can be attributed to the homogeneity of disease presentation, as many neurodegenerative diseases that arise from damage to the midbrain, such as progressive supranuclear palsy and multiple systems atrophy, share several neurological deficits. Moreover, it is also imperative to delineate a spectrum of disorders within a single disease, for example PD from DLB and PD with dementia. The identification of specific and sensitive biomarkers to help differentiate between multiple diseases, especially at pre-clinical stages, will greatly enhance the diagnostic power of the clinician, which will translate into the initiation of more personalized therapeutic interventions. The use of CSF for the discovery of disease-specific biomarkers has proved to be advantageous owing to its approximation to the brain and the site of pathology, providing a more accurate representation of the state of the brain under normal and disease conditions. Furthermore, CSF samples can be extracted from an individual over time, allowing for longitudinal evaluation of molecular changes throughout the course of the disease. Finally, CSF has the potential to be a useful tool in identifying unique proteins and pathways that may provide insight into the pathogenesis of the disease. While the use of CSF, plasma/serum and blood cells have proven to be especially informative in these endeavors, we will focus the remainder of our discussion on studies that have utilized CSF to evaluate various aspects of PD pathogenesis.
Protein identification
The presence and overlap of so many brain-derived proteins in the CSF suggest an intimate interaction between these two regions. Evidence has suggested that CSF may be involved in mediating cellular signaling between two brain regions through volume transmission or synaptic exocytosis [113–115]. In order for this to occur, it is necessary for structures and mechanisms involved in the transport, protection from degradation, targeting and transduction of the signal to be in place. Extensive CSF protein profiling has revealed more than 3000 proteins, including those involved structurally and/or functionally in the CNS, as well as those related to immune processes [116,117]. A recent study by Harrington et al. reported the presence of subcellular structures, approximately 30–200-nm spheres, further characterized as constituents of synaptic vesicles and exosomes, such as polyunsaturated fatty acids, synaptobrevin, synaptotagmin and syntaxin [115]. The presence of these proteins suggests that neurotransmission and signal transduction is occurring in the CSF. Interestingly, exosomes have been suggested to function within the endosomal pathway as a means for the cell to remove unwanted or unnecessary proteins through fusion with the plasma membrane and subsequent release into the extracellular space [118,119]. Once released, exosomes could interact with other neighboring cells and transfer their contents. In regard to neurodegeneration, exosomes have been shown to sequester amyloid precursor protein and the prion protein, which are involved in the pathogenesis of AD and prion disease, respectively [120,121]. Although abundantly localized intracellularly, mounting evidence has suggested that α-synuclein is also present in the extracellular space, where it could potentially interact with and damage other neurons [122–124]. The mechanism by which α-synuclein is extruded from the cell remains to be fully elucidated. However, we have recently shown a role for Rab11a, an endosomal protein in the secretion of α-synuclein [122]. However, while yet to be demonstrated, exosomal incorporation may also represent a possible release mechanism for α-synuclein into the extracellular compartment.
Protein quantification
In addition to the identification of proteins in CSF, we have also employed several unbiased quantitative proteomic approaches to the discovery of unique protein markers that will allow for the delineation of multiple disease states, particularly, AD, PD and DLB from each other, as well as controls. Utilizing iTRAQ labeling of CSF samples collected from these groups followed by tandem MS we identified over 1500 proteins, which can be separated into several biological categories, including neuronal activities/signal transduction, cell cycle/death and cellular transport, among others [116]. More importantly, we found 136, 72 and 101 proteins that were uniquely altered in AD, PD and DLB, respectively. Finally, validation of several proteins and their use in combination could reliably differentiate between AD, PD and DLB with high sensitivity and specificity. Several of these proteins have undergone further validation and then evaluated as a multianalyte panel for their utility in separating PD and AD from each other and controls [125]. Although more work is needed to elaborate upon these findings and to validate our current knowledge, the generation of this dataset provides an extensive platform from which future biomarker discovery attempts can be initiated, while providing further insight into molecular targets and cellular pathways disrupted or involved in PD and other neurodegenerative diseases.
Caveats of CSF proteomics
The utilization of CSF for proteomic analysis is not immune from its own set of drawbacks, as discussed above in brain tissue proteomics. While several of these have been extensively discussed in previous publications [126], we will briefly appraise a few of the most important points. First and foremost, the proteomic technique is inherently biased towards the identification of proteins of high abundance, a problem greatly enhanced in proteomic analysis of CSF, where approximately 70% of the protein content is attributed to the presence of albumin and immunoglobulins. Since most proteins secreted from the brain into the CSF are in low concentrations (~150 μg/ml), this makes detection extremely difficult. As mentioned, there are ways to selectively enhance the detection of these low-abundance proteins, most notably through exclusion of albumin and immunoglobulins by sequential fractionation of CSF using organic solvents. A further concern when using CSF is the presence of blood contamination in the sample. As the CSF protein profile significantly overlaps with that of plasma, even minor blood contamination can have a major effect on CSF protein concentration. Therefore, particular care should be taken to establish the concentration of red blood cells contained in the CSF sample. Ideally, samples should contain less than ten red blood cells per microliter of CSF.
Metabolomics
Overview of metabolomic techniques
As the development of targeted assays is well described in the literature, we will focus on summarizing the methodology involved in fingerprinting metabolomics. Interested readers are directed to the original publications and more detailed reviews on the topics addressed here [127,128]. Advances in analytical methodology, particularly separation science and more sensitive instrumentation, coupled with sophisticated software for data processing, statistical analysis and metabolite identification have enhanced the ease with which a ‘molecular signature’ can be described. Despite the various methodologies, there are underlying similarities to all metabolomic experiments: sample acquisition, sample preparation, sample analysis to generate data (using Nuclear magnetic resonance [NMR] or a spectrometry-based technique), data analysis and metabolite identification (Figure 3).
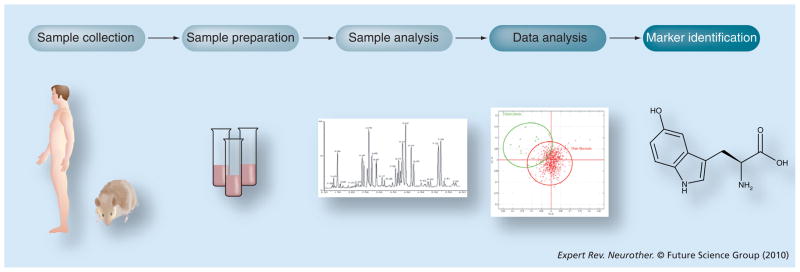
Figure 3. Overview of metabolomics experiment.
Samples are collected from normal and diseased subjects or from treated and untreated groups. The samples are prepared for nuclear magnetic resonance imaging or mass spectrometry-based analysis. Data are generated by the analytical instrument and software is used to discriminate significant markers that correlate with disease or treatment. Databases are used to search for the identity of the markers.
First, the collection of the samples depends on the study question, design and population. For example, although the ultimate tissue of interest may be the brain, it is difficult to collect brain specimens from living subjects and, as such, more easily accessible biofluids, such as blood or urine, are typically used in metabolomics studies. It is critical that the samples are collected using a uniform procedure to prevent spurious associations with changes to the collection methodology during the course of the study (e.g., changes in anticoagulants used to isolate plasma or collection of urine samples at different times of day). Moreover, samples should be aliquoted, stored at −80°C and multiple freeze–thaw cycles should be avoided. Sample preparation is even more important in metabolomics (than in transcriptomics and proteomics) because metabolites are liable and subject to alterations in seconds or even milliseconds.
Second, because of the diversity of small molecules in the samples, there is no one perfect sample preparation method to maximize the detection of all compounds for all biological fluids or tissues. For instance, specific analyses involving lipids will necessitate different sample preparation than polar compounds. The preparation will depend in part upon the analytical method subsequently used. For NMR, samples can be diluted, the pH adjusted or subjected to ultrafiltration to remove proteins. For liquid chromatography MS (LCMS), protein precipitation is usually performed by the addition of organic solvents and/or centrifugation. Gas chromatography MS (GCMS) will detect volatile compounds and treatment of samples with derivatization agents is often necessary. In all cases, the use of an internal standard is recommended to monitor for any sample-to-sample differences in recovery.
As discussed previously, multiple methodologies are available to analyze the biological samples. NMR, LCMS and GCMS are the most popular analytical methods and capillary electrophoresis and capillary electochromatography are the newer techniques. NMR spectroscopy has a lower sensitivity than MS, but has the advantages of high reproducibility and nondestruction of the sample. LCMS methods are usually performed on TOF instruments and have the ability to detect metabolites with high mass accuracy and high sensitivity. However, LCMS techniques are highly dependent upon the chromatographic conditions, such as mobile phase constitution and gradients, column selection (reverse-phase, normal-phase or hydrophilic interaction liquid chromatography ‘HILIC’ columns) and system set-up. In other words, the time at which a particular metabolite elutes from the column may not correspond between different instruments or laboratories, rendering the identification of compounds more difficult. By contrast, the detection of the compounds is much more uniform with GCMS, but may be limited to compounds that possess the functional groups that will react with the derivatization agent.
Metabolomics software is greatly improved, offering ease-of-use and more functionality, but is generally associated with particular instruments (NMR vs MS) or vendors (e.g., Waters, Agilent). Nonetheless, the data analysis protocol is relatively similar: generation of a list of compounds detected using peak alignment for MS data or binning and spectral fitting for NMR data; normalization to the internal standard or other compounds (e.g., creatinine in urine); and statistical analysis. The statistical approaches are more similar to microarray analyses than proteomics and borrow from the rich experience of expression analysis. As such, the identification of significant metabolites typically involves procedures such as principal component analysis, to cluster important differences to yield supposed differences between groups, univariate and multivariate statistics, supervised learning methods and cross validation methods. Significant compounds can be selected for further investigation.
Findings in metabolomics
To date, few studies have utilized metabolomic platforms to evaluate biomarkers associated with molecular signatures and pathways involved in PD pathology. The studies that have capitalized on this technology have focused their evaluation on peripheral body fluids, such as blood/plasma and CSF with the goal of identifying metabolic pathways that are perturbed in PD compared with control subjects. Current analysis of metabolic dysfunction in PD has been focused on the association between levels of urate in the serum or CSF and PD progression. Several studies have determined an inverse correlation between urate concentration and clinical progression of PD, with reduced urate levels suggesting an increase in dopaminergic neurodegeneration and advanced PD symptomology [129–132]. Furthermore, urate levels have been determined to be a sensitive indicator of risk for development of PD, with higher urate levels predicting a significantly lower risk of PD. These findings are very interesting as urate is considered to have important functions as an endogenous antioxidant [133]. Given the prominent role of oxidative stress as a pathogenic pathway involved in PD, these results provide further evidence for the involvement of this pathway in dopaminergic degeneration and PD.
Two studies have further explored the disruption of metabolic pathways in PD through the application of metabolomic platforms to analyze human blood for metabolic signatures. The first study by Bogdanov et al. determined significant alterations to a few metabolites associated with the oxidative stress pathway [134]. In particular, as previously reported, they demonstrated a reduction in uric acid in subjects with clinically defined PD, compared with controls. By contrast, they found an increase in the levels of glutathione as well as 8-hydroxy-2′-deoxyguanosine. Taken together, these data further support the role of oxidative stress in PD and demonstrate the ability to identify metabolic markers that are capable of reliably delineating PD from control subjects.
A more recent study by the same group elaborated upon their previous findings and used metabolomics to evaluate the metabolic profiles of patients with idiopathic PD, PD associated with the G2019S LRRK2 mutation, asymptomatic LRRK2 G2019S carriers and normal controls [135]. As previously seen, they were able to distinguish idiopathic PD cases from controls, based on levels of metabolic signatures. Even more interestingly, they reported a clear delineation between idiopathic PD and the LRRK2 PD group. Furthermore, a partial separation of LRRK2 PD patients and asymptomatic relatives who are carrying the LRRK2 mutation was also seen, although a substantial overlap between the two groups was present. While specific metabolites were not presented in this article, the authors suggest an alteration in the purine pathway, of which urate is an important participant.
Caveats in metabolomics
One of the major challenges of metabolomics is the identification of the metabolites, which starts with comparing the analytical signatures (e.g., NMR or LCMS spectra) of the metabolite against databases of known compounds. At this time, public and commercial databases contain compounds such as lipids, amino acids, fatty acids, amines, alcohols, sugars, organic phosphates, hydroxyl acids, aromatics, purines and other high abundance or clinically important molecules. Nevertheless, these databases are limited or incomplete as secondary metabolites, drugs and environmental compounds are less represented. Another issue associated with metabolomic studies is the potential introduction of sample variation that can be attributed to differences in sample collection and preparation. In this regard, stringent parameters should be set forth that address not only the constituents of the subject’s diet, which may interfere with the sample analysis, but also the time of sample collection, addition of anticoagulants for serum collection and the appropriate preservation measures. Adherence to standardized guidelines related to these issues will significantly reduce inter-individual variability and facilitate the generation of more comprehensive publicly available metabolite databases.
Common findings between transcriptomics, proteomics & metabolomics
Ideally, the analysis of parkinsonian and normal SNpc by independent -omics techniques would yield concordant findings between the different platforms; meaning, if gene A was observed to increase, then its protein product would also demonstrate a similar increase, followed by an appropriate alteration in the corresponding metabolic profile. Unfortunately, as we have seen, there exists a general lack of overlap in results from one study to another, even when using the same platform. Furthermore, this discordance is compounded when attempting to compare findings from tissue versus those obtained from CSF or plasma/serum. While some variability is biological, for example, some genes are not translated to proteins, the overall heterogeneity of the human population and the lack of presence of CNS-specific proteins in peripheral fluids, a significant contribution to the inconsistent results relates to variables involved in experimentation, including the evaluation of different regions or the use of different technologies. In terms of brain regions studied, it is not always clear which compartment of the substantia nigra was utilized, particlularly whether only the pars compacta was studied, which is the most vulnerable to dopaminergic degeneration, or whether it was combined with the pars reticulata, which is relatively spared in PD. Furthermore, a differential vulnerability is seen within the pars compacta, with the lateral tier exhibiting a greater loss of dopamine neurons than the medial component. Similarly, the use of different techniques and equipment introduces a substantial amount of complexity to the interpretation and integration of several studies, as these technologies may have different detection and resolution capabilities. Thus, in order to create a unified understanding of the alterations that are occurring in the SNpc throughout PD it is imperative to establish a standardized manner of defining and evaluating these changes. A few of these caveats were touched upon when discussing the use of transcriptomics in PD research. However, as the use of proteomics and metabolomics in PD increases, it is anticipated that similar issues will come to light. Indeed, when comparing the human midbrain proteome to the human CSF proteome, only approximately 20% of the proteins identified are shared between the two media [75], which can be attributed, at least in part, to constant changes in the human database.
Some of these issues can be resolved when less focus is put on the agreement of specific products and instead the entirety of the pathway within which particular genes/proteins/metabolites reside is considered. Indeed, when considering transcriptomics and proteomics, similar pathways have been shown to change between control and PD subjects, including synaptic transmission, mitochondrial function, protein degradation and oxidative stress. Interestingly, when this approach is applied it appears that alteration to the oxidative stress pathway is a common feature discovered in transcriptomic, proteomic, as well as metabolomic studies of PD. Moreover, while the transcriptomic and proteomic studies discovered this alteration in human midbrain tissue, metabolomics uncovered this pathway in human serum. Given the agreement of this finding through three independent means, in addition to different body tissues, these data provide a strong argument for the role of oxidative stress in the pathogenesis of PD. Finally, we feel that the redundant identification of specific pathways, such as oxidative stress, by independent research groups, techniques and biofluids is an important demonstration because it not only provides validation for these specific mechanisms being involved in PD pathogenesis but also an assurance that newly developed -omics do provide meaningful data, such as shedding light on novel targets and pathways. Indeed, multiple novel genes/proteins, such as mortalin, GATA-2 and ST13, revealed by -omics, have been confirmed to be biologically important in PD pathogenesis/progression.
Expert commentary
Although extensive effort has been put forth in order to identify and explain the precise mechanisms and molecular cascades that initiate and propagate neurodegeneration in PD, our understanding of the disease process is still unsatisfactory. A recent focus in PD research has been on the multidiscipline approach to elucidating pathways and targets that are perturbed in PD. It is believed that the application of transcriptomics, proteomics and metabolomics will provide an integrative approach to the problem of neurodegeneration in the SNpc as well as other affected regions, creating a more methodologically comprehensive pursuit of an understanding of the disease process. Through these techniques we have been able to confirm previously held beliefs about the role of mitochondrial dysfunction, proteasomal alteration and disruption of dopamine neurotransmission in the pathogenesis of PD. In addition, we have been able to identify and bring attention to unique and novel pathways, such as axon guidance, that may hold promise as a key mediator of neuronal loss.
A more important question that should be put forth is: what are the implications of these findings for our overall understanding of PD pathogenesis and the potential for the development of therapeutic interventions? As mentioned previously, the shared alteration to the oxidative stress pathway, as demonstrated by different -omics techniques and within different biofluids, suggests that this pathway and the myriad of biological components involved may represent the final common pathway through which all other pathological mechanism associated with PD eventually feed into and perpetuate/potentiate the pathogenesis. Given that each disease is so unique, however, this hypothesis would necessitate a further concentration of effort in understanding this pathway and the integration of other novel components and circuits revealed by -omics, such as mortalin or GATA-2, that are specifically related to PD.
However, given our current advances, there are still several issues that need to be addressed and resolved. In particular, the relative incompleteness of the proteomic, metabolomic and, to a lesser degree, transcriptomic databases continues to be a hindrance in the identification of these respective molecules. Indeed, an increased deposition of identified genes, proteins and metabolites into their respective databases would greatly enhance the analysis of specific pathways and proteins involved in PD, which could provide more insight into the pathogenesis of the disease. In addition, it would provide a platform for meta-analysis to be performed, which could assuage much of the between-study variability currently encountered when analyzing multiple studies. Furthermore, a more conscious effort needs to be made by each research group to confirm genes, proteins and metabolites discovered through each platform. This confirmation would provide assistance in the further biological validation of each in order to assess their relative biological contribution/significance to PD pathogenesis.
Five-year view
As the complexity of the human brain is further realized, research investigations are moving towards a more holistic approach to the understanding of the pathological cascades that underlie the disease process associated with PD. Dissection of these molecular pathways and mechanisms at multiple biological levels will be imperative to our understanding of these processes. The -omics tools with which to conduct these analyses are already in place and being implemented in the study of PD and other neurodegenerative disorders. As these techniques are further employed to address the pathogenesis of PD, we will find an elaboration of the genetic, proteomic and metabolomic databases available for target identification, allowing for a more powerful and extensive evaluation of pathways involved in the manifestation of PD. Furthermore, investigators will be able to take advantage of the capabilities of each platform, integrate their findings and thus take a more systems biology-based approach to the investigation of PD. This approach would assist in reducing the relative complexity of the brain and the pathways involved in PD, which would allow for a more thorough evaluation of not only the general components of these pathways, but also the effects of the interactions that occur between them. This will require a more conscious effort to coordinate these data, techniques and, more importantly, sample collection and preparation in order to uncover a potentially unifying disease pathway(s). In this regard, it should be kept in mind that several disease states, such as PD, AD and amyotrophic lateral sclerosis share similar pathological pathways, such as the oxidative stress pathway. This suggests that they may also demonstrate commonalities in their restorative pathways. Being mindful of these similarities will benefit not only our general knowledge of PD, but allow for the identification of more specific therapeutic targets aimed at ameliorating or abrogating the disease process.
Collaborative efforts aimed at characterizing the genetic, proteomic and metabolomic state from the same control and PD cases would enhance our holistic understanding of the pathology driving PD pathogenesis. In this vein of thought, the application of these integrative efforts to longitudinal studies of various human biofluids would allow for the delineation of the disease progression from a genetic, proteomic and metabolomic context, again, providing comprehensive insight into the pathways and mechanisms that may be altered as the pathologic state progresses from non-symptomatic to symptomatic and beyond. Furthermore, analysis of these samples would potentially contribute to identification of markers of progression that are critical to clinical trials.
Key issues.
Focus has been placed on the use of -omics, particularly transcriptomics, proteomics and metabolomics, in order to elucidate many of the pathogenic aspects of Parkinson’s disease (PD).
Transcriptomics is concerned with the identification of genes and gene expression, while proteomics is focused on recognizing and measuring respective changes in proteins. Finally, metabolomics is concerned with the identification and quantification of metabolites to provide a signature of the metabolic state at that point in time.
In PD, these techniques have been applied to various biological media, including brain tissue, cerebrospinal fluid, blood and blood constituents.
Transcriptomics has reliably identified alterations in pathways in the substantia nigra pars compacta and blood of PD subjects associated with mitochondrial and proteasomal function, dopamine neurotransmission and oxidative stress. In addition, it has uncovered the axon guidance pathway as a potential contributor to dopaminergic neurodegeneration.
The use of proteomics has provided a comprehensive characterization of the human midbrain, as well as the protein composition of human cerebrospinal fluid. This platform has been further utilized to identify specific proteins and pathways that are altered in the biofluids of PD compared with controls.
Metabolomics-based studies of blood from PD and control patients have further uncovered the role of oxidative stress in the pathogenesis of PD. Furthermore, a differential metabolic profile was observed between patients with idiopathic PD, PD associated with the G2019S LRRK2 mutation, asymptomatic LRRK2 G2019S carriers and normal controls, allowing for the separation of these groups from each other.
Despite multiple caveats associated with each -omics technique, several processes critical to PD pathogenesis, such as mitochondrial dysfunction and synaptogenesis, have been commonly indentified by different methods. Furthermore, appraisal of the findings from each of the -omics-based studies found the oxidative stress pathway to be uniquely altered between control and PD patients, independent of the technique or type of tissue sample used.
Further integration of these techniques into a systems biology approach will lead to a more comprehensive characterization of the molecular mechanisms and pathways perturbed in PD.
In addition to the identification of alterations to specific genes, proteins and metabolites associated with PD pathogenesis, transcriptomics, proteomics and metabolomics are exquisitely suited for the identification of sensitive biomarkers to help differentiate between neurodegenerative disorders with similar clinical phenotypes to PD, as well as assist in the preclinical diagnosis of PD and other related movement disorders.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
The effort of the authors was supported by the National Institutes of Health grants (ES004696, NS057567, AG025327, AG033398, NS060252, ES016873 and NS062684) as well as grants from the Michael J Fox Foundation. W Michael Caudle is supported by award number K99ES017477 from the National Institute of Environmental Health Science. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Fahn S. Description of Parkinson’s disease as a clinical syndrome. Ann NY Acad Sci. 2003;991:1–14. doi: 10.1111/j.1749-6632.2003.tb07458.x. [DOI] [PubMed] [Google Scholar]
- 2.DeLong MR, Wichmann T. Circuits and circuit disorders of the basal ganglia. Arch Neurol. 2007;64(1):20–24. doi: 10.1001/archneur.64.1.20. [DOI] [PubMed] [Google Scholar]
- 3.Ehringer H, Hornykiewicz O. Distribution of noradrenaline and dopamine (3-hydroxytyramine) in the human brain and their behavior in diseases of the extrapyramidal system. Klin Wochenschr. 1960;38:1236–1239. doi: 10.1007/BF01485901. [DOI] [PubMed] [Google Scholar]
- 4.Jellinger K. New developments in the pathology of Parkinson’s disease. Adv Neurol. 1990;53:1–16. [PubMed] [Google Scholar]
- 5.Braak H, Braak E, Yilmazer D, et al. Nigral and extranigral pathology in Parkinson’s disease. J Neural Transm Suppl. 1995;46:15–31. [PubMed] [Google Scholar]
- 6.Forno LS. Neuropathology of Parkinson’s disease. J Neuropathol Exp Neurol. 1996;55(3):259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Schlossmacher MG, Frosch MP, Gai WP, et al. Parkin localizes to the Lewy bodies of Parkinson disease and dementia with Lewy bodies. Am J Pathol. 2002;160(5):1655–1667. doi: 10.1016/S0002-9440(10)61113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenner P, Olanow CW. Understanding cell death in Parkinson’s disease. Ann Neurol. 1998;44(3 Suppl 1):S72–S84. doi: 10.1002/ana.410440712. [DOI] [PubMed] [Google Scholar]
- 9.Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):S26–S36. doi: 10.1002/ana.10483. discussion S36–S28. [DOI] [PubMed] [Google Scholar]
- 10.Olanow CW, Tatton WG. Etiology and pathogenesis of Parkinson’s disease. Annu Rev Neurosci. 1999;22:123–144. doi: 10.1146/annurev.neuro.22.1.123. [DOI] [PubMed] [Google Scholar]
- 11.Blandini F, Porter RH, Greenamyre JT. Glutamate and Parkinson’s disease. Mol Neurobiol. 1996;12(1):73–94. doi: 10.1007/BF02740748. [DOI] [PubMed] [Google Scholar]
- 12.Beal MF. Excitotoxicity and nitric oxide in Parkinson’s disease pathogenesis. Ann Neurol. 1998;44(3 Suppl 1):S110–S114. doi: 10.1002/ana.410440716. [DOI] [PubMed] [Google Scholar]
- 13.Wersinger C, Sidhu A. An inflammatory pathomechanism for Parkinson’s disease? Curr Med Chem. 2006;13(5):591–602. doi: 10.2174/092986706776055760. [DOI] [PubMed] [Google Scholar]
- 14.Schapira AH. Mitochondrial dysfunction in neurodegenerative disorders. Biochim Biophys Acta. 1998;1366(1–2):225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- 15.Schapira AH, Mann VM, Cooper JM, et al. Mitochondrial function in Parkinson’s disease. The Royal Kings and Queens Parkinson’s Disease Research Group. Ann Neurol. 1992;32(Suppl):S116–S124. doi: 10.1002/ana.410320720. [DOI] [PubMed] [Google Scholar]
- 16.Olanow CW, McNaught KS. Ubiquitin-proteasome system and Parkinson’s disease. Mov Disord. 2006;21(11):1806–1823. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- 17.McNaught KS, Jackson T, JnoBaptiste R, Kapustin A, Olanow CW. Proteasomal dysfunction in sporadic Parkinson’s disease. Neurology. 2006;66(10 Suppl 4):S37–S49. doi: 10.1212/wnl.66.10_suppl_4.s37. [DOI] [PubMed] [Google Scholar]
- 18.Petrucelli L, Dawson TM. Mechanism of neurodegenerative disease: role of the ubiquitin proteasome system. Ann Med. 2004;36(4):315–320. doi: 10.1080/07853890410031948. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch EC. Mechanism and consequences of nerve cell death in Parkinson’s disease. J Neural Transm Suppl. 1999;56:127–137. doi: 10.1007/978-3-7091-6360-3_7. [DOI] [PubMed] [Google Scholar]
- 20.Tatton NA, Maclean-Fraser A, Tatton WG, Perl DP, Olanow CW. A fluorescent double-labeling method to detect and confirm apoptotic nuclei in Parkinson’s disease. Ann Neurol. 1998;44(3 Suppl 1):S142–S148. doi: 10.1002/ana.410440721. [DOI] [PubMed] [Google Scholar]
- 21.Tatton NA. Increased caspase 3 and Bax immunoreactivity accompany nuclear GAPDH translocation and neuronal apoptosis in Parkinson’s disease. Exp Neurol. 2000;166(1):29–43. doi: 10.1006/exnr.2000.7489. [DOI] [PubMed] [Google Scholar]
- 22.Jellinger KA. Cell death mechanisms in neurodegeneration. J Cell Mol Med. 2001;5(1):1–17. doi: 10.1111/j.1582-4934.2001.tb00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 24.Schapira AH. Etiology of Parkinson’s disease. Neurology. 2006;66(10 Suppl 4):S10–S23. doi: 10.1212/wnl.66.10_suppl_4.s10. [DOI] [PubMed] [Google Scholar]
- 25.Allen W. Inheritance of the shaking palsy. Arch Int Med. 1937;60:424–436. [Google Scholar]
- 26.Leroux P-D. PhD Thesis. Imprimeur de la Faculte de Medecine; Paris, France: 1880. Contributions to the reasons and causes of shaking palsy. [Google Scholar]
- 27.Mjones H. Paralysis agitans: a clinical and genetic study. Acta Psychiatr Neurol Scand Suppl. 1949;54:1–195. [Google Scholar]
- 28.Tanner CM, Goldman SM. Epidemiology of Parkinson’s disease. Neurol Clin. 1996;14(2):317–335. doi: 10.1016/S0733-8619(05)70259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner CM, Langston JW. Do environmental toxins cause Parkinson’s disease? A critical review. Neurology. 1990;40(10 Suppl 3):17–30. discussion 30–11. [PubMed] [Google Scholar]
- 30.Ascherio A, Chen H, Weisskopf MG, et al. Pesticide exposure and risk for Parkinson’s disease. Ann Neurol. 2006;60(2):197–203. doi: 10.1002/ana.20904. [DOI] [PubMed] [Google Scholar]
- 31.Corrigan FM, Wienburg CL, Shore RF, Daniel SE, Mann D. Organochlorine insecticides in substantia nigra in Parkinson’s disease. J Toxicol Environ Health A. 2000;59(4):229–234. doi: 10.1080/009841000156907. [DOI] [PubMed] [Google Scholar]
- 32.Hubble JP, Cao T, Hassanein RE, Neuberger JS, Koller WC. Risk factors for Parkinson’s disease. Neurology. 1993;43(9):1693–1697. doi: 10.1212/wnl.43.9.1693. [DOI] [PubMed] [Google Scholar]
- 33.Priyadarshi A, Khuder SA, Schaub EA, Shrivastava S. A meta-analysis of Parkinson’s disease and exposure to pesticides. Neurotoxicology. 2000;21(4):435–440. [PubMed] [Google Scholar]
- 34.Semchuk KM, Love EJ, Lee RG. Parkinson’s disease and exposure to rural environmental factors: a population based case–control study. Can J Neurol Sci. 1991;18(3):279–286. doi: 10.1017/s0317167100031826. [DOI] [PubMed] [Google Scholar]
- 35.Semchuk KM, Love EJ, Lee RG. Parkinson’s disease and exposure to agricultural work and pesticide chemicals. Neurology. 1992;42(7):1328–1335. doi: 10.1212/wnl.42.7.1328. [DOI] [PubMed] [Google Scholar]
- 36.Steenland K, Hein MJ, Cassinelli RT, 2nd, et al. Polychlorinated biphenyls and neurodegenerative disease mortality in an occupational cohort. Epidemiology. 2006;17(1):8–13. doi: 10.1097/01.ede.0000190707.51536.2b. [DOI] [PubMed] [Google Scholar]
- 37.Fleming L, Mann JB, Bean J, Briggle T, Sanchez-Ramos JR. Parkinson’s disease and brain levels of organochlorine pesticides. Ann Neurol. 1994;36(1):100–103. doi: 10.1002/ana.410360119. [DOI] [PubMed] [Google Scholar]
- 38.Ho SC, Woo J, Lee CM. Epidemiologic study of Parkinson’s disease in Hong Kong. Neurology. 1989;39(10):1314–1318. doi: 10.1212/wnl.39.10.1314. [DOI] [PubMed] [Google Scholar]
- 39.Le Couteur DG, McLean AJ, Taylor MC, Woodham BL, Board PG. Pesticides and Parkinson’s disease. Biomed Pharmacother. 1999;53(3):122–130. doi: 10.1016/S0753-3322(99)80077-8. [DOI] [PubMed] [Google Scholar]
- 40.Norris EH, Uryu K, Leight S, et al. Pesticide exposure exacerbates α-synucleinopathy in an A53T transgenic mouse model. Am J Pathol. 2007;170(2):658–666. doi: 10.2353/ajpath.2007.060359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev. 2002;3(8):579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- 42.Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotech. 2004;22(6):656–658. doi: 10.1038/nbt0604-656b. author reply 658. [DOI] [PubMed] [Google Scholar]
- 43••.Sutherland GT, Matigian NA, Chalk AM, et al. A cross-study transcriptional analysis of Parkinson’s disease. PLoS One. 2009;4(3):e4955. doi: 10.1371/journal.pone.0004955. Reanalyzes transcriptional data from control and Parkinson’s disease (PD) substantia nigra pars compacta (SNpc) to create an unbiased evaluation of gene changes that are independent of loss of dopaminergic neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller RM, Federoff HJ. Altered gene expression profiles reveal similarities and differences between Parkinson disease and model systems. Neuroscientist. 2005;11(6):539–549. doi: 10.1177/1073858405278330. [DOI] [PubMed] [Google Scholar]
- 45.Miller RM, Kiser GL, Kaysser-Kranich TM, et al. Robust dysregulation of gene expression in substantia nigra and striatum in Parkinson’s disease. Neurobiol Dis. 2006;21(2):305–313. doi: 10.1016/j.nbd.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Duke DC, Moran LB, Kalaitzakis ME, et al. Transcriptome analysis reveals link between proteasomal and mitochondrial pathways in Parkinson’s disease. Neurogenetics. 2006;7(3):139–148. doi: 10.1007/s10048-006-0033-5. [DOI] [PubMed] [Google Scholar]
- 47.Duke DC, Moran LB, Pearce RK, Graeber MB. The medial and lateral substantia nigra in Parkinson’s disease: mRNA profiles associated with higher brain tissue vulnerability. Neurogenetics. 2007;8(2):83–94. doi: 10.1007/s10048-006-0077-6. [DOI] [PubMed] [Google Scholar]
- 48.Mandel S, Grunblatt E, Riederer P, et al. Gene expression profiling of sporadic Parkinson’s disease substantia nigra pars compacta reveals impairment of ubiquitin-proteasome subunits, SKP1A, aldehyde dehydrogenase, and chaperone HSC-70. Ann NY Acad Sci. 2005;1053:356–375. doi: 10.1196/annals.1344.031. [DOI] [PubMed] [Google Scholar]
- 49.Grunblatt E, Mandel S, Jacob-Hirsch J, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J Neural Transm. 2004;111(12):1543–1573. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 50.Hauser MA, Li YJ, Xu H, et al. Expression profiling of substantia nigra in Parkinson disease, progressive supranuclear palsy, and frontotemporal dementia with parkinsonism. Arch Neurol. 2005;62(6):917–921. doi: 10.1001/archneur.62.6.917. [DOI] [PubMed] [Google Scholar]
- 51•.Simunovic F, Yi M, Wang Y, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132(Pt 7):1795–1809. doi: 10.1093/brain/awn323. Comprehensive identification of gene alterations in the SNpc of PD subjects using laser capture microdissection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Przedborski S, Jackson-Lewis V, Djaldetti R, et al. The parkinsonian toxin MPTP: action and mechanism. Restor Neurol Neurosci. 2000;16(2):135–142. [PubMed] [Google Scholar]
- 53.Betarbet R, Sherer TB, MacKenzie G, et al. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3(12):1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 54.Jackson-Lewis V, Jakowec M, Burke RE, Przedborski S. Time course and morphology of dopaminergic neuronal death caused by the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Neurodegeneration. 1995;4(3):257–269. doi: 10.1016/1055-8330(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 55.Schapira AH. Human complex I defects in neurodegenerative diseases. Biochim Biophys Acta. 1998;1364(2):261–270. doi: 10.1016/s0005-2728(98)00032-2. [DOI] [PubMed] [Google Scholar]
- 56.Olanow CW, McNaught KS. Ubiquitin-proteasome system and Parkinson’s disease. Mov Disord. 2006;21(11):1806–1823. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- 57.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 58.Mata IF, Lockhart PJ, Farrer MJ. Parkin genetics: one model for Parkinson’s disease. Hum Mol Gen. 2004;13(Spec 1):R127–R133. doi: 10.1093/hmg/ddh089. [DOI] [PubMed] [Google Scholar]
- 59.McNaught KS, Bjorklund LM, Belizaire R, et al. Proteasome inhibition causes nigral degeneration with inclusion bodies in rats. Neuroreport. 2002;13(11):1437–1441. doi: 10.1097/00001756-200208070-00018. [DOI] [PubMed] [Google Scholar]
- 60.Galvin JE, Ginsberg SD. Expression profiling in the aging brain: a perspective. Ageing Res Rev. 2005;4(4):529–547. doi: 10.1016/j.arr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Nair-Roberts RG, Chatelain-Badie SD, Benson E, et al. Stereological estimates of dopaminergic, GABAergic and glutamatergic neurons in the ventral tegmental area, substantia nigra and retrorubral field in the rat. Neuroscience. 2008;152(4):1024–1031. doi: 10.1016/j.neuroscience.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olanow CW. The pathogenesis of cell death in Parkinson’s disease – 2007. Mov Disord. 2007;22(Suppl 17):S335–S342. doi: 10.1002/mds.21675. [DOI] [PubMed] [Google Scholar]
- 63.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics (Oxford, England) 2006;22(13):1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 64.Bossers K, Meerhoff G, Balesar R, et al. Analysis of gene expression in Parkinson’s disease: possible involvement of neurotrophic support and axon guidance in dopaminergic cell death. Brain Pathol (Zurich, Switzerland) 2009;19(1):91–107. doi: 10.1111/j.1750-3639.2008.00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lesnick TG, Papapetropoulos S, Mash DC, et al. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Gen. 2007;3(6):e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66••.Lin L, Lesnick TG, Maraganore DM, Isacson O. Axon guidance and synaptic maintenance: preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 2009;32(3):142–149. doi: 10.1016/j.tins.2008.11.006. Very nice appraisal of the role of axon guidance in neurodegeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67•.Zhang J, Goodlett DR, Montine TJ. Proteomic biomarker discovery in cerebrospinal fluid for neurodegenerative diseases. J Alzheimers Dis. 2005;8(4):377–386. doi: 10.3233/jad-2005-8407. Thorough review of the application of proteomics to the discovery of cerebrospinal fluid (CSF) biomarkers. [DOI] [PubMed] [Google Scholar]
- 68.Scherzer CR, Eklund AC, Morse LJ, et al. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc Natl Acad Sci USA. 2007;104(3):955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scherzer CR, Grass JA, Liao Z, et al. GATA transcription factors directly regulate the Parkinson’s disease-linked gene α-synuclein. Proc Natl Acad Sci USA. 2008;105(31):10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. Hsp70 reduces α-synuclein aggregation and toxicity. J Biol Chem. 2004;279(24):25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 71.Castensson A, Emilsson L, Preece P, Jazin EE. High-resolution quantification of specific mRNA levels in human brain autopsies and biopsies. Genome Res. 2000;10(8):1219–1229. doi: 10.1101/gr.10.8.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 73.Pan S, Rush J, Peskind ER, et al. Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 2008;7(2):720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- 74.Pan S, Shi M, Jin J, et al. Proteomics identification of proteins in human cortex using multidimensional separations and MALDI tandem mass spectrometer. Mol Cell Proteomics. 2007;6(10):1818–1823. doi: 10.1074/mcp.M700158-MCP200. [DOI] [PubMed] [Google Scholar]
- 75•.Kitsou E, Pan S, Zhang J, et al. Identification of proteins in human substantia nigra. Proteomics Clin Appl. 2008;2:776–782. doi: 10.1002/prca.200800028. Comprehensive identification of proteins in the human substantia nigra by proteomics. [DOI] [PubMed] [Google Scholar]
- 76.Tribl F, Gerlach M, Marcus K, et al. “Subcellular proteomics” of neuromelanin granules isolated from the human brain. Mol Cell Proteomics. 2005;4(7):945–957. doi: 10.1074/mcp.M400117-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Tribl F, Marcus K, Bringmann G, et al. Proteomics of the human brain: sub-proteomes might hold the key to handle brain complexity. J Neural Transm. 2006;113(8):1041–1054. doi: 10.1007/s00702-006-0513-7. [DOI] [PubMed] [Google Scholar]
- 78.Tribl F, Marcus K, Meyer HE, et al. Subcellular proteomics reveals neuromelanin granules to be a lysosome-related organelle. J Neural Transm. 2006;113(6):741–749. doi: 10.1007/s00702-006-0452-3. [DOI] [PubMed] [Google Scholar]
- 79.Fedorow H, Tribl F, Halliday G, et al. Neuromelanin in human dopamine neurons: comparison with peripheral melanins and relevance to Parkinson’s disease. Prog Neurobiol. 2005;75(2):109–124. doi: 10.1016/j.pneurobio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 80.Double KL, Rowe DB, Carew-Jones FM, et al. Anti-melanin antibodies are increased in sera in Parkinson’s disease. Exp Neurol. 2009;217(2):297–301. doi: 10.1016/j.expneurol.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Xia Q, Liao L, Cheng D, et al. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–3856. doi: 10.2741/2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Braak H, Del Tredici K, Rub U, et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 83.Leverenz JB, Umar I, Wang Q, et al. Proteomic identification of novel proteins in cortical lewy bodies. Brain Pathol (Zurich, Switzerland) 2007;17(2):139–145. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basso M, Giraudo S, Corpillo D, et al. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteomics. 2004;4(12):3943–3952. doi: 10.1002/pmic.200400848. [DOI] [PubMed] [Google Scholar]
- 85.Jin J, Hulette C, Wang Y, et al. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol Cell Proteomics. 2006;5(7):1193–1204. doi: 10.1074/mcp.M500382-MCP200. [DOI] [PubMed] [Google Scholar]
- 86.Shi M, Jin J, Wang Y, et al. Mortalin: a protein associated with progression of Parkinson disease? J Neuropathol Exp Neurol. 2008;67(2):117–124. doi: 10.1097/nen.0b013e318163354a. [DOI] [PubMed] [Google Scholar]
- 87.Jin J, Li GJ, Davis J, et al. Identification of novel proteins associated with both α-synuclein and DJ-1. Mol Cell Proteomics. 2007;6(5):845–859. doi: 10.1074/mcp.M600182-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Shi M, Bradner J, Bammler TK, et al. Identification of glutathione S-transferase pi as a protein involved in Parkinson disease progression. Am J Pathol. 2009;175(1):54–65. doi: 10.2353/ajpath.2009.081019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelada SN, Stapleton PL, Farin FM, et al. Glutathione S-transferase M1, T1, and P1 polymorphisms and Parkinson’s disease. Neurosci Lett. 2003;337(1):5–8. doi: 10.1016/s0304-3940(02)01286-7. [DOI] [PubMed] [Google Scholar]
- 90.Smeyne M, Boyd J, Raviie Shepherd K, et al. GSTpi expression mediates dopaminergic neuron sensitivity in experimental parkinsonism. Proc Natl Acad Sci USA. 2007;104(6):1977–1982. doi: 10.1073/pnas.0610978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Damier P, Hirsch EC, Agid Y, Graybiel AM. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain. 1999;122(Pt 8):1437–1448. doi: 10.1093/brain/122.8.1437. [DOI] [PubMed] [Google Scholar]
- 92.Fearnley JM, Lees AJ. Ageing and Parkinson’s disease: substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 93.Pakkenberg B, Moller A, Gundersen HJ, Mouritzen Dam A, Pakkenberg H. The absolute number of nerve cells in substantia nigra in normal subjects and in patients with Parkinson’s disease estimated with an unbiased stereological method. J Neurol Neurosurg Psychiatry. 1991;54(1):30–33. doi: 10.1136/jnnp.54.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McGeer PL, Itagaki S, Akiyama H, McGeer EG. Rate of cell death in parkinsonism indicates active neuropathological process. Ann Neurol. 1988;24(4):574–576. doi: 10.1002/ana.410240415. [DOI] [PubMed] [Google Scholar]
- 95.McRitchie DA, Cartwright HR, Halliday GM. Specific A10 dopaminergic nuclei in the midbrain degenerate in Parkinson’s disease. Exp Neurol. 1997;144(1):202–213. doi: 10.1006/exnr.1997.6418. [DOI] [PubMed] [Google Scholar]
- 96.German DC, Dubach M, Askari S, Speciale SG, Bowden DM. 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced parkinsonian syndrome in Macaca fascicularis: which midbrain dopaminergic neurons are lost? Neuroscience. 1988;24(1):161–174. doi: 10.1016/0306-4522(88)90320-x. [DOI] [PubMed] [Google Scholar]
- 97.Varastet M, Riche D, Maziere M, Hantraye P. Chronic MPTP treatment reproduces in baboons the differential vulnerability of mesencephalic dopaminergic neurons observed in Parkinson’s disease. Neuroscience. 1994;63(1):47–56. doi: 10.1016/0306-4522(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 98.Waters CM, Hunt SP, Jenner P, Marsden CD. An immunohistochemical study of the acute and long-term effects of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in the marmoset. Neuroscience. 1987;23(3):1025–1039. doi: 10.1016/0306-4522(87)90178-3. [DOI] [PubMed] [Google Scholar]
- 99.German DC, Nelson EL, Liang CL, et al. The neurotoxin MPTP causes degeneration of specific nucleus A8, A9 and A10 dopaminergic neurons in the mouse. Neurodegeneration. 1996;5(4):299–312. doi: 10.1006/neur.1996.0041. [DOI] [PubMed] [Google Scholar]
- 100.Rodriguez M, Barroso-Chinea P, Abdala P, Obeso J, Gonzalez-Hernandez T. Dopamine cell degeneration induced by intraventricular administration of 6-hydroxydopamine in the rat: similarities with cell loss in Parkinson’s disease. Exp Neurol. 2001;169(1):163–181. doi: 10.1006/exnr.2000.7624. [DOI] [PubMed] [Google Scholar]
- 101.Yamada T, McGeer PL, Baimbridge KG, McGeer EG. Relative sparing in Parkinson’s disease of substantia nigra dopamine neurons containing calbindin-D28K. Brain Res. 1990;526(2):303–307. doi: 10.1016/0006-8993(90)91236-a. [DOI] [PubMed] [Google Scholar]
- 102.Schein JC, Hunter DD, Roffler-Tarlov S. Girk2 expression in the ventral midbrain, cerebellum, and olfactory bulb and its relationship to the murine mutation weaver. Dev Biol. 1998;204(2):432–450. doi: 10.1006/dbio.1998.9076. [DOI] [PubMed] [Google Scholar]
- 103.Karschin C, Dissmann E, Stuhmer W, Karschin A. IRK(1–3) and GIRK(1–4) inwardly rectifying K+ channel mRNAs are differentially expressed in the adult rat brain. J Neurosci. 1996;16(11):3559–3570. doi: 10.1523/JNEUROSCI.16-11-03559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beyer K. α-synuclein structure, posttranslational modification and alternative splicing as aggregation enhancers. Acta Neuropathol. 2006;112(3):237–251. doi: 10.1007/s00401-006-0104-6. [DOI] [PubMed] [Google Scholar]
- 105.Choi J, Levey AI, Weintraub ST, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279(13):13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 106.Choi J, Rees HD, Weintraub ST, et al. Oxidative modifications and aggregation of Cu, Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J Biol Chem. 2005;280(12):11648–11655. doi: 10.1074/jbc.M414327200. [DOI] [PubMed] [Google Scholar]
- 107.Choi J, Sullards MC, Olzmann JA, et al. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem. 2006;281(16):10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3(3):556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 109.Shimura H, Schlossmacher MG, Hattori N, et al. Ubiquitination of a new form of α-synuclein by parkin from human brain: implications for Parkinson’s disease. Science (NY) 2001;293(5528):263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 110.Farrer M, Maraganore DM, Lockhart P, et al. α-synuclein gene haplotypes are associated with Parkinson’s disease. Hum Mol Gen. 2001;10(17):1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- 111.Hwang H, Zhang J, Chung KA, et al. Glycoproteomics in neurodegenerative diseases. Mass Spect Rev. 2010;29(1):79–125. doi: 10.1002/mas.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zecca L, Stroppolo A, Gatti A, et al. The role of iron and copper molecules in the neuronal vulnerability of locus coeruleus and substantia nigra during aging. Proc Natl Acad Sci USA. 2004;101(26):9843–9848. doi: 10.1073/pnas.0403495101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vigh B, Manzano e Silva MJ, Frank CL, et al. The system of cerebrospinal fluid-contacting neurons. Its supposed role in the nonsynaptic signal transmission of the brain. Histol Histopathol. 2004;19(2):607–628. doi: 10.14670/HH-19.607. [DOI] [PubMed] [Google Scholar]
- 114.Nilsson C, Lindvall-Axelsson M, Owman C. Neuroendocrine regulatory mechanisms in the choroid plexus-cerebrospinal fluid system. Brain Res Brain Res Rev. 1992;17(2):109–138. doi: 10.1016/0165-0173(92)90011-a. [DOI] [PubMed] [Google Scholar]
- 115••.Harrington MG, Fonteh AN, Oborina E, et al. The morphology and biochemistry of nanostructures provide evidence for synthesis and signaling functions in human cerebrospinal fluid. Cerebrospinal Fluid Res. 2009;6:10. doi: 10.1186/1743-8454-6-10. Identification of synaptic components in the CSF, indicating a role for the CSF in intercellular signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abdi F, Quinn JF, Jankovic J, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9(3):293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 117••.Pan S, Zhu D, Quinn JF, et al. A combined dataset of human cerebrospinal fluid proteins identified by multi-dimensional chromatography and tandem mass spectrometry. Proteomics. 2007;7(3):469–473. doi: 10.1002/pmic.200600756. Discusses the identification of 2500 proteins in human CSF using proteomics. [DOI] [PubMed] [Google Scholar]
- 118.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- 119.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 120.Ecroyd H, Sarradin P, Dacheux JL, Gatti JL. Compartmentalization of prion isoforms within the reproductive tract of the ram. Biol Reprod. 2004;71(3):993–1001. doi: 10.1095/biolreprod.104.029801. [DOI] [PubMed] [Google Scholar]
- 121.Vingtdeux V, Hamdane M, Loyens A, et al. Alkalizing drugs induce accumulation of amyloid precursor protein by-products in luminal vesicles of multivesicular bodies. J Biol Chem. 2007;282(25):18197–18205. doi: 10.1074/jbc.M609475200. [DOI] [PubMed] [Google Scholar]
- 122.Liu J, Zhang JP, Shi M, et al. Rab11a and HSP90 regulate recycling of extracellular α-synuclein. J Neurosci. 2009;29(5):1480–1485. doi: 10.1523/JNEUROSCI.6202-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.El-Agnaf OM, Salem SA, Paleologou KE, et al. α-synuclein implicated in Parkinson’s disease is present in extracellular biological fluids, including human plasma. FASEB J. 2003;17(13):1945–1947. doi: 10.1096/fj.03-0098fje. [DOI] [PubMed] [Google Scholar]
- 124.Lee SJ. Origins and effects of extracellular α-synuclein: implications in Parkinson’s disease. J Mol Neurosci. 2008;34(1):17–22. doi: 10.1007/s12031-007-0012-9. [DOI] [PubMed] [Google Scholar]
- 125.Zhang J, Sokal I, Peskind ER, et al. CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am J Clin Pathol. 2008;129(4):526–529. doi: 10.1309/W01Y0B808EMEH12L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shi M, Caudle WM, Zhang J. Biomarker discovery in neurodegenerative diseases: a proteomic approach. Neurobiol Dis. 2009;35(2):157–164. doi: 10.1016/j.nbd.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. 2006;6(17):4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- 128.Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD. Analytical and statistical approaches to metabolomics research. J Sep Sci. 2009;32(13):2183–2199. doi: 10.1002/jssc.200900152. [DOI] [PubMed] [Google Scholar]
- 129.Ascherio A, LeWitt PA, Xu K, et al. Urate as a predictor of the rate of clinical decline in Parkinson disease. Arch Neurol. 2009;66(12):1460–1468. doi: 10.1001/archneurol.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schwarzschild MA, Schwid SR, Marek K, et al. Serum urate as a predictor of clinical and radiographic progression in Parkinson disease. Arch Neurol. 2008;65(6):716–723. doi: 10.1001/archneur.2008.65.6.nct70003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131•.Schlesinger I, Schlesinger N. Uric acid in Parkinson’s disease. Mov Disord. 2008;23(12):1653–1657. doi: 10.1002/mds.22139. Review of the role of uric acid in PD. [DOI] [PubMed] [Google Scholar]
- 132.Weisskopf MG, O’Reilly E, Chen H, Schwarzschild MA, Ascherio A. Plasma urate and risk of Parkinson’s disease. Am J Epidemiol. 2007;166(5):561–567. doi: 10.1093/aje/kwm127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci USA. 1981;78(11):6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bogdanov M, Matson WR, Wang L, et al. Metabolomic profiling to develop blood biomarkers for Parkinson’s disease. Brain. 2008;131(Pt 2):389–396. doi: 10.1093/brain/awm304. [DOI] [PubMed] [Google Scholar]
- 135.Johansen KK, Wang L, Aasly JO, et al. Metabolomic profiling in LRRK2-related Parkinson’s disease. PLoS One. 2009;4(10):e7551. doi: 10.1371/journal.pone.0007551. [DOI] [PMC free article] [PubMed] [Google Scholar]