Abstract
Topoisomerases maintain the proper topological state of DNA. Human topoisomerase I removes DNA supercoils by clamping a duplex DNA segment, nicking one strand at a phosphodiester bond, covalently attaching to the 3′ end of the nick, and allowing the DNA downstream of the cut to rotate around the intact strand. Using molecular dynamics simulations and umbrella sampling free energy calculations, we show that the rotation of downstream DNA in the grip of the enzyme that brings about release of positive or negative supercoils occurs by thermally assisted diffusion on ratchet energy profiles. The ratchetlike free-energy-versus-rotation profile that we compute provides a model for the function of topoisomerase in which the periodic maxima along the profile modulate the rate of supercoil relaxation, while the minima provide metastable conformational states for DNA religation. The results confirm previous experimental and computational work, and suggest that relaxation of the two types of supercoils involves distinct protein pathways. Additionally, simulations performed with the ternary complex of topoisomerase, DNA, and the chemotherapeutic drug topotecan show important differences in the mechanisms for supercoil relaxation when the drug is present, accounting for the relative values of relaxation rates measured in single-molecule experiments. Good agreement is found between rate constants from tweezer experiments and those calculated from simulations. Evidence is presented for the existence of semiopen states of the protein, which facilitate rotations after the initial one, as a result of biasing the protein into a conformation more favorable to strand rotation than the closed state required for nicking of the DNA.
Introduction
Topoisomerases are enzymes with a crucial role in the genetic transactions of the cells: they modify the supercoiling topology of DNA, which in turn affects transcription, replication, recombination, nucleosome dynamics, and a host of other processes vital to cell proliferation (1–4). They are divided into two classes: type I enzymes, which relax supercoils by nicking one phosphodiester backbone strand and passing the other strand through the nick (5,6); and type II enzymes, which concurrently break both strands and, with energy provided through ATP binding and/or hydrolysis, may increase or decrease the DNA linking number (7). Both classes are further subdivided into types A or B, depending on whether they covalently attach to the 5′- or 3′ end.
Human topoisomerase I (topo1), a type IB enzyme, consists of a single chain of 765 amino acids. Its structure in complex with duplex DNA (8) consists of four domains (see Fig. 1): the N-terminal domain; a core domain (further divided into three subdomains); an α-helical linker domain; and the C-terminal domain. The N-terminal domain is likely a disordered region that contains nuclear localization signals, and its first 190 amino acids are not required for activity in vitro (9–11). Core subdomains (CS) I and II form a cap positioned onto the DNA duplex (Fig. 1), and contain a pair of α-helices forming the so-called nose cone that sits close to the bound DNA. The cap is connected to core subdomain III by a short hinge helix (termed such because on it hinges the opening motion of the lips, see below), whereas the core subdomain III and the C-terminal domain, which contain the catalytic site, are connected by the linker domain (the latter protruding roughly parallel to the DNA helical axis (see Fig. 1 B)) and form a lower base.
Figure 1.
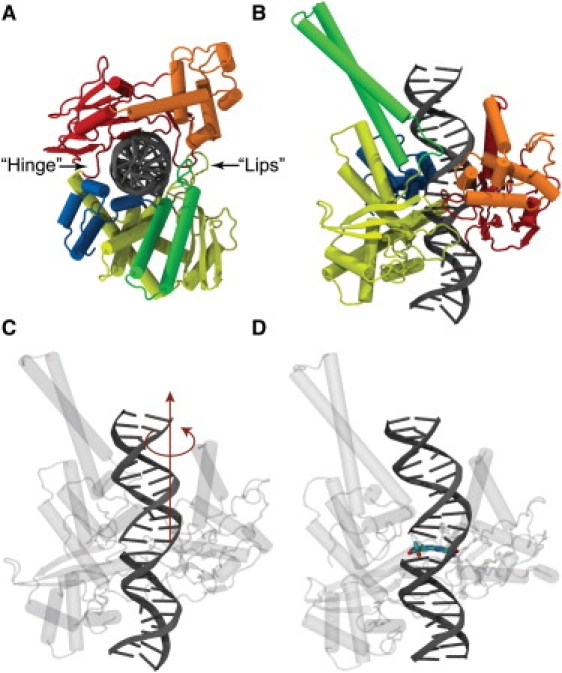
Human topoisomerase I in complex with DNA. Protein is composed of four domains: the N-terminal (not resolved in crystal structures); the core domain (which is divided into three subdomains shown in red, orange, and yellow); the linker domain (shown in green); and the C-terminal domain (in blue). (A) View down the DNA axis. Protein resembles a Pac-Man with an upper cap and a lower base: subdomain I and II of the core form the upper cap, and subdomain III, together with the C-terminal domain, form the lower base of the Pac-Man. The Pac-Man-like enzyme opens its lips to remove positive supercoils and stretches its hinge when removing negative ones (see text). (B) View of the complex perpendicular to the DNA axis. A 22-basepair DNA segment is shown in gray. (C) The swiveling axis about which the DNA duplex downstream of the cut is rotated; the downstream DNA is actually running upward in this snapshot. (D) Same snapshot as in panel c, with the drug topotecan shown in its crystal-structure position, intercalating between the two DNA basepair stack flanking the nick, which displaces the rotating DNA part one flight up and imposes steric constraints during DNA rotation.
Together with the upper cap, the lower base forms a Pac-Man-like clamp that grips around DNA, with hydrogen bonds between CS I and III forming between the lips of the protein (8,12) (the portion of the protein that dynamically opens and closes to allow for clamping about the DNA upon binding). The DNA-topo complex reveals the structural basis of a swivel mechanism (proposed as early as 1972 (13)) for the enzyme's function. It involves the protein clamping around DNA, nicking it, and forming a covalent bond between the active-site tyrosine and the 3′ end of the DNA upstream of the cut, while hydroxyl-capping the 5′ end of the downstream DNA. After this enzymatic step, supercoils are relaxed by rotation of the DNA duplex downstream of the cut around the intact strand; the drive to rotate is provided by the energy stored in the supercoils and the number of full rotations of the downstream DNA equals the number of supercoils that are eliminated. Subsequently, the DNA backbone is religated, and the protein releases the DNA.
The importance of human topoisomerase I for cell replication has made it a prime chemotherapeutic target. The camptothecin class of compounds has been found to selectively inhibit topo1 (14,15). A crystal structure was solved in which the camptothecin analog topotecan (TPT) was complexed with a topo1 molecule and a 22-basepair DNA segment (8). The structure revealed that TPT, a five-ringed molecule which mimics a DNA basepair, intercalates between the DNA segments upstream and downstream (see Fig. 1 D and Fig. S5 in the Supporting Material), creating base-stacking interactions with basepairs immediately up and down relative to the cut, and forcing the free 5′-OH group further away from the active-site tyrosine. It was proposed that this increased separation would substantially decrease the probability for religation of the backbone, trapping the topo/DNA/TPT ternary complex in a covalently attached state that would prohibit interaction of the DNA with cellular machinery for replication and transcription.
Once clamped around the DNA, the various structural components of the topo/DNA complex, i.e., the two Pac-Man lobes (see Fig. 3) formed by the upper cap and the lower base, decorated by the linker, lips, and hinge, are assembled such that they grip the DNA and control its swiveling inside the protein (16). This controlled rotation model for a related topo IB enzyme, vaccinia topo IB, was confirmed in real-time measurements by a remarkable single molecule tweezer study (17) that used optical tweezers to twist supercoils into DNA, supercoils that were subsequently relaxed when topoisomerases were allowed to bind and perform their function. By measuring the rate of DNA swiveling, the study showed that DNA does not rotate freely, but that there is friction between the DNA and the engulfing vaccinia topo IB enzyme as the DNA rotates. This friction is expected to be even more prevalent in the case of the human topo IB, because the human form of the enzyme clamps by surrounding DNA more substantially than the vaccinia form (8,17,18). However, inspection of the structure of the human topo1 in the closed form shows that the clamping of the protein onto the DNA duplex is so tight that rotation of the DNA inside of it is impossible without some degree of protein structural rearrangement. This is because the DNA duplex downstream of the cut does not rotate around the helical axis (a steric impossibility), but off-centrically, i.e., around an axis parallel to the double-helical axis but passing through the periphery of the upstream DNA duplex (see Fig. 1 C).
Figure 3.
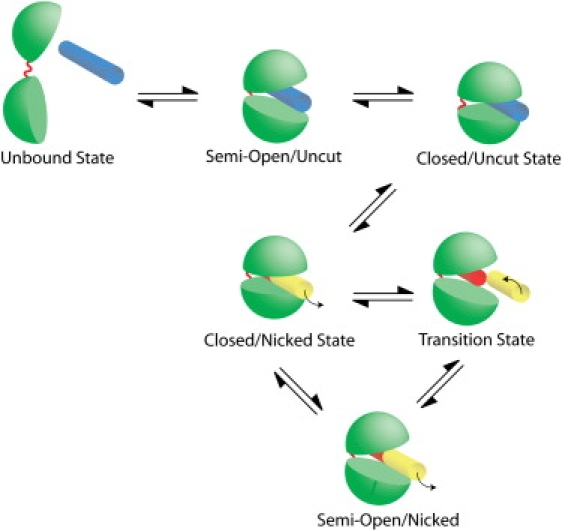
Schematic mechanism for the relaxation of DNA supercoils by human topoisomerase I which incorporates a semiopen configuration of the protein. Protein is represented in Pac-Man-like form with upper cap and lower base (in green) connected by a springlike hinge. (Yellow) DNA downstream of the cut; (red) DNA upstream of it. Note that positive supercoil relaxation is showcased here. For the relaxation of negative supercoils, the overall mechanism remains the same; however, the transition and semiopen states are replaced by configurations in which the hinge is opened as opposed to the lips, and the free energy profiles are distinct (see text).
Concurrent with the single molecule work on vaccinia topo IB, in computational work on human topoisomerase IB (19), we suggested, based on evidence from biased molecular dynamics, i.e., from simulations which forced the rotation of the DNA inside the protein's grip, that the protein undergoes opening of the upper cap and lower base. Results were compared with two independent disulphide-clamping studies (20,21) in which a cysteine cross-link was engineered such as to suture the upper and lower lips so that their opening was hindered. The locations of the Cys residues involved in the cross-link were, in the two studies, in relatively close proximity of one to another. The results from the study by Woo et al. (20) demonstrated an inhibition of DNA rotation when the disulphide clamp was proximal to the DNA rotation axis, whereas the more distal clamp engineered by Carey et al. (21) had little effect on the rotation rates. The contradictory results suggested that subtle protein changes to the protein's conformation in the unrotated state may result in larger-than-expected deviations from the enzymatic pathway of the wild-type. More recently, it has been shown that the disparity in these studies may be a result of the difference in whether the N-terminal domain was actually present, and on its role in vivo versus in vitro (22).
Importantly, both disulphide linking studies above were done with positive supercoils only. By rotating the DNA in both clockwise and anticlockwise directions—as required to relax supercoils of both signs—we found in our simulations that the relaxation of positive and negative DNA supercoils progressed along different pathways. When positive supercoils were present, DNA rotation induced an opening of the lips region of the protein by 10–14 Å to create space for downstream DNA swiveling, while to allow relaxation of negative supercoils, the hinge region stretched by ∼12 Å. Evidence supporting our model of distinct pathways for positive-versus-negative supercoil relaxation was presented in experimental work by Frøhlich et al. (9), which showed that mutations to Trp205 confer selective camptothecin resistance to the relaxation of positive supercoils but not negative ones, suggesting distinct mechanisms for relaxing positive and negative supercoils. The asymmetry between positive and negative supercoils is also expected based on theoretical grounds. For example, statistical-mechanical arguments have been proposed (23) in a theory that can describe relaxation of supercoiling by enzymes which permit friction-controlled rotational relaxation of linking number. In this theory, such enzymes must display a breaking of symmetry between relaxations driven by equal magnitude but opposite direction torques. As we will show below, from our calculated free energy profiles, this asymmetry has to do with the different barrier width in the positive-versus-negative direction.
The single molecule tweezer experiment done on vaccinia topoisomerase DNA complex (17) was subsequently repeated for the human topo1-DNA complex, as well as for the human topo1-DNA-TPT complex (24). Although the measured rate for DNA supercoil relaxation appeared to be similar for positive (overtwisted) and negative (undertwisted) supercoils in the absence of TPT, the combination of single-molecule and in vivo experiments suggested that, remarkably, TPT inhibited the relaxation of positive supercoils substantially more than that of negative ones (24). A 20-fold selective inhibition in the rate of positive supercoil relaxation (relative to the absence of TPT) by topo1 was measured, as compared to negative supercoils being inhibited only by fourfold; this resulted not only in the retardation of enzymatic function, but also the buildup of positive supercoils in poisoned cells. The single molecule observation, the fact that positive uncoiling is faster than the negative one, was confirmed by the in vivo probes, which showed a buildup of positive supercoils in yeast cells during G1 and S phase as topo1 preferentially removed negative supercoils (24).
In our work, we asked ourselves the following questions:
What are the mechanisms and energetics underlying the protein-controlled rotation of DNA?
Why do the rates differ significantly when TPT is bound?
How can we reconcile the hypothesis put forth in our previous molecular dynamics work (the existence of different pathways for positive and negative supercoils) with the values of rotation rates measured in the single molecule experiments probing DNA swiveling in the grip of the enzyme?
To offer a quantitative analysis, in this article we compute free energy profiles for the rotation of DNA downstream of the cut in the central pore of human topoisomerase I to mimic the relaxation of positive and negative supercoils. These calculations are done both in the presence and in the absence of TPT, i.e., on a ternary complex that contains DNA, topoisomerase, and TPT, and on a binary DNA/topoisomerase complex. The calculations show that DNA supercoil relaxation can be modeled along a reaction coordinate involving DNA rotation over a ratchetlike free energy profile, with thermally assisted rotation-relaxation by passage over free energy barriers and with metastable minima after full DNA rotations in DNA conformations from which religation can occur. The free energy profiles are used in the context of Kramer's rate theory to calculate rotation rate constants, and the results are used to determine how TPT alters the mechanisms and energies of supercoil relaxation.
Comparison of the single-molecule experiments by Koster et al. (24,25) on the DNA-topoisomerase complex with and without TPT bound reveal two striking features: 1), with TPT, rotation is slower; and 2), that no religation takes place. We here show that the first feature is explained by rotation in a semiopen state with different free energy barriers compared to the absence of TPT, and that the second feature may be explained by a large barrier to go from semiopen to closed in the positive case. Using umbrella sampling simulations, we calculate a free energy profile along the DNA rotation coordinate as it swivels inside the grip of topo1 to release positive and negative supercoils, with and without TPT. For both positive and negative torques, the calculation yields a unidirectional, rotating ratchet energy surface, and we identify the torque-dependent metastable states and the free-energy barrier heights connecting them. The main points, detailed in Results, are as follows.
First, a hypothesis results from the simulation and involves the existence of semiopen, kinetic intermediates in the form of two semiopen states for the two different sign supercoils. Then, as indirect evidence, there is the fact that the rates computed from the free energy profiles for DNA relaxation from semiopen states, rather than from closed states, are more in agreement with single molecule experimental data—which lends credence to a semiopen state hypothesis.
Methods
Umbrella sampling for free energy profiles
Details about system construction, MD simulations, and umbrella sampling have been placed in the Supporting Material. To summarize: the MD package NAMD (26) was used in four sets of umbrella sampling simulations, two simulating overtwisting and two simulating undertwisting, and with and without the inhibitor topotecan. Windows were sampled in 10° increments from −10° to 560° (or from 10° to −560° in the case of negative supercoils) and were simulated for a minimum of 10 ns using NAMD (26) (longer for those windows which had not fully equilibrated in the binary/positive supercoil system) for a minimum of 10 ns per window, with the final 4 ns being used in the WHAM algorithm (27).
First-passage times from Smoluchowski diffusion
We calculated rate constants for topo-modulated DNA rotation by numerically integrating the Smoluchowski equation describing diffusion on the computed free energy profile. Rates were computed as the inverse of the mean first-passage time (28) from a location x to one revolution beyond x, x ± 360 (depending on the sign of the supercoils being released) as
| (1) |
where U(x) represents the free energy at x and D is the rotational diffusion coefficient, which was position-dependent and calculated from the individual umbrella windows (29,30). The rotational diffusion coefficient, D0, of the DNA explicitly modeled in our simulation depended on the reaction coordinate Q according to
This calculation was performed for each window, with Δ(Q)2 representing the variance of the reaction coordinate observed therein, and τcorr the correlation time from that window, as calculated from the statistical inefficiency of the data (31). The rotational diffusional coefficient for DNA is dependent on the length of the DNA sequence (32,33). Thus, while simulations were performed by rotating nine restrained basepairs, a correction factor for the diffusion coefficient calculated from simulations, D0, was required to calculate an experimentally observable value of D for a DNA duplex that is as long as the persistence length,
with l the persistence length (50 nm), l0 the length of the nine restrained basepairs in the simulation (2.97 nm), and r the DNA radius (1.4 nm). DBN represents the functional form of the rotational diffusion coefficient of a rigid-rod as derived by Newman et al. (32).
The Marko model (23) for torque was derived through a statistical mechanical approach in which the free energy of the plectonemic and elongated states was minimized for a given tension. Accordingly, the critical torque is (23)
| (2) |
where g is the free energy of the extended-state, Cs the twist stiffness of the extended state, and P the stiffness of the plectonemic state. The first two quantities are given as functions of the tension, and an estimate is made for the third. To fit the data of Forth et al. (34), we have introduced a linearly variable P (because linear dependence provided the best fit) of the form P(f) = 35.3–3.4f. As this creates values for P beyond the estimated 21–27 nm, the overall fit is improved for the range where experimental data exists.
Results
Ratchetlike free energy profiles for DNA rotation
In Fig. 2 we showcase the central result of our simulations, the free energy profiles for positive and negative supercoil relaxation, i.e., with the positive and negative driving torque on the rotating DNA duplex from the torsional strain built up through supercoils added to the computed potential of mean force U,
| (3) |
where θ is the rotation angle and Γc the applied torque, which is constant (for a given tension) when sufficient torque has been applied such that additional twist energy is transformed into writhe (that is, the DNA backbone coils about itself) to form plectonemes (supercoiled DNA structures with highly nonlinear axis). Three overall features of the profiles are salient:
-
1.
The tilting of the rotation energy landscape (toward positive or negative rotation angles) is brought about by the torque stored in the DNA positive or negative supercoils, respectively. This, in turn, results in unidirectional rotation downhill on a ratchetlike energy landscape.
-
2.
The minima along the rotation profile occurs after complete rotations, when the nick returns close to the prenicked position. These are positions in which DNA is prone to religation, after sufficiently many supercoils are relaxed; see schematic in Fig. 3.
-
3.
The rotation energy maxima occur at nearly half-turn rotation (where the diameter of the rotation cylinder is maximal); thermally assisted diffusion over these barriers modulate the rate for rotation, hence for supercoil relaxation.
Figure 2.
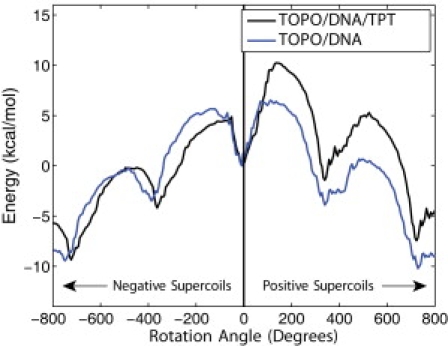
Free energy profiles along the DNA swiveling angle for the binary and ternary complexes reveal a thermally activated rotational diffusion over the barriers of a ratchetlike surface. The torque (positive or negative) tilts the surfaces to the right (clockwise rotation) or the left (anticlockwise), and drives relaxation of supercoils of the two signs. Minima on the surface (occurring after full-circle rotations) are conformations from where DNA backbone religation can occur; energy maxima modulate thermally assisted rotational diffusion rate for supercoil relaxation. The absolute value of the torque results in 5.8 kcal/mol/supercoil (corresponding to a tension of 0.2 pN; see text).
In the tweezer experiments, DNA is not only twisted (to induce a supercoiling torque), but also pulled; the applied tension modulates the torque Γc. An analytic formula relating the dependence of the torque on tension applied to a supercoiled DNA molecule was proposed by Marko (23) based on the stiffness of the plectonemic and extended states. Recently, this torque was experimentally measured for tensions >1 pN and compared to the Marko model (34,35). As presented in Methods, we use a modification to the Marko model which was performed to fit the experimental data of Forth et al. (34). Whereas a range of 0.2–3 pN have been applied in studies on vaccinia topo1B, the work with human TopoIB was performed with a tension of 0.2 pN, yielding a value for Γc corresponding to an approximate free energy change of 5.8 kcal/mol/supercoil relieved. Table 1 shows how this affects the energy barriers of the systems, decreasing them by ∼2–4 kcal/mol and lowering the energy of the first minima to below that of the initial closed state.
Table 1.
Free energy barriers and the local minima after the first revolution for all four topoisomerase systems, and the effect of adding a constant torque Γc in Eq. 3, corresponding to a free energy change of 5.8 kcal/mol/supercoil released (for a tension of 0.2 pN, see text)
| Torque-free system |
Tilted system |
|||||
|---|---|---|---|---|---|---|
| System | Barrier 1 | Minimum 1 | Barrier 2 | Barrier 1 | Minimum 1 | Barrier 2 |
| Topo/DNA (+) | 8.9 ± 0.27 | 1.7 ± 0.35 | 7.4 ± 0.29 | 6.8 ± 0.27 | −3.9 ± 0.35 | 4.57 ± 0.29 |
| Topo/DNA/TPT (+) | 12.6 ± 0.23 | 4.0 ± 0.35 | 9.6 ± 0.25 | 10.2 ± 0.23 | −1.3 ± 0.35 | 6.7 ± 0.25 |
| Topo/DNA (−) | 8.3 ± 0.28 | 2.6 ± 0.37 | 5.1 ± 0.24 | 5.6 ± 0.28 | −3.5 ± 0.37 | 3.1 ± 0.24 |
| Topo/DNA/TPT (−) | 6.2 ± 0.25 | 1.5 ± 0.37 | 6.1 ± 0.27 | 4.7 ± 0.25 | −4.2 ± 0.37 | 4.0 ± 0.25 |
Although the semiopen and deformed states are unfavorable in relaxed systems, the energy provided by the release of supercoils makes them favorable and also reduces the second energy barriers to the range of 3–7 kcal/mol.
The single molecule work demonstrated that multiple supercoils were relieved per nicking event in these enzymes, and that the change in linking number has a probability distribution that can be modeled as a decaying exponential that is modulated by the tension in the downstream DNA (17). These findings are inconsistent with a protein-assisted strand-passage mechanism (the model suggested by experiments for type IA and II topoisomerase) but is fully consistent with a swivel mechanism as modeled here.
Relaxation of positive supercoils
Simulations of the relaxation of positive supercoils by the topo/DNA complex in the absence of TPT showed an opening of the lips region by as much as 18.5 Å in some windows, similar to our previous results (see the Supporting Material). Surprisingly, in contrast, the calculations for the ternary DNA-topo-TPT complex show that the hydrogen bonds between the lips are not broken by rotation of the downstream DNA. The potential of mean force for these processes shows less energy input is initially required to rotate the DNA in the ternary complex, but, at ∼80°, rotations become more prohibitive due to the lack of opening of the lips. Because TPT forces DNA out of the protein binding site, there is less protein-DNA rotational friction initially (hence the lower energy), but this results in the DNA not providing enough push on the protein to open the lips, which results in increasing the energy barrier for the initial rotations.
When the DNA has completed its first revolution, the protein does not return to its original closed conformation. Rather, in the binary complex, it assumes a state where the lips are still separated from one another by ∼8 Å; we term this intermediate the semiopen state (see Fig. 5 C). In the absence of the biasing torque, this state has a free energy 1.7 kcal/mol higher than that of the closed state, and calculations show that the second DNA rotation (begun in the semiopen state) has an energy barrier of 7.2 kcal/mol. The ternary complex also has a higher energy after one revolution (by 4.1 kcal/mol); however, the protein's conformation has not changed dramatically (the protein root-mean-square deviation (RMSD) when not including the linker region is ∼3.25 Å from its initial state compared to ∼5.3 Å for the binary complex). The higher energy is a result of slight modifications to the system structure (such as the downstream DNA having an RMSD of 3 Å rather than 2 Å). However, in general, we note that the state is close to that of the original closed conformation. These slight deformations do nonetheless decrease the energy barrier for a second rotation to 9.2 kcal/mol.
Figure 5.
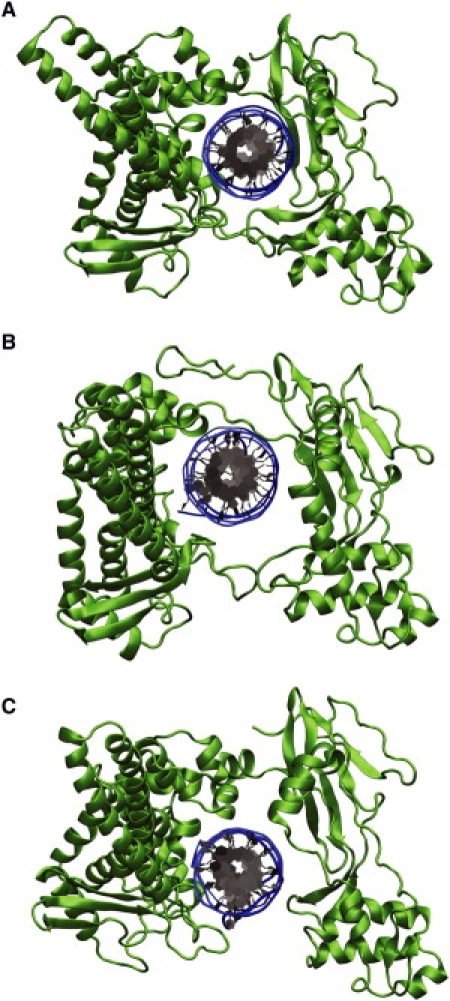
Topoisomerase-DNA structures. (A) The initial closed conformation as observed in crystal structures. (B) A semiopen conformation observed for the relaxation of negative supercoils in which the hinge of the protein has opened to allow room for DNA rotation. (C) A semiopen conformation for the relaxation of positive supercoils in which the lips remain open after the first rotation. These semiopen conformations lower the free energy barrier for rotation compared to the initial closed conformation, thus providing a more favorable mechanism for supercoil relaxation. The semiopen states differ among themselves and from the closed state only by the overall placement of the upper cap relative to the lower base (as defined in the text); structures of the protein-composing domains are otherwise mainly unchanged.
To characterize the overall concerted motion of the entire protein structure as DNA rotates inside, we computed the position-position cross-correlation functions for the Cα atoms of topo1. Data is presented in Fig. 4 B, with the top-half representing the motions of the atoms in the native system and the bottom-half those in the inhibited one (with TPT). There are three regions which have motions highly correlated to one another: CS I and II (the upper cap) move as one unit, CS III and the C-terminal (the lower base) as another, and the linker domain as a third. There is also a negative correlation between the motions of the cap and that of the CS III/C-terminal region indicating that, as the lips separate, these two regions, which are on opposite sides of the protein, move apart from one another. Interestingly, we note a slight positive correlation between the motions of the cap and the linker domain. The presence of TPT (bottom half of Fig. 4 B) shows a dampening of correlations between amino acids that are not in the same domain as each other. These results further indicate that intercalation of TPT into the DNA helix fundamentally disrupts the protein motions that allow for efficient supercoil relaxation.
Figure 4.
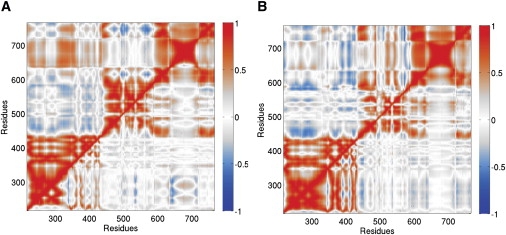
Correlation functions of protein Cα atoms for positive and negative DNA supercoil relaxation, both for the free (top half) and inhibited (with TPT, bottom half) rotations. We observe that intercalation of TPT into the DNA helix fundamentally disrupts the protein motions that allow for efficient supercoil relaxation, and this disruption is more prevalent for negative supercoils than for positive ones.
Relaxation of negative supercoils
Calculations of negative supercoil relaxation reveal that the hinge region of topoisomerase appears to open by ∼11 Å in the absence of TPT, and, akin to the case of positive supercoils, the presence of TPT prevents the hinge from opening (see the Supporting Material). Unlike the case of positive supercoils, the lack of enzyme opening does not increase the energy barrier to a point higher than that of the binary free energy barrier (Fig. 2).
Akin to the case of positive supercoil relaxation, for which the lips did not return to their closed conformations after a complete rotation, the hinge region of the protein remains open after a single rotation, creating a different semiopen state (as shown in Fig. 5 B). In the absence of the driving torque, the energy change is 2.6 kcal/mol and the barrier for a second rotation is 5.0 kcal/mol. There is also a deformed state for the ternary complex with an energy of 1.5 kcal/mol and which has an energy for a second rotation of 5.8 kcal/mol. Calculations show that in this deformed state the cap region has an RMSD of ∼5.5 Å (compared to ∼3 Å in the binary case) but that the internal structure of CS I and CS II do not change significantly (their RMSD are <3 Å), indicating that it is the interaction geometry between the two subdomains that creates this deformed state.
The cross-correlation maps for negative supercoils (Fig. 4 A) show a similar interdomain communication structure to that of positive supercoils: the cap region has strong internal correlations as do the CS III and the C-terminal domains of the lower base. The motions of the cap are much more strongly correlated to those of the linker (being mostly positive whereas in the positive supercoil case it is mostly negative) and the motions are still negatively correlated to those of the base (CS III and the C-terminal domains). Topotecan appears to have a much stronger effect on decorrelating the motions between domains than in the positive supercoil case, and in fact, it even decorrelates some of the motions between CS I and II.
DNA rotation rates
Integration of the Smoluchowski equation (as discussed in Methods) allowed for the derivation of rate constants for both the first and second rotations, which are shown in Table 2 (along with the values from the single-molecule experiments (24)). It is to be noted that the estimated rates from the Kramer theory correspond to a simple one-dimensional profile, which, even if physical, brings together tremendous complexity. Nonetheless, reasonable accord with experiments is found. The rate constants for relaxing the first supercoil show a very strong inhibition by topotecan of positive supercoils (∼232-fold) and an increase in rate by 38-fold for the addition of TPT to systems that relax negative supercoils. For systems beginning in a semiopen (or deformed) state, the effects of TPT on rotation rates are much closer to experimental values. TPT provides an inhibition of 25.5-fold relative to positive supercoils (compared to the ratio of 20 in the experiments) and for negative supercoils the rates are similar to one another (topotecan rates are higher by 30%) as is seen in experiments where the inhibition by TPT is minimal (24). These calculations provide further evidence that it is the second barrier, where the system starts in a semiopen or deformed state, that is repeatedly traversed during in vivo multiple relaxation-enabling rotations; the in vitro experiments indicate that there are ∼10–20 supercoils removed per nicking event. Although we have induced a closed-to-semiopen conformational change by the first DNA rotation imposed during umbrella sampling, a formal possibility also exists for a conformational change from the closed-to-semiopen state before any rotation and triggered solely by DNA binding and protein fluctuations. The lack of a proper reaction coordinate for this possible change precludes, however, any quantitative estimation of the barrier height that would be involved. It is also of interest to note that the rotational diffusion coefficients (as described in Methods) show a dramatic increase as the DNA begins its rotation, levels out when the DNA has completed half of a revolution, and drops to its initial value upon returning to its initial configuration for all four systems studied (see Fig. S1).
Table 2.
Calculated and experimental rate constants in Hz for an applied torque Γc, corresponding to a free energy cost of 5.8 kcal/mol/supercoil at a tension of 0.2 pN
| Rate constants | Experimental value | First rotation | Second rotation |
|---|---|---|---|
| Topo/DNA (+) | ∼75 | 6.4 | 34.8 |
| Topo/DNA/TPT (+) | ∼3.6 | 0.012 | 1.4 |
| Topo/DNA (−) | ∼61 | 4.7 | 147 (61) |
| Topo/DNA/TPT (−) | ∼12 | 268 | 192 |
Rates for the relaxation of the second supercoil (from a semiopen state) show good agreement for positive supercoil relaxation, whereas negative supercoils show less agreement but also appear less strongly affected by the presence of TPT, in accord with experiments. For Topo/DNA(−), changing plectoneme stiffness to 21 nm yields the value in parentheses, in accord with experiment. Rates for the first relaxation show a poor agreement with experiments, furthering our hypothesis that rotations from a semiopen state predominate.
Rate calculations for negative supercoil relaxation do show a higher quantitative value than those for positive supercoils by 4.2-fold for a given tension, whereas experiment shows little discrepancy between the two. This is likely due to an incomplete understanding of the correlation between tension and torque for negative supercoils. Although formalisms such as those provided by Marko provide a fair description for positively supercoiled molecules, there is the possibility that, for a given choice of parameters, they do not provide an accurate description for negative supercoils (in fact it is known that mechanical properties of positive and negative supercoils are quite distinct from one another (36–38)). As an example, when we lower the plectonemic stiffness of negative supercoils to 21 nm, the free energy change is reduced by 2.2 kcal/mol/supercoil and the rate of negative supercoil relaxation decreases by 2.4-fold, to yield the value of 62 Hz, which is close to the experimentally determined one (∼61 Hz).
TPT during DNA rotation: relaxation inhibition
It is conceivable that TPT, once bound at the nick site, inhibits supercoil relaxation by either slowing down rotation or by impeding religation, but which of the two mechanisms is predominant is unknown. Also unknown are the different details of inhibiting the positive vs. negative supercoil relaxation. Mutations to several amino acids have been implicated in imparting resistance to camptothecin (an analog of topotecan) in its inhibition of topoisomerase activity (39,40). Two amino acids in particular, Asp533 and Asn722, have been deemed especially important: the first forms the only direct contact between the ligand and the protein, whereas the second forms a water-mediated interaction (41,42). The position of Asp533 is stabilized by Arg364 on the opposing lip, therefore mutations to residues that affect the conformation of Arg364 also tend to induce resistance. Crystal structures of a ternary topo-DNA-camptothecin complex have advanced models for resistance which focus on the ligand stabilization in the pre- and postrotation complexes (42). However, another important factor in these mutations, unaddressed previously, is their effect on drugs while rotation is occurring. This is important to gauge inasmuch as an unstable protein/drug complex may result in the compound separating from the active site while it is exposed to solution during rotation of the downstream DNA. Our simulations of the ternary complex for the release of positive supercoils show a disruption of the Asp533-TPT interaction (see Fig. S5). As the DNA begins to rotate, it passes over Asp533 and Arg364, disrupting both of their positions and breaking the hydrogen bonds with the TPT (see Fig. S5 b). After this, TPT diffuses around the active site and may form direct hydrogen bonds (i.e., without an intermediate water molecule) with Asn722 through either its carbonyl or hydroxyl group (see Fig. S5, c and d). In contrast, for simulations of the relaxation of negative supercoils, DNA does not pass over Asp533 and Arg364 until the end of the rotation; and even then, it does not disrupt their positions, resulting in the position of TPT remaining virtually unchanged throughout the simulations that mimic relaxation of negative supercoils. Although these observations are results of single trajectories, we postulate that Asn722 plays a different role in stabilizing TPT during the relaxation of positive-versus-negative supercoils, and we hypothesize that the 722 mutation will render a TPT-resistant topo enzyme for which the drug can dissociate more readily in the presence of positively supercoiled substrates. The simulation results in the position of TPT during DNA rotation, which, taken together with the free energy profiles in the two directions, suggests that TPT inhibits predominantly by slowing down religation, and that, in the case of positive supercoils, the mode of drug binding during rotation is more flexible. However, a complete understanding of this process would require numerous independent trajectories of the relaxation of positive and negative supercoils in the ternary complex to check for consensus.
Concluding Discussions
Results suggest the following overall mechanism for DNA supercoil relaxation by human topoisomerase IB (which is shown in Fig. 3). Initially, supercoiled DNA presents itself to an open-state topoisomerase (modeled in (43)). Topoisomerase recognizes DNA and binds by clamping around it, then a nicking step occurs that cuts the DNA backbone, after which strand rotation ensues. Using umbrella sampling, we have computed the free energy around this rotation coordinate, for both positive and negative supercoils, with and without the inhibitor topotecan with good agreement to measures of DNA relaxation obtained from single-molecule tweezer experiments. What results from our simulations is a rotational ratchet energy surface that is the basis of a quantitative model involving thermally assisted barrier hopping. Torsional stress drives the DNA over the first (high) energy barrier to a transition state, whereby the protein may enter the semiopen conformation or may return to the closed one (while relieving one supercoil). If the protein enters the semiopen conformation, the DNA would not be in position for religation to occur, and assuming sufficient torsional stress remains, the DNA will again rotate, coming again to a transition state after which it may sample the semiopen or closed state. There exists a nonzero probability that the system, once in a free energy minimum (roughly after full-circle rotations), switches to a different pathway. This pathway would be perpendicular to the rotational one we have simulated (one in which the topoisomerase returns to a closed conformation after release of a supercoil), and if this occurs, then religation of the backbone may result, with the release of the DNA by the protein following. Alternatively, it is conceivable that there exist pathways between the closed and semiopen states which do not require the release of supercoils. In the presence of topotecan, the semiopen state undergoes additional deformation at the nick site and the rate of religation is substantially decreased (41). Additionally, for positive supercoils, the barriers to rotation are substantially increased, creating a stronger inhibition of supercoil relaxation with respect to the negative case.
Direct observation of the topo1 conformational changes correlated with DNA supercoil extension/rotation can likely be achieved in single molecule fluorescence experiments. For example, in the case of eukaryotic topo II, single molecule FRET has been used successfully to measure the opening and closing of the protein gate through which DNA passes (44). One can also imagine combining single molecule fluorescence with optical tweezer techniques. For example, a hybrid experiment, monitoring the energy transfer between FRET pairs attached to either side of the lips region while optical tweezers are used to report on the supercoiling state of DNA, may experimentally test the presence of a semiopen state. In practice, however, several technical challenges will have to be addressed, such as finding appropriate spatial and temporal resolutions, and tuning the localization of distinct signals (45,46). Nevertheless, with the advancement of single molecule techniques, future studies should be able to observe concomitantly orthogonal molecular motions (47).
Acknowledgments
I.A. acknowledges support from the National Science Foundation (CMMI-0941741 and CAREER award program CHE-0918817). This research used resources provided by the National Science Foundation through TeraGrid provided at the Texas Advanced Computing Center under grant No. TG-MCB080026N and the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under contract No. DE-AC02-05CH11231.
Footnotes
Jeff Wereszczynski's present address is The Department of Chemistry and Biochemistry, University of California, San Diego, CA.
Supporting Material
References
- 1.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Schoeffler A.J., Berger J.M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 5.Leppard J.B., Champoux J.J. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 6.Dekker N.H., Rybenkov V.V., Croquette V. The mechanism of type IA topoisomerases. Proc. Natl. Acad. Sci. USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger J.M., Gamblin S.J., Wang J.C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 8.Redinbo M.R., Stewart L., Hol W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 9.Frøhlich R.F., Veigaard C., Knudsen B.R. Tryptophane-205 of human topoisomerase I is essential for camptothecin inhibition of negative but not positive supercoil removal. Nucleic Acids Res. 2007;35:6170–6180. doi: 10.1093/nar/gkm669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart L., Ireton G.C., Champoux J.J. Reconstitution of human topoisomerase I by fragment complementation. J. Mol. Biol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- 11.Frøhlich R.F., Andersen F.F., Knudsen B.R. Regions within the N-terminal domain of human topoisomerase I exert important functions during strand rotation and DNA binding. J. Mol. Biol. 2004;336:93–103. doi: 10.1016/j.jmb.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Stewart L., Ireton G.C., Champoux J.J. The domain organization of human topoisomerase I. J. Biol. Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 13.Champoux J.J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA—a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay) Proc. Natl. Acad. Sci. USA. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanhoefer U., Harstrick A., Rustum Y.M. Irinotecan in the treatment of colorectal cancer: clinical overview. J. Clin. Oncol. 2001;19:1501–1518. doi: 10.1200/JCO.2001.19.5.1501. [DOI] [PubMed] [Google Scholar]
- 15.Fujimori A., Harker W.G., Pommier Y. Mutation at the catalytic site of topoisomerase I in CEM/C2, a human leukemia cell line resistant to camptothecin. Cancer Res. 1995;55:1339–1346. [PubMed] [Google Scholar]
- 16.Stewart L., Redinbo M.R., Champoux J.J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 17.Koster D.A., Croquette V., Dekker N.H. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 18.Cheng C., Kussie P., Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 19.Sari L., Andricioaei I. Rotation of DNA around intact strand in human topoisomerase I implies distinct mechanisms for positive and negative supercoil relaxation. Nucleic Acids Res. 2005;33:6621–6634. doi: 10.1093/nar/gki935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo M.H., Losasso C., Bjornsti M.A. Locking the DNA topoisomerase I protein clamp inhibits DNA rotation and induces cell lethality. Proc. Natl. Acad. Sci. USA. 2003;100:13767–13772. doi: 10.1073/pnas.2235886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carey J.F., Schultz S.J., Champoux J.J. DNA relaxation by human topoisomerase I occurs in the closed clamp conformation of the protein. Proc. Natl. Acad. Sci. USA. 2003;100:5640–5645. doi: 10.1073/pnas.1031537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palle K., Pattarello L., Bjornsti M.A. Disulfide cross-links reveal conserved features of DNA topoisomerase I architecture and a role for the N-terminus in clamp closure. J. Biol. Chem. 2008;283:27767–27775. doi: 10.1074/jbc.M804826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marko J.F. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys. Rev. E. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 24.Koster D.A., Palle K., Dekker N.H. Antitumor drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 25.Koster D.A., Czerwinski F., Dekker N.H. Single-molecule observations of topotecan-mediated TopoIB activity at a unique DNA sequence. Nucleic Acids Res. 2008;36:2301–2310. doi: 10.1093/nar/gkn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S., Bouzida D., Rosenberg J. The weighted histogram analysis method for free-energy calculations on biomolecules. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 28.Zwanzig R. Oxford University Press; London, UK: 2001. Nonequilibrium Statistical Mechanics. [Google Scholar]
- 29.Chahine J., Oliveira R.J., Wang J. Configuration-dependent diffusion can shift the kinetic transition state and barrier height of protein folding. Proc. Natl. Acad. Sci. USA. 2007;104:14646–14651. doi: 10.1073/pnas.0606506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Socci N., Onuchic J., Wolynes P. Diffusive dynamics of the reaction coordinate for protein folding funnels. J. Chem. Phys. 1996;104:5860–5868. [Google Scholar]
- 31.Allen M., Tildesley D. Oxford University Press; London, UK: 1987. Computer Simulation of Liquids. [Google Scholar]
- 32.Newman J., Swinney H.L., Day L.A. Hydrodynamic properties and structure of fd virus. J. Mol. Biol. 1977;116:593–603. doi: 10.1016/0022-2836(77)90086-9. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y., Weers B., Stellwagen N.C. DNA persistence length revisited. Biopolymers. 2001–2002;61:261–275. doi: 10.1002/bip.10151. [DOI] [PubMed] [Google Scholar]
- 34.Forth S., Deufel C., Wang M.D. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheinin M.Y., Wang M.D. Twist-stretch coupling and phase transition during DNA supercoiling. Phys. Chem. Chem. Phys. 2009;11:4800–4803. doi: 10.1039/b901646e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strick T., Allemand J., Croquette V. Twisting and stretching a DNA molecule leads to structural transitions. Biol. Phys. 1999;487:249–270. [Google Scholar]
- 37.Bryant Z., Stone M.D., Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 38.Wereszczynski J., Andricioaei I. On structural transitions, thermodynamic equilibrium, and the phase diagram of DNA and RNA duplexes under torque and tension. Proc. Natl. Acad. Sci. USA. 2006;103:16200–16205. doi: 10.1073/pnas.0603850103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura H., Kohchi C., Nishikawa K. Molecular cloning of a cDNA of a camptothecin-resistant human DNA topoisomerase I and identification of mutation sites. Nucleic Acids Res. 1991;19:69–75. doi: 10.1093/nar/19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pourquier P., Pommier Y. Topoisomerase I-mediated DNA damage. Adv. Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- 41.Staker B.L., Hjerrild K., Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chrencik J.E., Staker B.L., Redinbo M.R. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004;339:773–784. doi: 10.1016/j.jmb.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 43.Chillemi G., Bruselles A., Desideri A. The open state of human topoisomerase I as probed by molecular dynamics simulation. Nucleic Acids Res. 2007;35:3032–3038. doi: 10.1093/nar/gkm199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smiley R.D., Collins T.R., Hsieh T.S. Single-molecule measurements of the opening and closing of the DNA gate by eukaryotic topoisomerase II. Proc. Natl. Acad. Sci. USA. 2007;104:4840–4845. doi: 10.1073/pnas.0700342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang M.J., Fordyce P.M., Block S.M. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hohng S., Zhou R., Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter N.G., Huang C.Y., Sobhy M.A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


