Abstract
We considered the issue of whether shifts in the metarhodopsin I (MI)-metarhodopsin II (MII) equilibrium from lipid composition are fully explicable by differences in bilayer curvature elastic stress. A series of six lipids with known spontaneous radii of monolayer curvature and bending elastic moduli were added at increasing concentrations to the matrix lipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and the MI-MII equilibrium measured by flash photolysis followed by recording UV-vis spectra. The average area-per-lipid molecule and the membrane hydrophobic thickness were derived from measurements of the 2H NMR order parameter profile of the palmitic acid chain in POPC. For the series of ethanolamines with different levels of headgroup methylation, shifts in the MI-MII equilibrium correlated with changes in membrane elastic properties as expressed by the product of spontaneous radius of monolayer curvature, bending elastic modulus, and lateral area per molecule. However, for the entire series of lipids, elastic energy explained the shifts only partially. Additional contributions correlated with the capability of the ethanolamine headgroups to engage in hydrogen bonding with the protein, independent of the state of ethanolamine methylation, with introduction of polyunsaturated sn-2 hydrocarbon chains, and with replacement of the palmitic acid sn-1 chains by oleic acid. The experiments point to the importance of interactions of rhodopsin with particular lipid species in the first layer of lipids surrounding the protein as well as to membrane elastic stress in the lipid-protein domain.
Introduction
Rhodopsin is the light receptor responsible for dim light vision in the rod photoreceptor cells of vertebrates. Within a few milliseconds after photon absorption, a metastable equilibrium is established between metarhodopsin II (MII), the conformation which binds and activates transducin, and its inactive precursor, metarhodopsin I (MI). Recent structural and spectroscopic data have provided important insights into MI and MII structure (1). The conformation of MI is closer to the conformation of rhodopsin in the ground state (2). The transition from MI to MII involves an outward radial movement of TM6 relative to a core composed of TM helices 1–4 (3). This large-scale conformational change has, most likely, two consequences: first, MII has a different shape than MI; and second, some amino acids previously buried in MI are now exposed to annular lipids in MII. Previous studies of lipid substitutions on rhodopsin function have shown that the MI-MII equilibrium is shifted toward MII in membranes containing phospholipids with phosphatidylethanolamine headgroups (PE) (4,5) or phospholipids with docosahexanoyl acyl (DHA) chains (22:6n3) (6,7). PE lipids have two remarkable properties. First, PE lipids are characterized by a negative spontaneous intrinsic curvature (C0). When forced into a flat bilayer of zero curvature, PE lipids are in a state of curvature stress. A protein conformation which induces negative monolayer curvature would be stabilized by a membrane containing PE lipids. Second, not only are the lipid-lipid interactions different, but interaction of the protein with the first layer of lipid surrounding it is different as well. Contrary to lipids with phosphatidylcholine headgroups (PC), PE lipids have the ability to establish hydrogen bonds (H-bonds) through their ammonium group. One can then imagine that a protein conformation exposing H-bond partners would be stabilized through direct interactions in a membrane containing PE lipids.
Phospholipids containing DHA acyl chains are structurally distinguished from phospholipids containing fewer unsaturated fatty acids by the presence of a repeating =CH-CH2-CH= unit, which produces an extremely flexible chain that rapidly converts between conformational states (8–10). Molecular simulations suggested that rhodopsin has a dramatic preference for solvation by the polyunsaturated DHA chains that may penetrate significantly deeper into the protein interface (11). Hence, one could predict that the energetic cost of wetting a protein after a conformational change should be lower when surrounded by polyunsaturated acyl chains. This hypothesis or the possibility that DHA chains might differentially engage the MI and MII conformations via polar interactions with the double bounds of DHA, are current lines of thought (12,13).
This raises the question: To what extent do the different mechanisms modulate the MI-MII equilibrium? Assuming that the conformational changes upon MII formation lead to an increase in protein hydrophobic thickness, Botelho et al. (4) and Gibson and Brown (14) introduced the flexible surface model (FSM). This model proposed that the increase in lipid/protein interfacial free energy, due to hydrophobic mismatch between the bilayer and the protein core upon MII formation, is compensated by a reduction in curvature elastic stress from the presence of lipids with a negative spontaneous curvature (such as PE), therefore shifting the equilibrium toward MII. Using this model, the Brown laboratory was able to fit the linear relationship observed experimentally between the amount of MII formed after photoactivation and the concentration of PE in PC-PE mixed bilayers. However, one can argue that the FSM totally leaves out any energetic contribution from lipid-specific interactions between rhodopsin and a first layer of lipids. Recently it was reported that H-bonding with lipids is critical for the regulation of two different membrane proteins. For instance, Hakizimana et al. (15) reported that the function of the secondary multidrug transporter LmrP of Lactococcus lactis does not depend on the elastic energy stored in the membrane but solely on the hydrogen bonding ability of interfacial headgroups. Similarly, Powl et al. (16) demonstrated that the activation of the mechanosensitive channel of large conductance MscL of Escherichia coli does not scale with membrane spontaneous curvature, but again with the ability of the annular lipid headgroups to form hydrogen bonds with the channel.
In this article, we assessed the role of PE lipids and DHA-containing lipids on rhodopsin function by reconstituting rhodopsin into 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membranes doped with increasing concentrations of PE- and methylated-PE lipids or DHA-containing PC and PE lipids. We show that the amount of MII formed after photoactivation partially scales with predictions based on changes of the elastic energy stored in the membrane. On the other hand, the ability of PE and methylated PEs to form H-bonds between the lipid headgroup and rhodopsin contributes to shifts toward MII as well. Oleic acid at glycerol sn-1 and polyunsaturation at sn-2 additionally favor MII formation, independent of lipid headgroups.
Materials and Methods
Preparation of reconstituted membranes
Sample preparation was carried out in complete darkness. The phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2 dioleoyl-sn-glycero-3-dimethyl-phosphoethanolamine (DOPE-Me2), 1,2-dioleoyl-sn-glycero-3-monomethyl-phosphoethanolamine (DOPE-Me1), 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphoethanolamine (SDPE), and 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine (SDPC) were obtained from Avanti Polar Lipids (Alabaster, AL) and used without further purification. Rhodopsin was purified from bovine retinas using procedures that were developed by the Litman laboratory (17).
One homogeneous fraction of rhodopsin in 3 wt % octylglucoside (OG), which gave a UV-vis absorption intensity ratio at 280:500 nm of 1.8, was used for the entire series of experiments to avoid an influence on results from a natural variability of the protein. For rhodopsin reconstitution, a glass round-bottom flask was coated with phospholipids by slow rotation and removal of solvent in a stream of pure nitrogen gas. The rhodopsin-OG solution was added to lipid-OG mixed micelles so that the OG/lipid molar ratio was 10:1 and the rhodopsin/phospholipid molar ratio was 1:250. The sample was vortexed to complete solubilization of the lipid and then equilibrated for 12 h under argon. Subsequently, this rhodopsin/lipid solution was added dropwise at a rate of 400 μL/min to deoxygenated piperazine-n,n′-bis(2-ethanesulfonic acid) (PIPES) buffer (10 mM PIPES, 100 mM NaCl, 50 μM DTPA, pH = 7.0) under rapid stirring, resulting in formation of unilamellar proteoliposomes. Typically, the final OG concentration was 7.2 mM, which is well below the critical micelle concentration. For all experiments, the proteoliposome dispersion (2.5–3 mL) was then dialyzed against 1 L of PIPES buffer (Slide-A-Lyzer membrane, 10 kDa cutoff; Pierce, Rockford, IL). The buffer was exchanged three times over 24 h. The final rhodopsin concentration in the samples was measured by light absorption at 500 nm assuming a molar extinction coefficient ɛ500 = 40,600 M−1 cm−1 (18). The concentration of residual OG in the lipid bilayers was <0.4 mol % of the lipid concentration as determined by high-resolution 1H NMR.
NMR experiments
Solid-state 2H NMR experiments were carried out on a model No. AV800 spectrometer equipped with a double-resonance probe with a 4-mm solenoid coil (both by Bruker Biospin, Billerica, MA) operating at a 2H NMR resonance frequency of 122.8 MHz. Data were acquired at 25°C with a quadrupolar echo pulse sequence, d1-90°x-τ-90°y-τ-acq, with a relaxation delay time d1 = 250 ms, a 4.5-μs 90° pulse, a delay time τ = 50 μs, and a 200 kHz spectral width. Typically, 16,000 transients were acquired.
Circular dichroism experiments
Far-UV circular dichroism spectra of rhodopsin were recorded at 21°C with a model No. J-810 spectropolarimeter (JASCO, Easton, MD). The rhodopsin concentration was ∼5 μM and the rhodopsin/lipid molar ratio was 1:250. The exact rhodopsin concentrations were measured by light absorption at 500 nm. Cells with an optical path length of 0.01 cm were used to minimize light scattering. Spectra were recorded from 260 to 190 nm with a data pitch of 0.2 nm, a bandwidth of 1 nm, and an integration time of 4 s.
MII/MI ratio
The equilibrium constant Keq = [MII]/[MI] was determined from rapidly acquired UV-vis spectra of the MI-MII equilibrium as previously described (19). Briefly, vesicles were diluted to a rhodopsin concentration of 0.2–0.25 mg/mL in pH 7.0 PIPES-buffered saline buffer and equilibrated at 37°C in a thermally regulated sample holder. A set of four absorption spectra were collected sequentially in a model No. 8453 diode array spectrophotometer (Agilent, Santa Clara, CA). These included:
-
1.
The spectra acquired after the sample was equilibrated in the dark at 37°C;
-
2.
The spectra acquired 3 s after the sample was 15–20% bleached by a 520-nm flash;
-
3.
The spectra acquired 10 min after addition of 30 mM hydroxylamine to convert bleached rhodopsin to opsin and retinal oxime; and
-
4.
The spectra acquired after a complete bleach of the sample.
Individual MI and MII spectra were deconvolved from spectra of their equilibrium mixture. The concentration of the photointermediates MI and MII were determined using extinction coefficients of 44,000 cm−1 and 38,000 cm−1 at their absorbance maxima of 478 nm and 365 nm, respectively.
Results and Discussion
Curvature elastic stress
Rhodopsin was reconstituted into binary mixtures of POPC doped with increasing concentrations of a second lipid that was DOPC, SDPC, DOPE-Me2, DOPE-Me1, DOPE, or SDPE. The lipids were selected to determine whether membrane curvature elastic stress is the sole energetic contribution to the change in free energy between the rhodopsin photointermediates MI and MII. The spontaneous intrinsic curvature, C0Lip = 1/r0Lip, represents the curvature that a lipid monolayer would assume if it were allowed to bend freely, independent of the constraints imposed by the bilayer. All selected lipids are zwitterionic characterized by a negative spontaneous intrinsic curvature, C0Lip, ranging from −0.11 nm−1 for DOPC to −0.35 nm−1 for DOPE (20,21). The C0Lip increases linearly with the loss of each methyl group from DOPE-Me2, to DOPE-Me1, to DOPE (22). The spontaneous intrinsic curvature of SDPE is identical to the values for DOPE (23).
Following the flexible surface model (FSM) of Botelho et al. (4), the free energy change for the MI-MII equilibrium can be written as
| (1) |
where ΔG0,L is the change of bilayer free energy due to elastic stress, ΔG0,LP is the change of bilayer free energy due to direct interactions between lipids and protein, i.e., the solvation energy of the hydrophobic protein surface, and ΔG0,P is the internal free energy change of the protein. Because the energetic changes are attributed to the lipids, ΔG0,P is independent of the membrane composition and provides a constant that does not affect the analysis. This leads to
| (2) |
Following Helfrich (24), the elastic free energy change is given by
| (3) |
where kcmono is the bending rigidity of the monolayer, CMII and CMI are the effective monolayer curvatures of all lipids when rhodopsin is in the MII or MI conformation, respectively, and C0 is the spontaneous curvature of the lipid monolayer. The assumption of a constant monolayer curvature for all lipids in the bilayer is a simplification that may be justified only at sufficiently low lipid/protein ratios. At the ratio of 250:1, as in this article, the rhodopsin molecules are surrounded by approximately five layers of lipid only, which is reasonably low. At larger lipid/protein ratios, models need to be considered that take into consideration a decay of lipid perturbation with increasing distances from the protein.
The second term in Eq. 2 is the change of the bilayer free energy upon MII formation, with
where γLP is the interfacial tension of the lipid-protein interface, and AMII and AMI are the hydrophobic surface of the MII and MI photointermediates, respectively. This yields
| (4) |
In the FSM, the amount of MII formed after photoactivation depends on the balance between the change in elastic free energy (first term in Eq. 4) and the change in solvation energy at the protein-lipid interface upon photoactivation (second term). Surface plasmon resonance experiments suggested that bilayer thickness increases upon MII formation (25). More-recent NMR studies using chain-deuterated lipids indicated that bilayer thickness remains constant upon MII formation (26). Still, the structural changes upon the MI → MII transition are expected to increase the hydrophobic surface area (AMII > AMI). It is assumed that it costs energy to form the lipid-MII interface (4). Therefore, the contribution from the last term of Eq. 4 is most likely positive. When AMII > AMI, the local bilayer curvature at the rhodopsin-lipid interface, CMII, changes in the direction of negative spontaneous curvature. Consequently, elastic stress of each monolayer is reduced for reconstitution into lipids with negative spontaneous curvature, such as PEs, yielding a negative first term in Eq. 4.
With
Eq. 4 can be rewritten as
| (5) |
where Alip is the lateral area per lipid, nL is the total number of lipids per rhodopsin, k is the Boltzmann constant, and T is the absolute temperature. If it is assumed that the last two terms in the bracket do not depend on membrane composition, the equation can be simplified to
| (6) |
This equation was used to compare theoretical predictions on the shift in the MI-MII equilibrium with experimental results. Lipid areas, Alip, were calculated according to
using the bilayer hydrophobic thicknesses, dPOPC, measured by 2H NMR on POPC (27–29) and assuming a lateral lipid area APOPC = 0.683 nm2 (30). The bending rigidities, kcmono, of DOPC, DOPE, and SDPE monolayers were obtained from literature (20,31). Bending rigidity values for DOPE-Me2, DOPE-Me1, and SDPE were calculated assuming a square dependence between the bending rigidity and membrane hydrophobic thickness with kc ∝ (dlipid)2 following Bermudez et al. (32). The elastic parameters of all lipids are provided in Table 1. Monolayer curvatures near rhodopsin in the MII and MI conformations (CMII and CMI) were assumed to be identical in all binary mixtures. This assumption is vaguely supported by the fact that the circular dichroism experiments did not yield any significant difference in rhodopsin helical content for all investigated lipid species.
Table 1.
Spontaneous curvature of lipid monolayers, C0, hydrophobic thickness of bilayers, d, lateral areas per lipid, A, and monolayer bending rigidities, kc, used for the calculations in this article
| Lipid | C0(nm)-1 | dNMRnm | A(nm)2 | kckBT |
|---|---|---|---|---|
| POPC | 0 | 2.54 | 0.683 | 9.5∗ |
| DOPC | −0.11 | 2.48 | 0.699 | 9† |
| SDPC | N/D | 2.46 | 0.686 | 9.1∗ |
| DOPE(Me2) | −0.19 | 2.60 | 0.667 | 9.4∗ |
| DOPE(Me1) | −0.27 | 2.64 | 0.657 | 9.7∗ |
| DOPE | −0.35 | 2.74 | 0.633 | 11† |
| SDPE | −0.35 | 2.68 | 0.647 | 9.7† |
Note that dNMR values were calculated using average 2H NMR chain order parameters of POPC-d31 in bilayers of POPC-d31 and in bilayers of equimolar mixtures of POPC-d31 with the other lipids. Thickness was calculated according to , where n is the number of acyl chain carbons and Sav is the average order parameter. For equimolar lipid mixtures (POPC/Lip2), it was assumed that .
Bending moduli from calculations taken here (see text).
Experiments were conducted on binary lipid mixtures with POPC as matrix lipid (lipid-1). It was assumed that the intrinsic curvature and binding rigidity of a lipid mixture sums in proportion to the mole fraction of the individual phospholipids. Hence, the intrinsic curvature and bending rigidity for a mixed bilayer composed of POPC and lipid-2 are
and
respectively. Furthermore, it was assumed that bilayer thickness of binary lipid mixtures is
In the following, the spontaneous intrinsic curvature of POPC, C0POPC, will be assumed to be zero yielding a spontaneous curvature
for POPC/lipid-2 binary mixtures. Because differences of kcmono and of dlip between the investigated lipids are small (see Table 1), it is straightforward to show that
and
which yields
| (7) |
in good approximation.
If membrane curvature stress is the only energetic contribution to the MI-MII equilibrium, one would expect that ln Keq is proportional to the spontaneous curvature of lipid-2, C0Lip2, and that ln Keq varies linearly with the mole fraction of the secondary lipid in the binary mixtures, XLip2. In other words, a plot of
should be linear. Finally, at XLip2 = 1, Eq. 7 becomes
| (8) |
To verify Eqs. 7 and 8 experimentally, we measured the equilibrium concentration of MII by UV-vis spectroscopy as a function of DOPC, SDPC, DOPE-Me2, DOPE-Me1, DOPE, and SDPE concentration in POPC bilayers and calculated ln Keq (see Table 2). Fig. 1 shows the dependence of ln Keq as a function of XLip2 obtained experimentally. As predicted, for each binary mixture, ln Keq rises linearly with increasing mole fraction of the secondary lipid in the bilayer, i.e., with increasing membrane curvature stress. Although a linear relationship between ln Keq and XLip2 is a necessary requirement for linking membrane curvature elastic energy to shifts in Keq, it does not eliminate the possibility that interactions different from elastic energy may contribute to shifts in the MI-MII equilibrium.
Table 2.
Keq values measured for rhodopsin reconstituted into membranes of various composition (lipid/rhodopsin molar ratio = 250/1, temperature 37°C, pH 7.0)
| Lipid 1 | Lipid 2 | Mole fractionof lipid 2 | Keq |
|---|---|---|---|
| POPC | N/A | N/A | 2.18 ± 0.15 |
| POPC | DOPC | 0.5 | 2.84 ± 0.16 |
| POPC | DOPC | 0.625 | 2.65 ± 0.2 |
| POPC | DOPC | 0.75 | 3.11 ± 0.19 |
| N/A | DOPC | 1 | 3.2 ± 0.18 |
| POPC | DOPE-Me2 | 0.5 | 3.4 ± 0.2 |
| POPC | DOPE-Me2 | 0.625 | 4.07 ± 0.2 |
| POPC | DOPE-Me2 | 0.75 | 5.25 ± 0.29 |
| N/A | DOPE-Me2 | 1 | 6.35 ± 0.2 |
| POPC | DOPE-Me1 | 0.5 | 3.56 ± 0.15 |
| POPC | DOPE-Me1 | 0.625 | 4.61 ± 0.18 |
| POPC | DOPE-Me1 | 0.75 | 5.62 ± 0.2 |
| N/A | DOPE-Me1 | 1 | 7.24 ± 0.23 |
| POPC | DOPE | 0.5 | 4.1 ± 0.19 |
| POPC | DOPE | 0.625 | 5.33 ± 0.23 |
| POPC | DOPE | 0.75 | 7 ± 0.15 |
| POPC | SDPE | 0.125 | 1.51 ± 0.12 |
| POPC | SDPE | 0.25 | 3.66 ± 0.2 |
| POPC | SDPE | 0.5 | 6.11 ± 0.25 |
| POPC | SDPE | 0.75 | 7.53 ± 0.27 |
| POPC | SDPC | 0.5 | 2.97 ± 0.17 |
| N/A | SDPC | 1 | 4.3 ± 0.25 |
Figure 1.
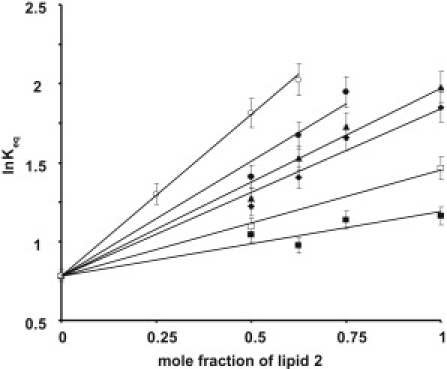
Dependence of ln Keq on the mole fraction of DOPC (■), SDPC (□), DOPE-Me2 (♦), DOPE-Me1 (▴), DOPE (•), and SDPE (○) in binary mixtures containing POPC as the base lipid.
To test whether membrane curvature stress is the only energetic contribution to the equilibrium, according to Eq. 8, at XLip2 = 1, one also needs to observe a linear relationship between ln Keq and ALip2 · kcLip2 · C0Lip2, the product of area per molecule, the bending elastic modulus, and the spontaneous intrinsic curvature of DOPC, SDPC, DOPE-Me2, DOPE-Me1, DOPE, and SDPE, respectively. Values of ln Keq at XLip2 = 1 were obtained by a linear fit of the data in Fig. 1. ALip2 was calculated from the order parameter changes of the perdeuterated sn-1 chain in POPC recorded as a function of concentration of lipid-2 in the mixture (see Fig. S1 in the Supporting Material).
The plot of ln Keq vs. ALip2 · kcLip2 · C0Lip2 is shown in Fig. 2. Interestingly, ln Keq values obtained for membranes containing DOPE-Me2, DOPE-Me1, and DOPE increase linearly, while the ln Keq values for recombinant membranes containing POPC or DOPC are clearly lower and the value for SDPE much higher than predicted. The discrepancy between theoretical predictions and experiments for some of the lipids indicates that the change of free energy due to a release of membrane curvature stress is not the sole determinant of the conformational energetics of the MI-MII equilibrium. Whatever those additional interactions are, Fig. 1 shows that their energetic contributions scale linearly with the mole fraction of lipid-2, XLip2. Therefore, those lipid-protein interactions must be sufficiently weak and transient such that no saturation with increasing concentrations of lipid-2 occurs. What could be the interactions that contribute to the additional shifts in Keq?
Figure 2.
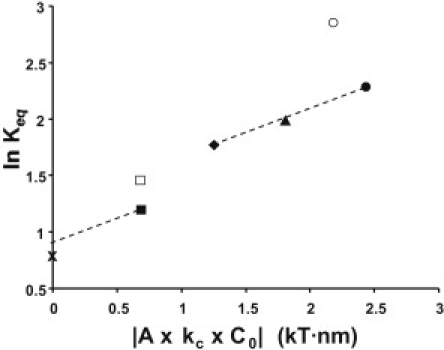
Dependence of ln Keq as a function of ALip2 · kcLip2 · C0Lip2 for POPC (×), DOPC (■), SDPC (□), DOPE-Me2 (♦), DOPE-Me1 (▴), DOPE (•), and SDPE (○).
Hydration
It was reported that the MI-MII equilibrium is sensitive to osmotic stress (33), suggesting that differences in hydration properties of lipids may shift the equilibrium. X-ray diffraction and NMR studies have shown that substituting one of the hydrogens of PE by a methyl group results in a strong increase of headgroup hydration (34–36). Phospholipids with PE-Me2 and PE-Me1 headgroups have hydration properties similar to that of phospholipids with PC headgroups. If headgroup hydration was key, one would expect the ln Keq values to be similar for membranes containing DOPC, DOPE-Me2, and DOPE-Me1, with a jump to occur to DOPE. In contrast, the experiments showed a jump from DOPC to DOPE-ME2, and the expected jump from DOPE-Me1 to DOPE was absent (Fig. 2). Therefore, one can safely exclude differences in hydration properties of the lipid matrix as a contributing mechanism. The sensitivity of the MI-MII equilibrium to osmotic stress must be related to differences in hydration of the core of rhodopsin, as was reported recently (37).
Hydrogen bonding
The lipids DOPE-Me2, DOPE-Me1, and DOPE have in common that their ammonium group may form a hydrogen bond with the protein, while POPC and DOPC may not. Therefore, if the ability of lipids to form a hydrogen bond with rhodopsin is a critical energetic contribution to the MI-MII equilibrium, similar ln Keq values are expected for membranes containing DOPE-Me2, DOPE-Me1, and DOPE and a jump should occur between DOPC and DOPE-Me2. This was confirmed experimentally, suggesting that the propensity of lipid ammonium groups to establish a hydrogen bond with the protein contributes actively to the MI-MII equilibrium.
Polyunsaturated hydrocarbon chains
To study the effect of DHA-containing phospholipids, rhodopsin was reconstituted into binary mixtures of POPC with increasing concentrations of the polyunsaturated SDPC or SDPE with DHA chains at the sn-2 position. Fig. 1 shows that more MII is formed in membranes with SDPC instead of DOPC, and with SDPE instead of DOPE. For both polyunsaturated lipids, ln Keq increases linearly with the mole fraction of SDPC or SDPE in the bilayers, indicating that interactions between rhodopsin and the polyunsaturated DHA chains are not saturable.
How do membranes that are rich in DHA differ from less unsaturated ones? Curvature elasticity and the spontaneous radius of curvature of SDPE monolayers were measured at our lab (23) and determined to be close to values of DOPE (Table 2), which eliminates membrane curvature stress as cause for the shift toward MII. However, NMR experiments of spin magnetization transfer between protein and lipids suggest that DHA chains interact preferentially with rhodopsin (38). NMR studies at our laboratory (8,9) as well as quantum chemical calculations and molecular dynamics simulations (8,10,39) revealed a high level of conformational flexibility of DHA chains. By analyzing 13C relaxation data, we could show that even the DHA chains near rhodopsin isomerize on a timescale of 1–100 ps, and that DHA chains explore their entire conformational space within 10 ns (9).
The quantum chemical and molecular mechanical calculations demonstrated that this flexibility is caused by extremely low potential barriers for changes of dihedral bond angles in vinyl bonds. We speculate that the low potential barriers for conformational changes permit the polyunsaturated chains to better adjust to the structure of the MII photointermediate, therefore lowering its free energy and tilting the MI-MII equilibrium toward MII. Although the difference in curvature elastic stress between SDPC and DOPC was not measured, if this difference is analogous to the minor differences between DOPE and SDPE, it is reasonable to assume that its energetic contribution to the shift in the MI-MII equilibrium is small. Therefore, the ability of the DHA chains in SDPC to adjust to the structure of MII is likely to be primarily responsible for the shift toward MII as well.
Energetic contributions to the MI-MII equilibrium
Fig. 2 allows a crude evaluation of the contributions from membrane curvature stress, hydrogen bonding with lipid headgroups, and of interactions with specific hydrocarbon chains to the total shift in ln Keq. The upper dashed line in Fig. 2 shows the slope of ln Keq vs. ALip2 · kcLip2 · C0Lip2 that is predicted for the series of methylated PEs. The shifts in the MI-MII equilibrium from DOPE-Me2 to DOPE-Me1 and to DOPE are faithfully predicted by the differences in the product
There are significant deviations from predictions for the transition from POPC (crosses) to DOPC (solid squares) and to DOPE-Me2 (solid diamonds), and from DOPE (solid circles) to 18:0-22:6n3-PE (open circles). POPC and DOPC favor more MI than predicted, and 18:0-22:6n3-PE more MII.
The prediction for ln Keq of SDPC (open squares) seems to be almost in agreement with expectations from elastic energy, but this is likely to be accidental. SDPC has a PC headgroup that lowers ln Keq and a DHA chain at sn-2 which raises ln Keq. Therefore, both effects may have canceled each other. Furthermore, C0 of SDPC was not measured but was assumed to be identical to DOPC—which may not be very accurate.
Finally, there is a small increase in ln Keq with increasing DOPC content that is not predicted from changes in ALip2 · kcLip2 · C0Lip2 (lower dashed line in Fig. 2). We tentatively assigned that shift to differences in direct lipid rhodopsin interactions from substituting the palmitic acid sn-1 chain to oleic acid.
In the light of those experimental observations, the free change of energy for the MI–MII equilibrium should be adjusted to
| (9) |
where ΔG0,HB is the change in free energy resulting from hydrogen bonding between lipid headgroups and rhodopsin and ΔG0,HC is the change in free energy from interaction with specific lipid hydrocarbon chains. As discussed earlier, the contributions from elastic energy, ΔG0,L is negative and inversely proportional to the spontaneous radius of curvature of lipid-2, the term ΔG0,LP is positive and identical for all lipid mixtures, and ΔG0,HB is zero for POPC, DOPC, and SDPC and a constant negative value for DOPE-Me2, DOPE-Me1, DOPE, and SDPE.
This allows a quantitative comparison of energetic contributions to shifts in ln Keq from the different energies shown in Fig. 3.
Figure 3.
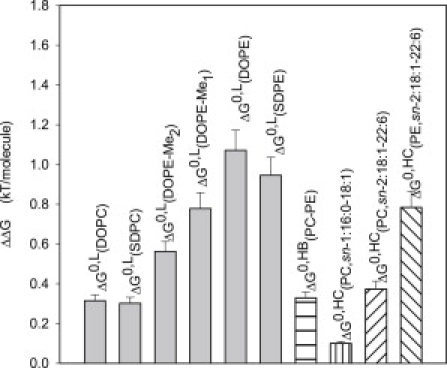
Magnitude of energetic contributions to the shift in ln Keq from POPC to DOPC, SDPC, DOPE-Me2, DOPE-Me1, DOPE, and SDPE at a lipid/rhodopsin molar ratio of 250:1. (Shaded bars: ΔG0,L, membrane elastic energy; hatched bars: ΔG0,HB, energy from hydrogen bonding between PEs and rhodopsin; and ΔG0,HC, energy from direct interactions of sn-1 oleic acid chains and sn-2 DHA chains.
Conclusions
We investigated the influence of PE-lipids with different levels of headgroup methylation and of PC- and PE-lipids with DHA chains on the MI-MII equilibrium of rhodopsin. The resulting shifts in ln Keq were compared with predictions from elastic theory, calculated by the FSM-model (4,14). It was observed that shifts in ln Keq among the lipids DOPE-Me2, DOPE-Me1, and DOPE were fully explicable by changes in membrane elastic properties. However, predictions for the transition from PCs to PEs, as well as from introduction of polyunsaturated DHA chains, poorly correlated with experimental observations. PEs induce a shift toward MII that correlates with the ability of PE headgroups to form a hydrogen bond with rhodopsin. Additional shifts toward MII were induced by the introduction of DHA chains at sn-2, which is additive to effects from the lipid headgroup, and a smaller shift from replacement of palmitic acid at sn-1 by oleic acid. It appears that the specific molecular properties of lipids in the first layer of lipids surrounding rhodopsin are as important for the MI-MII equilibrium as are changes in membrane elastic properties.
Acknowledgments
This work was supported by the Intramural Research Program of National Institute on Alcohol Abuse and Alcoholism, of the National Institutes of Health, Bethesda, Maryland.
Supporting Material
References
- 1.Hubbell W.L., Altenbach C., Khorana H.G. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv. Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 2.Ruprecht J.J., Mielke T., Schertler G.F.X. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altenbach C., Kusnetzow A.K., Hubbell W.L. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botelho A.V., Gibson N.J., Brown M.F. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 5.Brown M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell D.C., Niu S.L., Litman B.J. Optimization of receptor-G protein coupling by bilayer lipid composition. I. Kinetics of rhodopsin-transducin binding. J. Biol. Chem. 2001;276:42801–42806. doi: 10.1074/jbc.M105772200. [DOI] [PubMed] [Google Scholar]
- 7.Niu S.L., Mitchell D.C., Litman B.J. Optimization of receptor-G protein coupling by bilayer lipid composition. II. Formation of metarhodopsin II-transducin complex. J. Biol. Chem. 2001;276:42807–42811. doi: 10.1074/jbc.M105778200. [DOI] [PubMed] [Google Scholar]
- 8.Eldho N.V., Feller S.E., Gawrisch K. Polyunsaturated docosahexaenoic vs docosapentaenoic acid-differences in lipid matrix properties from the loss of one double bond. J. Am. Chem. Soc. 2003;125:6409–6421. doi: 10.1021/ja029029o. [DOI] [PubMed] [Google Scholar]
- 9.Soubias O., Gawrisch K. Docosahexaenoyl chains isomerize on the sub-nanosecond time scale. J. Am. Chem. Soc. 2007;129:6678–6679. doi: 10.1021/ja068856c. [DOI] [PubMed] [Google Scholar]
- 10.Feller S.E., Gawrisch K., MacKerell A.D. Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J. Am. Chem. Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 11.Feller S.E., Gawrisch K., Woolf T.B. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J. Am. Chem. Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 12.Gawrisch K., Soubias O. Structure and dynamics of polyunsaturated hydrocarbon chains in lipid bilayers-significance for GPCR function. Chem. Phys. Lipids. 2008;153:64–75. doi: 10.1016/j.chemphyslip.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gawrisch K., Soubias O., Mihailescu M. Insights from biophysical studies on the role of polyunsaturated fatty acids for function of G-protein coupled membrane receptors. Prostaglandins Leukot. Essent. Fatty Acids. 2008;79:131–134. doi: 10.1016/j.plefa.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson N.J., Brown M.F. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 1993;32:2438–2454. doi: 10.1021/bi00060a040. [DOI] [PubMed] [Google Scholar]
- 15.Hakizimana P., Masureel M., Govaerts C. Interactions between phosphatidylethanolamine headgroup and LmrP, a multidrug transporter: a conserved mechanism for proton gradient sensing? J. Biol. Chem. 2008;283:9369–9376. doi: 10.1074/jbc.M708427200. [DOI] [PubMed] [Google Scholar]
- 16.Powl A.M., East J.M., Lee A.G. Importance of direct interactions with lipids for the function of the mechanosensitive channel MscL. Biochemistry. 2008;47:12175–12184. doi: 10.1021/bi801352a. [DOI] [PubMed] [Google Scholar]
- 17.Litman B.J., Lester P. Purification of rhodopsin by concanavalin A affinity chromatography. Methods Enzymol. 1982;81:150–153. doi: 10.1016/s0076-6879(82)81025-2. [DOI] [PubMed] [Google Scholar]
- 18.Wald G., Brown P.K. The molar extinction of rhodopsin. J. Gen. Physiol. 1953;37:189–200. doi: 10.1085/jgp.37.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straume M., Mitchell D.C., Litman B.J. Interconversion of metarhodopsin-I and metarhodopsin-II—a branched photointermediate decay model. Biochemistry. 1990;29:9135–9142. doi: 10.1021/bi00491a006. [DOI] [PubMed] [Google Scholar]
- 20.Fuller N., Rand R.P. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys. J. 2001;81:243–254. doi: 10.1016/S0006-3495(01)75695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z., Rand R.P. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys. J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamai C., Yang T., Musser S.M. Effect of average phospholipid curvature on supported bilayer formation on glass by vesicle fusion. Biophys. J. 2006;90:1241–1248. doi: 10.1529/biophysj.105.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teague W.E., Fuller N.L., Gawrisch K. Polyunsaturated lipids in membrane fusion events. Cell. Mol. Biol. Lett. 2002;7:262–264. [PubMed] [Google Scholar]
- 24.Helfrich W. Elastic properties of lipid bilayers—theory and possible experiments. Z. Naturforsch. [C] 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 25.Salamon Z., Wang Y., Tollin G. Surface plasmon resonance spectroscopy studies of membrane proteins: transducin binding and activation by rhodopsin monitored in thin membrane films. Biophys. J. 1996;71:283–294. doi: 10.1016/S0006-3495(96)79224-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soubias O., Niu S.L., Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J. Am. Chem. Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindler H., Seelig J. Deuterium order parameters in relation to thermodynamic properties of a phospholipid bilayer. A statistical mechanical interpretation. Biochemistry. 1975;14:2283–2287. doi: 10.1021/bi00682a001. [DOI] [PubMed] [Google Scholar]
- 28.Seelig A., Seelig J. The dynamic structure of fatty acyl chains in a phospholipid bilayer measured by deuterium magnetic resonance. Biochemistry. 1974;13:4839–4845. doi: 10.1021/bi00720a024. [DOI] [PubMed] [Google Scholar]
- 29.Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q. Rev. Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 30.Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta Rev. Biomembr. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teague W.E., Petrache H.I., Gawrisch K. Membrane curvature elasticity of polyunsaturated phosphatidylethanolamine monolayers. Biophys. J. 2004;86:198A. [Google Scholar]
- 32.Bermudez H., Hammer D.A., Discher D.E. Effect of bilayer thickness on membrane bending rigidity. Langmuir. 2004;20:540–543. doi: 10.1021/la035497f. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell D.C., Litman B.J. Effect of protein hydration on receptor conformation: decreased levels of bound water promote metarhodopsin II formation. Biochemistry. 1999;38:7617–7623. doi: 10.1021/bi990634m. [DOI] [PubMed] [Google Scholar]
- 34.Rand R.P., Fuller N., Rau D.C. Variation in hydration forces between neutral phospholipid bilayers—evidence for hydration attraction. Biochemistry. 1988;27:7711–7722. doi: 10.1021/bi00420a021. [DOI] [PubMed] [Google Scholar]
- 35.Gruner S.M., Tate M.W., Turner D.C. X-ray diffraction study of the polymorphic behavior of n-methylated dioleoylphosphatidylethanolamine. Biochemistry. 1988;27:2853–2866. doi: 10.1021/bi00408a029. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh C.H., Wu W.G. Molecular order and hydration property of amine group in phosphatidylethanolamine and its n-methyl derivatives at subzero temperatures. Biophys. J. 1995;69:2521–2530. doi: 10.1016/S0006-3495(95)80123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossfield A., Pitman M.C., Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J. Mol. Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soubias O., Teague W.E., Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J. Biol. Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 39.Rabinovich A.L., Ripatti P.O. On the conformational, physical properties and functions of polyunsaturated acyl chains. Biochim. Biophys. Acta. 1991;1085:53–62. doi: 10.1016/0005-2760(91)90231-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


