Abstract
In ubiquitination, cullin-RING E3 ubiquitin ligases (CRLs) assist in ubiquitin transfer from ubiquitin-conjugating enzyme E2 to the substrate. Neddylation, which involves NEDD8 transfer from E2 to E3-cullin, stimulates ubiquitination by inducing conformational change in CRLs. However, deneddylation, which removes NEDD8 from cullin, does not suppress ubiquitination in vivo, raising the question of how neddylation/deneddylation exerts its effects. Using molecular-dynamics simulations, we demonstrate that before neddylation occurs, the linker flexibility of Rbx1, a CRL component, leads to conformational changes in CRLs that allow neddylation and initiation of ubiquitination. These large NEDD8-induced conformational changes are retained after deneddylation, allowing both initiation of the ubiquitination process and ubiquitin chain elongation after deneddylation. Furthermore, mutation of lysine, the cullin residue to which NEDD8 covalently attaches, dramatically reduces CRL conformational changes, suggesting that the acceptor lysine allosterically regulates CRLs. Thus, our results imply that neddylation stimulates ubiquitination by CRL conformational control via lysine modification.
Introduction
The ubiquitin-proteasome system (UPS) degrades malfunctioning or damaged proteins in cells (1). Ubiquitination of a target substrate protein is carried out by a three-enzyme cascade beginning with covalent bond formation between ubiquitin-activating enzyme (E1) and ubiquitin, followed by ubiquitin transfer from E1 to ubiquitin-conjugating enzyme (E2), and ending with ubiquitin transfer from E2 to a substrate protein that is recruited by E3 ubiquitin ligases (2). The polyubiquitin-labeled substrate is degraded by the proteasome (3).
Cullin-RING E3 ubiquitin ligases (CRLs) constitute the largest and one of the most studied E3 superfamilies (4,5). A CRL has two arms. One binds to substrates via the substrate binding protein, and the other binds to E2 through the RING-Box protein (Rbx), which contains a RING-finger domain. The two arms are connected by cullin and an adaptor protein, such as Skp1, ASK1, or Elongin C/Elongin B. CRLs play an important role in ubiquitination by binding to both E2 and the substrate and bringing them into proximity, thereby facilitating ubiquitin transfer. However, crystal structures of CRL and E2 indicate that there is a 50–60 Å gap between the E2 and the substrate (6–8) (Fig. 1 A). A key mechanistic question, then, is how the CRL E3 machine brings the E2 and substrate into proximity to enable efficient ubiquitin transfer.
Figure 1.

Overviews of the CRL E3 ligase machine. A schematic illustration of the Ub-E2-E3-substrate machine shows ubiquitination (A) and neddylation (B). The distances to be bridged are marked. On the left arm, S stands for substrate. The substrate-binding protein has two domains: the substrate-binding domain (SBD) and the Box domain. The adaptor protein mediates between the substrate-binding protein and the cullin. Cullin (Cul) has two domains: Cul NTD and Cul CTD, which contains the Cul CTD 4HB-α/β and Cul CTD WHB subdomains. The Rbx has two domains. The active residues in neddylation (C and K) are marked.
In our previous studies (9,10), we reported that all substrate-binding proteins with available crystal structures have flexible linkers that can serve as hinges to rotate the substrate-binding proteins toward E2. We performed molecular-dynamics (MD) simulations for these substrate-binding proteins in four forms: unbound, bound to substrate only, bound to adaptor only, and bound to both substrate and adaptor. We observed rotations in all. For the bound-to-both form, the 50–60 Å gap between E2 and the substrate can be shortened by 7∼12 Å by the rotation; however, our simulations showed that there is still a 39∼49 Å distance between E2 and the tip of the substrate peptide. This suggests that additional flexible linkers may exist in the CRL E3 machine.
It was recently reported that there is a flexible linker in Rbx1, on the second arm of CRL E3 (11). This observation was based on x-ray crystal structures with different conformations in the same unit cell. Rbx1 was complexed with both cullin and NEDD8, which is a CRL regulator. This raises the question of whether the Rbx1 linker flexibility is NEDD8-dependent. It is known that NEDD8 forms a covalent bond with a lysine residue in cullin, and that it can stimulate (but is not required for) ubiquitination (12,13). It was also reported that ubiquitination is significantly reduced when the cullin acceptor lysine is mutated to arginine in the presence or absence of NEDD8 (13). Clearly, in the presence of NEDD8, Arg can block the covalent linkage of NEDD8 to cullin. However, it is not clear why the mutant reduces ubiquitination in the absence of NEDD8. Further, neddylation involves NEDD8 transfer from the E2 catalytic cysteine to cullin's acceptor lysine; however, based on the model of CRL and E2 that was constructed by docking the crystal structures (Fig. 1 A), there is a 25∼35 Å gap between these two sites (11), raising the question of how the CRLs change the conformation to bridge this gap and facilitate neddylation. Still more intriguingly, NEDD8 is known to be removed from cullin by COP9/signalosome (CSN) (14,15), in a process known as deneddylation; paradoxically, however, deneddylation does not suppress ubiquitination in vivo (15,16). How the CRLs facilitate ubiquitination after deneddylation is unclear.
Here, we use MD simulations to address these key mechanistic questions. We simulated the Rbx1-Cul5 complex in three states: a closed conformation, representing the conformation before neddylation, taken from the Rbx1-Cul5CTD complex structure; and two open conformations, representing the conformation after deneddylation, taken from the Rbx1-Cul5CTD∼NEDD8 structure (11) with NEDD8 removed. We observed large conformational changes in all three conformations, suggesting that the Rbx1 linker is flexible before neddylation and after deneddylation. We further simulated the same three conformations with cullin acceptor lysine mutated to arginine. Our covariance analysis suggests that the lysine residue in cullin to which NEDD8 becomes covalently attached (dubbed cullin acceptor lysine below) can allosterically control the conformational change of Rbx1-Cul5. Our work provides a mechanistic framework to address key questions regarding ubiquitination in CRLs: 1), how are the distances that need to be overcome in neddylation and ubiquitination reactions spanned before neddylation; and 2), how does ubiquitination occur after deneddylation?
Materials and Methods
System setup
The starting structures of all simulated complexes were constructed from crystal structures. For the closed-conformation simulations, the crystal structure of Rbx1 complexed with Cul5CTD alone (PDB code: 3dpl) was used. For the open-conformation simulations, the Rbx1-Cul5 structures were extracted from the Rbx1-Cul5CTD∼NEDD8 complex structures (PDB code: 3dqv) with NEDD8 removed. The initial structures of the mutants were built from the wild-type (WT) structures, with the point mutation performed by VMD (17). The missing residues in Rbx1 and Cul5 were added as random coils and minimized for 1000 steps with the rest of the protein fixed. All models were solvated in a TIP3P water box with a minimum distance of 10 Å from the edge of the box to any protein atom. The system charges were neutralized by the addition of chloride or sodium ions. Tests confirmed that the water box was large enough to ensure that no atom in the protein would be within 12 Å of any atoms in any periodic image during the simulation.
Simulation protocol
We performed MD simulations using the CHARMM 27 force field (18) and the NAMD program (19). To eliminate residual unfavorable interactions between the solvent and the protein, the solvated systems were first minimized for 3000 steps with the protein restrained, followed by another 3000 steps of minimization with all atoms free to move. The systems were heated from 0 K to 300 K (or 340 K for closed conformation) in 100 ps, with the protein backbone atoms constrained to allow relaxation of the solvent, and then equilibrated for 600 ps without any constraints. For each state, two trajectories of production simulations were performed for 40 ns with the NPT ensemble. The temperature was controlled at 300 K (or 340 K for the closed conformation) with a Langevin thermostat. A Nose-Hoover Langevin piston barostat was used to maintain pressure at 1 bar. During the production simulations, the time step was 2 fs, with a SHAKE constraint on all bonds containing hydrogen atoms. Short-range nonbonded interactions employed a switch function with a switching distance of 10 Å and a cutoff of 12 Å. Long-range electrostatic interactions were treated with particle mesh Ewald summation. During the equilibration and production simulations, the distances of the zinc atom from the Rbx1 RING finger and its neighboring atoms were restrained. The structural alignments and figure rendering were performed by VMD (17). The angle-rotation analyses during the simulation were performed using Hingefind (20).
Model setup
The model for neddylation was built with E2 UbcH7 (PDB code: 1fbv) docking into the Rbx1 RING subdomain. The model for ubiquitination was built with E2 UbcH7 docking into the Rbx1 RING subdomain, 4HB-α/β subdomains of Cul5 superimposing with Cul1 (PDB code: 1ldk), and F-box of β-TrCP1 (PDB code: 1p22) superimposing with Skp2 (PDB code: 1ldk).
Results
The Rbx1 linker is flexible before neddylation and after deneddylation
X-ray crystal structures are available for the Rbx1-Cul5CTD complex in three conformations: closed, open 1, and open 2. The closed conformation, with the Rbx1 RING subdomain and Cul5 WHB subdomain packed together, was crystallized in the absence of NEDD8, representing the Rbx1-Cul5 structure before neddylation. The two open conformations, with the Rbx1 RING rotating away from Cul5 WHB, were crystallized when Cul5 was bound to NEDD8. We removed NEDD8 to make them represent the conformations after deneddylation. We carried out two trajectories of 40-ns explicit-solvent MD simulations for these three conformations.
During the simulations of the two open conformations, the RING subdomain of Rbx1 rotated dramatically toward and away from Cul5 WHB. Snapshots with the maximum Rbx1 rotation angles for each trajectory are shown in Fig. 2, B and C. Rbx1 has a flexible linker between the Rbx1 RING subdomain and the Cul5-binding subdomain, and Cul5 has a flexible linker between the WHB domain and the α/β domain. In open 1, the flexible linker rotated the Rbx1 RING subdomain away from Cul5 WHB to a maximum of 105° compared with the starting conformation, and the Cul5 linker rotated the WHB domain toward Rbx1 with a maximum of 84°. In open 2, the major rotation direction of Rbx1 was toward the Cul5 WHB, with the maximal rotation reaching 170°. The Cul5 rotation was relatively small compared to the open 1 conformation, with a maximal rotation of 39°.
Figure 2.

WT conformational changes during the simulation. Snapshots at the beginning of the simulation, the maximum rotations obtained in the two trajectories, as well as the rotation angles for Rbx1 and Cul5 during simulations of the (A and D) closed conformation at 340 K, (B and E) open 1 conformation, and (C and F) open 2 conformation are illustrated. The Cul5 WHB subdomain, Cul5 4HB-α/β subdomains, and Rbx1 are shown in red, gold, and blue, respectively.
The closed conformations changed little during the simulations at 300 K (see Fig. S1 in the Supporting Material). We raised the temperature to 340 K. Fig. 2 A presents snapshots at 0 ns and with the maximum Rbx1 rotation angles for each trajectory. Cul5 WHB rotated toward Rbx1, pushing Rbx1 in the same direction as the two open conformations. The rotation angles of Rbx1 reached 63° during the simulation (Fig. 1 D). The Cul5 WHB and Rbx1 RING remained compact during the simulations, with no trend to adopt the open conformation.
The distance gap for NEDD8 transfer shortens before neddylation
In neddylation, NEDD8 is transferred from the E2 catalytic cysteine to the Cul5 acceptor lysine. We built a model by docking E2 to Rbx1 in the Rbx1-Cul5 complex, and measured the distance between the E2 Cys sulfur atom and the catalytic Cul5 Lys nitrogen atom during the simulation of the WT closed conformation at 340 K. Fig. 3 shows the Cys-Lys distances and the conformations with the largest and smallest distances. The distance gap reaches a maximum of 33 Å. However, due to the dramatic conformational change, E2 Cys and Cul5 Lys are brought into proximity with a minimum distance of 6 Å during the simulation. The distribution of the distance (Fig. 3 D) shows that the distance with the highest probability is within 10–20 Å.
Figure 3.

Distances between the E2 Cys donor and Cul5 Lys acceptor shorten during simulation of the WT closed conformation at 340 K, allowing the NEDD8 transfer. (A) The snapshot with the smallest distance during the simulation. (B) The snapshot with the largest distance during the simulation. (C) The distance changes as a function of time during the simulation. (D) Normalized histogram for the distribution of the distances. The Cul5 WHB subdomain, Cul5 4HB-α/β subdomains, Rbx1, and E2 are shown in red, gold, blue, and cyan, respectively; Lys is in green, and Cys is in yellow.
The distance gap for ubiquitin transfer shortens before neddylation and after deneddylation
The last step in ubiquitination is the transfer of ubiquitin from E2 cysteine to substrate acceptor lysine. We built a model based on the crystal structures of CRL by superimposing the F-box of substrate-binding protein β-TrCP1 and Skp2, superimposing the C-terminal 4HB-α/β subdomains of Cul1 and Cul5, and docking E2 to Rbx1. Since the substrate acceptor lysine is not present in the crystal structure, we measured the distance between the E2 catalytic cysteine and the tip of the β-TrCP1 substrate for three conformations (closed, open 1, and open 2) during the simulations. The results are shown in Fig. 4. For the closed conformation, the maximum and minimum distances are 66 and 19 Å, respectively. The distance for open 1 ranges from 25 to 87 Å. The open 2 conformation has the most dramatic distance change between E2 cysteine and the tip of the substrate, ranging from a maximum of 98 Å to a minimum of 9 Å. The distances during the simulation as a function of time are shown in Fig. 5, A–C. We also measured the distribution of the distances, as shown in Fig. 5 D. For the closed conformation, the distance distributes between 19 and 66 Å. Open 1 has two distribution peaks: one between 25 and 45 Å, and the other between 65 and 85 Å. Open 2 is relatively evenly distributed in the 90 Å range, from 9 Å to 98 Å.
Figure 4.

Distances between E2 Cys and the substrate fluctuate during the simulations. The snapshot with the largest or shortest distance for the (A and B) WT closed conformation at 340 K, (C and D) open 1 conformation, and (E and F) open 2 conformation. The substrate peptide is in purple, the substrate-binding protein in pink, the adaptor protein in green, the Cul NTD in red, and the Cul CTD, Rbx1, and E2 in gold, blue, and cyan, respectively.
Figure 5.
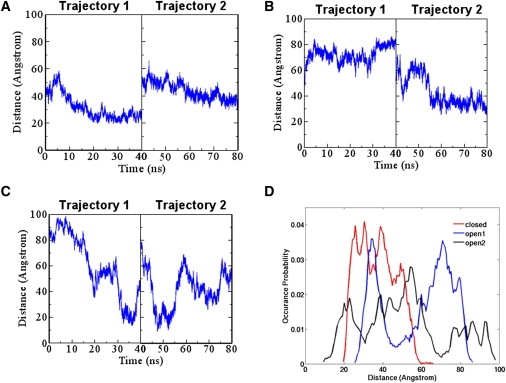
E2-substrate distances during simulations for the closed, open 1, and open 2 conformations. (A) Closed conformation. (B) Open 1 conformation. (C) Open 2 conformation. (D) Normalized histogram for the distribution of the distances.
The Cul5 K724R mutation allosterically changes the distance distribution before neddylation and after deneddylation
The Cul1 K720R mutation, corresponding to the Cul5 K724R mutation, can reduce ubiquitination (13). We performed two MD simulation trajectories of 40 ns for the three conformations with the Cul5 K724R mutation. For the closed conformation, the MD simulations were run at 340 K for consistency with the WT closed conformation. As in the case of the WT, we measured the distance between the E2 catalytic cysteine and the tip of the β-TrCP1 substrate for these three conformations during the simulations, as shown in Fig. 6, A–C. Compared to the WT, the distance range is narrower for each form of the mutants, as shown in Fig. 6 D. The distances for the closed form peak at 38 Å, and the most significant peaks for open 1 and open 2 are at 60 Å and 83 Å, respectively.
Figure 6.

Cul5 K724R mutants change the E2-substrate distance distribution during the simulation. (A) Closed conformation. (B) Open 1 conformation. (C) Open 2 conformation. (D) Normalized histogram for the distribution of the distances.
Cul5 Lys724 regulates the correlation of the motion between Rbx1 and Cul5
The motion correlations between Rbx1 and Cul5 are illustrated in Fig. 6 for the three conformations of the WT and the K724R mutants. For the closed conformation, the WT at 300 K, with a rather rigid conformation, has strong positive motion correlations between Rbx1 and Cul5 (Fig. S2). As expected, increasing the temperature to 340 K reduces the correlation with freer motion (Fig. 7 A); however, for the K724R mutant, surprisingly, the correlation between the Rbx1 RING domain and the Cul5 α/β domain changed from positive to negative (circled area in the middle panel of Fig. 7 A). That is, the correlation between these domains in the WT is more positive than in the K724R mutant, as shown in the right panel of Fig. 6 A. Of even more interest, the same trend is observed for both the open 1 and open 2 forms (Fig. 7, B and C).
Figure 7.

Mutation of the Lys cullin acceptor residue to Arg changes the motion correlations between Rbx1 and Cul5. Starting conformations for the covariance maps: (A) closed conformation, (B) open 1 conformation, and (C) open 2 conformation. The WT is shown on the left, the K724R mutant is shown in the middle, and the difference between the WT and mutant is shown on the right. The more red, the stronger the positive correlation; the more blue, the stronger the negative (anti-) correlation. The bar provides the scale. (D) The open 2 K724R conformation showing the domains with the largest correlation change. The Rbx1 RING domain is shown in blue, the Cul5 α/β domain is in orange, and the rest of system is in green. The K724R mutation is shown in red.
Discussion
Ubiquitination tags proteins for degradation. CRLs facilitate the transfer of ubiquitins from the E2 enzyme to the target protein. CRL works as a two-arm molecular machine: one arm binds substrates, and the other has the Rbx protein, which binds E2. The cullin scaffold protein connects the two arms. Neddylation can stimulate ubiquitination, but ubiquitination is not suppressed after deneddylation. A key requirement for efficient monoubiquitin transfer is that the two reactive residues on the donor E2 and on the acceptor substrate must be sufficiently close to each other; yet, at the same time, polyubiquitin tagging requires a space large enough to accommodate the ubiquitin tail. How the two arms operate to promote the first ubiquitin transfer and at the same time facilitate polyubiquitin tagging, and why neddylation helps are still open questions. In this work, we used simulations to address three challenging questions: 1) Is the Rbx1 linker flexible both before neddylation and after deneddylation? 2) How are the active sites of E2 and cullin brought into proximity for neddylation to take place? 3) Can CRLs bridge the distance gap between E2 and the substrate in ubiquitination both before neddylation and after deneddylation? MD simulations provide a powerful tool to address mechanistic questions on the molecular level. In our previous studies (9,10), we reported that flexible linkers in substrate-binding proteins may serve as a hinge to bring a substrate toward E2. Here, our simulations suggest that the other arm also contains flexible linkers that can function as a hinge to not only rotate E2 toward the substrate for ubiquitination, but also to rotate E2 toward the cullin for neddylation. Together, these results lead us to propose a mechanism for neddylation and ubiquitination, and explain why ubiquitination is not suppressed after deneddylation.
We first simulated the Rbx1-Cul5 complex in the closed conformations, representing the before-neddylation conformation. Conformational change was observed only when the temperature was raised to 340 K, suggesting a local trap. When the barrier is overcome, the Rbx1 linker is flexible, with the largest rotation angle during the simulation being 63°. The flexible Rbx1 linker leads to an ensemble of Rbx1-Cul5 conformations, which may help explain the enigma of the distance gap in neddylation. Neddylation involves transfer of NEDD8 from E2 cysteine to Cul5 Lys724. Our model, which is based on the docking of crystal structures, indicates a 27 Å distance between these two sites. During our simulation of the before-neddylation conformation, this distance shrank to 6 Å because of the conformational change brought about by the flexible Rbx1 linker; thus, the flexible Rbx1 linker can help neddylation by bringing the E2 cysteine and the cullin acceptor lysine into proximity.
The ensemble of conformations obtained as a result of the Rbx1 linker flexibility can also help elucidate the functionally required distance between E2 and the substrate for initiation of ubiquitination, for which the E2 and substrate active sites need to be sufficiently close. We propose that CRL works as a two-arm machine to facilitate the initiation of ubiquitination. Our results show that the rotation of the Rbx1 arm can shorten the distance gap from ∼50–60 Å to 20 Å, and this distance can be further shortened to ∼10 Å by the rotation of the other arm of the CRL machine. This second arm contains the substrate-binding protein, whose flexible linker can rotate the substrate toward E2 and shorten the distance gap, as shown in our previous studies (9,10). Considering that this distance is from the E2 cysteine site to the tip of the substrate, and the substrate tip is 10 residues away from the substrate lysine acceptor, the 10 Å distance gap could be small enough for the first ubiquitin transfer. Thus, the Rbx1 flexible linker can help the initiation of ubiquitination before neddylation by bringing the E2 and substrate active sites into proximity.
After the initiation of ubiquitination, a chain of at least four ubiquitins is required to tag the substrates for degradation. These multiple ubiquitins are sequentially transferred and space is needed to accommodate them. For ubiquitin chain elongation to occur, Gly76 and Lys48 of two consecutive ubiquitins must be connected. The distance between these two residues is ∼20 Å. If we assume that linear chain elongation is in the direction of E2, there must be at least ∼80 Å between E2 and the substrate to allow formation of a chain of at least four ubiquitins. Our before-neddylation simulations show that the distances between E2 and the substrate are mostly distributed in the 20–60 Å range, with a maximal distance of 66 Å, which may not be enough for chain elongation with four ubiquitins. This suggests that before neddylation, the conformational change induced by the Rbx1 flexible linker can allow the initiation of ubiquitination, and may also help in di-ubiquitin formation to some extent, but the conformational change may not be enough for ubiquitination chain elongation.
Neddylation has been reported to help ubiquitin chain elongation. Saha and Deshaies (21) showed that NEDD8 increases the rate of chain elongation of a ubiquitinated substrate, and Read et al. (13) reported that neddylation can increase the formation of high-molecular-weight ubiquitin conjugations. We speculate that the presence of NEDD8 shifts the distribution of the preexisting ensembles, populating the open states (22–25), which could derive from the Rbx1 linker. This population shift model, which was proposed a decade ago (22–25), was recently validated by a body of consistent experimental data (26).
Could the redistributed ensemble following a population shift be retained after deneddylation, when NEDD8 is removed from cullin? Our simulations of the after-deneddylation conformations, taken from the Rbx1-Cul5CTD∼NEDD8 structure (11) with NEDD8 removed, indicate that the Rbx1 linker flexibility is larger than in the before-neddylation conformation, implying that the population shift toward the open states caused by neddylation may remain after deneddylation. In addition, after deneddylation, the distances between E2 and the substrate are relatively evenly distributed in a ∼90 Å range, which is much larger than the ∼46 Å of the before-neddylation range. This also suggests that the large conformational change remains after deneddylation.
There is increasing evidence that CSN promotes deneddylation (14,15); however, CSN also enhances ubiquitination and substrate degradation (15,16). The question is then, what is the mechanism of ubiquitination after deneddylation? Our results indicate that the after-deneddylation conformation assists in the initiation of ubiquitination by shortening the distance gap between E2 and the substrate to a minimum distance of 9 Å, which is 10 Å smaller than what is observed for the before-neddylation conformation. Saha and Deshaies (21) reported that neddylation facilitates the initiation of ubiquitination. Here we report that after deneddylation, the conformational ensembles that were enhanced by neddylation still facilitate initiation of ubiquitination. The ubiquitin chain elongation can also be facilitated by conformational change after deneddylation. The maximal distance for the after-deneddylation conformation during the simulations between E2 and the substrate is 98 Å, which could accommodate at least a five-ubiquitin chain. If neddylation can promote chain elongation (13,21) by redistributing the Rbx1-Cul5 ensemble, subsequent to deneddylation, the existing Rbx1-Cul5 ensemble could facilitate both the initiation and chain elongation of ubiquitination. This would explain why ubiquitination is not suppressed by NEDD8 removal.
What regulates the Rbx1 linker's flexibility? It is known that mutating the cullin acceptor lysine to arginine can reduce ubiquitination regardless of the presence of NEDD8 (13). We performed MD simulations for this mutant and observed a significantly narrowed distance distribution for all three conformations, which implies that this single residue mutation allosterically affects the conformational change of the whole system. The less evenly distributed distances and smaller distance range also suggest that this mutation reduces the likelihood that Cul5 will undergo ubiquitin transfer initialization and chain elongation, both before neddylation and after deneddylation. Of interest, this Cul5 K724R is far away from both the Rbx1 RING domain and the Cul5 α/β domain, but it dramatically changes the correlations between these two domains, further suggesting that the highly conserved Lys724 allosterically regulates the conformational change of the Rbx1-Cul5 complex (27–31). We thus propose an allosteric mechanism for the Cul5-Rbx1 complex. We propose that an allosteric communication could exist among Lys724, the Rbx1 RING domain, and the Cul5 α/β domain, with the Cul5 α/β domain acting with Lys724 and the Rbx1 RING domain to control the conformational change of the Rbx1-Cul5 complex. Since Lys724 is the NEDD8 acceptor, we speculate that Lys724 may further control the conformational change through the perturbation that follows covalent bond formation between Lys724 and NEDD8. Such an allosteric effect is similar to that of the substrate-binding protein pVHL (32), suggesting that Lys724 could be an allosteric drug target. The role of the Cul5 α/β domain is unclear, and to our knowledge no study has been performed on this domain. Further investigation of this domain is needed to test our allosteric model. As for the Rbx1 RING domain, Yamoah et al. (33) reported that the K89A mutation in the Rbx1 RING domain can significantly enhance the ubiquitination activities of Cul1-Rbx1 complexes, implying a Cul1-Rbx1 conformational change. The interface of Cul5-Rbx1 is slightly different from that of Cul1-Rbx1; therefore, we propose that Gln59 and Ser62 at the Cul5-Rbx1 interface may be critical for conformational control.
To conclude, we have demonstrated that the flexibility of the Rbx1 linker plays an important role in facilitating CRL neddylation and ubiquitination, before neddylation and after deneddylation. Before neddylation, the intrinsic flexible linkers of the two CRL arms containing the substrate-binding protein and Rbx1 present a broad conformational ensemble that allows the initiation of ubiquitination. The flexible Rbx1 linker facilitates NEDD8 transfer from the E2 enzyme to cullin. The neddylation of cullin shifts the landscape. The flexibility of the Rbx arm increases. The higher flexibility stimulates both the initiation of ubiquitination and chain elongation. Deneddylation removes the NEDD8, but the shifted fluctuating ensembles still exist. We further observe that cullin Lys724, to which NEDD8 is transferred from E2, allosterically regulates the conformational change of the Rbx1-Cul5 complex. Overall, our results suggest a mechanism for neddylation and ubiquitination before neddylation and after deneddylation. We further propose an allosteric mechanism that implies that Cul5 Lys724, the Rbx1 RING domain, and the Cul5 α/β domain could be critical for conformational control, and could serve as allosteric drug targets, in similarity to allosteric residues in pVHL (32).
Acknowledgments
This project was funded in whole or in part by federal funds from the National Cancer Institute, National Institutes of Health (NIH), under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Support was also provided by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH. This study used the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov).
Supporting Material
References
- 1.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Capili A.D., Lima C.D. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr. Opin. Struct. Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotton S.K., Callis J. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- 5.Merlet J., Burger J., Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell. Mol. Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng N., Schulman B.A., Pavletich N.P. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 7.Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 8.Zheng N., Wang P., Pavletich N.P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Nussinov R. The mechanism of ubiquitination in the cullin-RING E3 ligase machinery: conformational control of substrate orientation. PLOS Comput. Biol. 2009;5:e1000527. doi: 10.1371/journal.pcbi.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Nussinov R. Molecular dynamics reveal the essential role of linker motions in the function of cullin-RING E3 ligases. J. Mol. Biol. 2010;396:1508–1523. doi: 10.1016/j.jmb.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duda D.M., Borg L.A., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morimoto M., Nishida T., Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem. Biophys. Res. Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- 13.Read M.A., Brownell J.E., Palombella V.J. Nedd8 modification of cul-1 activates SCF(β(TrCP))-dependent ubiquitination of IκBα. Mol. Cell. Biol. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyapina S., Cope G., Deshaies R.J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 15.Cope G.A., Suh G.S., Deshaies R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 16.Miyauchi Y., Kato M., Iwai K. The COP9/signalosome increases the efficiency of von Hippel-Lindau protein ubiquitin ligase-mediated hypoxia-inducible factor-α ubiquitination. J. Biol. Chem. 2008;283:16622–16631. doi: 10.1074/jbc.M710599200. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 18.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 19.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wriggers W., Schulten K. Protein domain movements: detection of rigid domains and visualization of hinges in comparisons of atomic coordinates. Proteins. 1997;29:1–14. [PubMed] [Google Scholar]
- 21.Saha A., Deshaies R.J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai C.J., Kumar S., Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai C.J., Ma B., Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proc. Natl. Acad. Sci. USA. 1999;96:9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurz T., Ozlü N., Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S., Ma B., Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunasekaran K., Ma B., Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 28.Tsai C.J., del Sol A., Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. J. Mol. Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai C.J., Del Sol A., Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol. Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.del Sol A., Tsai C.J., Nussinov R. The origin of allosteric functional modulation: multiple pre-existing pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozkan E., Yu H., Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J., Nussinov R. Allosteric effects in the marginally stable von Hippel-Lindau tumor suppressor protein and allostery-based rescue mutant design. Proc. Natl. Acad. Sci. USA. 2008;105:901–906. doi: 10.1073/pnas.0707401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamoah K., Oashi T., Pan Z.Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


