Abstract
Interleukin-1β (IL-1β) induces the expression of a variety of proteins responsible for acute inflammation and chronic inflammatory diseases. However, the molecular regulation of IL-1β expression in myeloid differentiation has not been elucidated. In this study the chromatin structure of the IL-1β promoter and the impact of methylation on IL-1β expression in monocytic development were examined. The results revealed that the IL-1β promoter was inaccessible in undifferentiated promyeloid HL-60 cells but highly accessible in differentiated monocytic cells which additionally acquired the ability to produce IL-1β. Accessibilities of differentiated cells were comparable to those of primary monocytes. Lipopolysaccharide (LPS) stimulation did not affect promoter accessibility in promyeloid and monocytic HL-60 cells, demonstrating that the chromatin remodelling of the IL-1β promoter depends on differentiation and not on the transcriptional status of the cell. Demethylation via 5-aza-2′-deoxycytodine led to the induction of IL-1β expression in undifferentiated and differentiated cells, which could be increased after LPS stimulation. Our data indicate that the IL-1β promoter is reorganized into an open poised conformation during monopoiesis being a privilege of mature monocytes but not of the entire myeloid lineage. As a second mechanism, IL-1β expression is regulated by methylation acting independently of the developmental stage of myeloid cells.
Keywords: cytokines, haematology, immunogenetics, inflammation, transcription factors/gene regulation
Introduction
Interleukin-1β (IL-1β) is one of the most potent pro-inflammatory cytokines and is essential for the immune system.1 The main producers of this cytokine are monocytes and macrophages, but IL-1β has also been detected in the supernatants of endothelia, fibroblasts, natural killer cells and T cells.2 Important functions are the induction of tumour necrosis factor-α (TNF-α), IL-6, colony-stimulating factors, interferons and prostaglandin E2. The link to the adaptive immune system is shown by its ability to stimulate antibody production in B lymphocytes.1 Concentration of IL-1β in the blood has to be tightly regulated because elevated levels can cause severe diseases such as joint destruction in rheumatoid arthritis and other autoimmune diseases.3,4
The human IL-1β gene is regulated by two regions.2,5 One of those is the proximal promoter, which is described as packed into a poised architecture.5 The transcription start is reported to be free of core histones in IL-1β-producing monocytes with transcription factors PU.1 and C/enhancer binding protein-β (EBPβ) already bound to the proximal promoter. The modification of both enables the recruitment of the transcription complex and the initiation of IL-1β expression.5,6 Recent papers show that the proximal promoter is inaccessible in IL-1β non-producing primary T and B lymphocytes,5,7 indicating that the poised structure is limited to only a subset of cell types. The second regulating region includes two co-operative enhancer regions located at positions − 2782 to − 2729 and − 2896 to − 2846 upstream from the transcription start.2,8 Stimulation with lipopolysaccharide (LPS) leads to post-translational modifications of the pre-formed transcription factors nuclear factor–IL-6 and cyclic adenosine monophosphate response element-binding protein, which then bind to the enhancer regions.2,6,8 Co-operative events at the proximal promoter and enhancer region guarantee a fast and efficient transcription in monocytes after stimulation.5–7
The influence of epigenetic mechanisms on the IL-1β expression has not been investigated in detail. Although Kovacs et al.9 showed that demethylation activates IL-1β expression in promyeloid U937 and monocytic THP-1 cells and Sato et al.10 described that de-acetylation affects IL-1β expression in choriodecidual tissue after LPS stimulation, the influence of methylation and acetylation on the IL-1β expression in different cell types and at diverse developmental stages have not been compared.
To analyse how monocytes gain their ability to secrete large amounts of IL-1β during monopoiesis the promyeloid acute myeloid leukaemia cell line HL-60 was used. HL-60 cells have widely been accepted as a cell line that can be differentiated into monocytic cells using 1,25-dihydroxyvitamin D3 (VD3).11 Differentiation enables HL-60 cells to express genes for TNF-α, toll-like receptor 4 (TLR4) and TLR2, and the surface marker for monocytes, CD14.12–14
In this report we elucidate the epigenetic changes occurring in the IL-1β gene during monocytic differentiation. We address the question whether the poised structure of the IL-1β promoter is a characteristic of the entire myeloid lineage or whether it is a privilege for monocytes. Hence, the accessibilities of the IL-1β promoter in promyeloid and monocytic differentiated HL-60 cells were investigated and compared with that in primary human monocytes. Furthermore, the influence of methylation in this differentiation model was analysed as another mechanism that might regulate IL-1β expression.
Material and methods
Cell culture and HL-60 differentiation conditions
The HL-60 and THP-1 cells were grown in RPMI-1640 (Cambrex, Verviers, Belgium) supplemented with 10% low-endotoxin fetal calf serum (PAA, Pasching, Austria), 2 mm l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (all Cambrex) in a 5% CO2 humidified atmosphere at 37°. Cell cultures were initiated at a density of 0·4 × 106 to 2 × 106 cells/ml. For monocytic differentiation 100 nm VD3 (Biomol, Hamburg, Germany) was added to 4 × 105 HL-60 cells/ml for 72 hr. Progress of differentiation was assessed morphologically by acquired CD14 expression and via production of the pro-inflammatory cytokines IL-1β, IL-6 and TNF-α.
The HL-60 and differentiated HL-60 cells were stained with phycoerythrin-conjugated CD14 monoclonal antibody (mAbs; BD Biosciences, Heidelberg, Germany) or with the isotypic control phycoerythrin-conjugated immunoglobulin IgG2bκ mAb (BD Biosciences) for 15 min. Washed cells were then analysed in a FACscan (BD Biosciences) using cellquest software 3·0. In experimental approaches, differentiated cell populations with > 50% CD14-positive cells were used.
HL-60 cells were incubated with 10 μm 5-aza-2′-deoxycytidine (AZA, decitabine; Sigma-Aldrich, Munich, Germany) for 48 hr in demethylation studies. Media were changed daily and supplied with freshly prepared AZA. For expression studies. 0·5 × 106 to 2 × 106 cells/ml were incubated with or without LPS from Escherichia coli serotype O111:B4 (250 ng/ml; Sigma-Aldrich) for 4 and 24 hr.
Enrichment of CD14+ HL-60 cells
After 3 days of differentiation with VD3, CD14+ HL-60 cells were separated using anti-human CD14 mAbs (BD Biosciences) and magnetic dynabeads labelled with anti-mouse antibodies (Invitrogen, Karlsruhe, Germany) following the manufacturers’ instructions. In brief, CD14+ cells were purified by incubation with anti-human CD14 mAbs (10 μg/107 cells) for 15 min followed by incubation with pre-washed antibody-labelled anti-mouse dynabeads (bead : target ratio 8 : 1) for 30 min under rotation at 4°. The positively enriched CD14+ cells were then separated using a magnetic particle concentrator reaching a mean purity of approximately 80%. The magnetically bound CD14+ cell fraction was lysed for the purpose of nucleus isolation and for further usage in subsequent chromatin accessibility by real-time (CHART) PCR assay.
Isolation of human peripheral blood mononuclear cells and enrichment of monocytes
Blood from healthy volunteers was obtained by venepuncture with informed consent and ethics committee approval (Medical Faculty, RWTH Aachen University, document No.: EK 023/05). Peripheral blood mononuclear cells were prepared by Ficoll (Biochrom, Berlin, Germany) gradient centrifugation as previously described.15 The pellet was washed twice with phosphate-buffered saline (Cambrex) and adjusted to the appropriate cell concentrations in the same medium as HL-60 cells. Monocytes were enriched after plastic-adherence on 10 ml Petri dishes (2 × 106 cells/ml for 1 hr at 37° in 5% CO2).
Enzyme-linked immunosorbent assay
Supernatants were harvested after 4 hr, stored at −80° until measurement, and only thawed once for cytokine detection. To measure IL-1β, IL-6 and TNF-α, enzyme-linked immunosorbent assay (ELISA) kits purchased from BD Biosciences were used. All ELISAs were quantified using the Magellan ELISA-reader (Tecan, Crailsheim, Germany).
CHART-polymerase chain reaction assay
Accessibility assays were performed as described previously.5 Briefly, nuclei isolated from 5 × 106 cells were digested with 200 U micrococcal nuclease (MNase) at 37° for 60 min. Reactions were terminated by addition of 80 μl MNase stop buffer (100 mm ethylenediaminetetraacettc acid and 10 mm ethyleneglycoltetraacetic acid). Protein digestion was performed through incubation with proteinase K (25 mg/ml) and 2% sodium dodecyl sulphate (SDS) overnight at 37°. A control without MNase was run in parallel to analyse endonuclease activity. The genomic DNA was isolated using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany). Real-time polymerase chain reaction (PCR) was performed in 25-μl reaction volumes in duplicates using Brilliant Sybr Green qPCR Master Mix (Stratagene, Waldbronn, Germany) including 100 ng DNA and primer pairs for IL-1β promoter regions −I, −II and −VIII as described elsewhere.5 To correlate the Ct values (threshold values) from the IL-1β CHART-PCR amplification plots to per cent accessibility, a standard curve was generated using serial dilutions of genomic DNA.5 For MNase accessibility, the data were plotted as a percentage of the accessibility observed in the unstimulated digested DNA sample using the following formula:
Apparent negative accessibility is a mathematical artefact that is probably the result of an error in DNA quantification and is neither reproducible nor biologically significant, and therefore was set to 0% accessibilities as described before.5,7
Reverse transcription and PCR
Total cellular RNA was isolated using a NucleoSpin RNA II-Kit (Macherey-Nagel, Düren, Germany) according to the users’ manual. One microgram RNA was reversed transcribed using qScript cDNA Synthesis Kit (Quanta Bioscience, Gaithersburg, MD). The IL-1β PCR was performed in a thermal cycler using a cycle programme of 40 seconds denaturation (95°), 60 seconds annealing (60°) and 3 min amplification (72°) for 30 cycles. As a control, all samples were amplified using β-actin primers. All primers used were as previously described.16
Cell extracts and Western blotting
A total of 2 × 106 cells were lysed in 100 μl buffer containing 0·5 m Tris–HCl (pH 6·8), 26·6% glycerin, 10% SDS and 100 mm Na3VO4. The cells were sonicated for 10 seconds, then boiled for 3 min at 95°. An SDS–polyacrylamide gel electrophoresis was performed using 15 μg protein and Western blot analysis was performed as described previously.17 Nitrocellulose membranes (Bio-Rad, Munich, Germany) were blocked for 1 hr with TBS-T [20 mm Tris–HCl, pH 7·6, 136 mm NaCl, 0·1% (volume/volume) Tween-20] containing 5% fat-free dry milk, and incubated with the primary antibody (biotin-anti-IL-1β; R&D Systems, Wiesbaden-Nordenstadt, Germany) at 1 : 500 dilution in TBS-T containing 5% bovine serum albumin for 2 hr. Subsequently, membranes were washed three times with 25 ml TBS-T and incubated for 1 hr with the secondary antibody (anti-biotin horseradish peroxidase-conjugated immunoglobulin; Cell Signaling Technology, Frankfurt am Main, Germany) followed by detection with LumiGlo Reagent (Cell Signaling Technology). The membrane was stripped and then reprobed for β-actin as described above (rabbit-anti-β-actin, anti-rabbit immunoglobulin G horseradish peroxidase-linked immunoglobulin; Cell Signaling Technology).
Statistical analysis
Significances of differences were analysed by Student’s t-test using the program spss version 15 (SPSS Inc., Chicago, IL).
Results
Comparison of promyeloid HL-60 and 1,25-dihydroxyvitamin D3-differentiated HL-60 cells
To characterize changes in the IL-1β promoter during monopoiesis, a HL-60 differentiation model was established. In comparison to untreated cells, cells treated with VD3 became adherent and had a greater cytoplasm/nucleus ratio (data not shown). Surface expression of CD14 on HL-60 cells was induced after 72 hr only in VD3-differentiated HL-60 cells whereas untreated cells did not show any CD14 expression (Fig. 1, inserted histogram). In addition, differentiated HL-60 cells secreted significantly higher amounts of pro-inflammatory cytokines IL-1β, IL-6 and TNF-α than undifferentiated cells (Fig. 1). Cytokine production in contrast to undifferentiated cells could also be significantly augmented in VD3-differentiated HL-60 cells after stimulation with LPS for 4 hr (Fig. 1). Based on these data, treatment of HL-60 cells with VD3 is an appropriate method for investigation of IL-1β regulation during monocytic differentiation.
Figure 1.
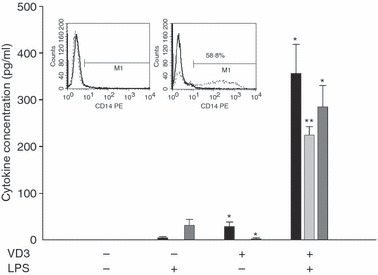
Characterization of undifferentiated and 1,25-dihydroxyvitamin D3 (VD3) differentiated HL-60 cells. The inserted histograms show one representative example out of 11 for the induction of CD14 expression on VD3-treated cells (right histogram) in contrast to CD14– untreated HL-60 cells (left histogram); black: isotypic control, dashed line: CD14. Secretion of interleukin-1β (IL-1β), IL-6 and tumour necrosis factor-α (TNF-α) after lipopolysaccharide (LPS) stimulation was significantly higher in VD3-treated HL-60 cells compared with controls. Means and standard error of n = 4 independent experiments are shown. Significant differences between the VD3-differentiated cells and the appropriate stimulated or unstimulated controls are indicated (*P < 0·05; **P < 0·01).
The IL-1β promoter opens during VD3-mediated HL-60 differentiation
To uncover whether there is a correlation between IL-1β promoter chromatin structure and acquired IL-1β expression of VD3-differentiated HL-60 cells, quantitative chromatin accessibility studies were performed. The MNase sensitivity of three different promoter regions was compared. Regions −I (− 107 to − 17) and −II (− 199 to − 109) proximal to the transcription start contain binding sites for the transcription factors PU.1 and C/EBPβ, which are involved in myelopoiesis.5,7,18 These regions have also been described to be highly accessible to MNase digestion in monocytic cell lines.5 Region −VIII (− 673 to − 583) upstream of the transcription start was used as an inaccessible negative control. As shown in Fig. 2, approximately 60% of IL-1β promoter regions −I and −II of undifferentiated HL-60 cells were accessible for MNase. In contrast, CHART assays performed with differentiated HL-60 cells showed significantly higher accessibility of about 80% in both of these regions. These results are in common with our finding that undifferentiated HL-60 cells were not able to produce IL-1β whereas the differentiated cells synthesize high levels of the cytokine, which were increased after LPS stimulation. Control region −VIII was not accessible in all tested cells (Fig. 2).
Figure 2.
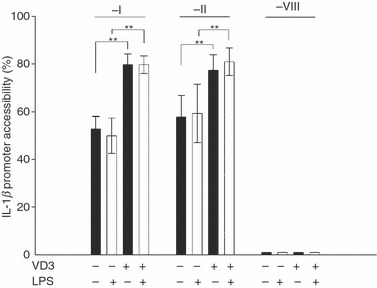
Regions −I and −II of the interleukin-1β (IL-1β) promoter are significantly more accessible in differentiated HL-60 than in undifferentiated HL-60 cells. Chromatin accessibility by real-time (CHART) polymerase chain reaction analysis of IL-1β promoter regions −I, −II and −VIII in human differentiated and undifferentiated HL-60 cells are shown. HL-60 cells were incubated with or without vitamin D3 (VD3) for 72 hr followed by stimulation with lipopolysaccharide (LPS; white bars). Control cultures remained unstimulated for the same time (black bars). CHART assay was performed using primer sets for promoter regions −I, −II and −VIII. The LPS did not influence accessibility. Mean calculated accessibilities and standard errors for n = 8 independent experiments are shown. (**P < 0·01).
The number of CD14+ cells after VD3 differentiation was 50% so CD14+ cells were enriched by magnetic positive cell selection to exclude the possibility that a higher content of CD14+ cells results in higher accessibilities. The IL-1β promoter structure of the enriched cells was compared with that of not enriched but VD3-differentiated cells. There were no significant differences in accessibilities in regions −I and −II when compared with undifferentiated cells (Fig. S1), suggesting that chromatin remodelling occurs earlier in differentiation to monocytes than expression of CD14.
To test the effect of stimulation on the accessibility of the IL-1β promoter, cells were incubated with LPS for 3 hr. Accessibility did not change upon LPS stimulation in undifferentiated and differentiated cells (Fig. 2). Taken together, these data confirm that the IL-1β promoter of monocytic cells is packaged in a poised chromatin architecture independent of the transcriptional status. In contrast, promyeloid cells have a MNase inaccessible chromatin most likely responsible for IL-1β gene suppression in a premature stage.
Accessibility of IL-1β promoter regions −I and −II of differentiated HL-60 cells is comparable with primary blood monocytes
To exclude the possibility that the structure of the IL-1β promoter is a characteristic of a single cell line, we analysed the structure of the IL-1β promoter in another human cell line, THP-1. We also compared the chromatin structure of the differentiated HL-60 cells and THP-1 cells with that of primary blood monocytes to endorse our results for primary blood cells. Therefore, by analysing monocytes from eight healthy donors using the CHART assay, it was observed that the IL-1β promoter regions −I and −II MNase accessibility of VD3-treated HL-60 cells was similar to those detected in monocytic THP-1 cells and primary monocytes (Fig. 3). This confirmed that the differentiated HL-60 cells reflect the monocyte-specific chromatin fine structure in regions −I and −II. Interestingly, region −VIII was not accessible in HL-60 and THP-1 cells whereas the accessibility in monocytes was 57% (Fig. 3). The difference in the accessibility of region −VIII between primary monocytes compared with HL-60 and THP-1 cells is probably the result of the genetic instability of the leukaemic cell lines.
Figure 3.
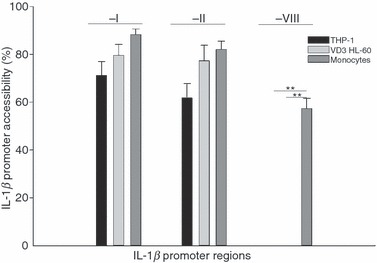
Primary human monocytes display equal accessibilities of the interleukin-1β (IL-1β) promoter as differentiated HL-60 cells and monocytic THP-1 cells. Primary human monocytes, differentiated HL-60 cells and THP-1 cells were analysed as described in Fig. 2. No significant differences could be detected comparing accessibilities of regions −I and −II, whereas region −VIII was more accessible in primary monocytes than in differentiated HL-60 and THP-1 cells (**P < 0·01). Means and standard error of n = 7 independent experiments are shown.
IL-1β expression is regulated by methylation
Demethylation is often described in context with gene activation.6,9,19 In addition, Kovacs et al.9 described methylation as playing a role in the regulation of pro-inflammatory cytokine expression in U937 cells. Therefore, the influence of the demethylase inhibitor AZA alone and in combination with LPS stimulation on IL-1β expression was compared in undifferentiated and differentiated HL-60 cells. We could not detect IL-1β messenger RNA (mRNA) in unstimulated, undifferentiated HL-60 cells before AZA treatment but in unstimulated, VD3-differentiated HL-60 cells (Fig. 4a). Treatment with AZA for 48 hr led to IL-1β transcription in undifferentiated and differentiated HL-60 cells. Pre-incubation with AZA followed by LPS stimulation also elevated IL-1β mRNA levels (Fig. 4a). The effects of AZA in combination with or without LPS on IL-1β mRNA expression were confirmed at the protein level in ELISA and Western blot (Fig. 4b, c). The highest levels of IL-1β expression were detected in differentiated HL-60 cells pre-incubated with AZA and additionally stimulated with LPS. The influence of AZA alone and in combination with LPS on the accessibility of the IL-1β promoter was also tested, but no changes could be detected in CHART assays (data not shown). Using the histone deacetylase inhibitor trichostatin A (TSA) no increase in IL-1β expression was detected in differentiated and undifferentiated HL-60 cells after treatment for 24 hr. Nor did the combination of AZA and TSA show any differences compared with the effects of one of the agents alone (data not shown), suggesting that acetylation is not involved in IL-1β transcription. These data suggest that demethylation acts independently from the IL-1β promoter structure or differentiation status on IL-1β regulation.
Figure 4.
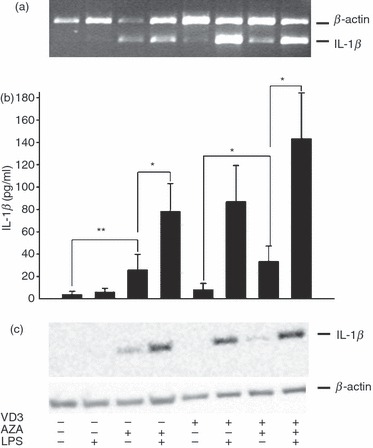
Influence of demethylation of DNA on interleukin-1β (IL-1β) expression of differentiated versus undifferentiated HL-60 cells. Undifferentiated and vitamin D3 (VD3) -differentiated HL-60 cells showed significant increase in IL-1β expression after pre-incubation with 5-aza-2′-deoxycytidine (AZA), additionally increased by lipopolysaccharide (LPS) stimulation. Differentiated or undifferentiated HL-60 cells (5 × 105 cells) were pre-incubated with AZA for 48 hr. For messenger RNA analysis (a, one representative example of n = 3) 5 × 105 cells were stimulated with LPS (250 ng/ml) for 4 hr; IL-1β transcription was determined using polymerase chain reaction with IL-1β and β-actin primer pair sets. The IL-1β concentrations in supernatants and lysates of 2 × 106 cells/ml after 24 hr with or without LPS stimulation were measured by enzyme-linked immunosorbent assay, (b, mean and standard error of n = 3 experiments are shown, *P < 0·05, **P < 0·01) and Western blot analysis (c, one representative example of n = 3), respectively.
Discussion
Our data demonstrate that the poised structure of the IL-1β promoter is established late in monopoiesis and is a feature of monocytes but not of the entire myeloid lineage. We could also show that the suppression of IL-1β expression via methylation in contrast to chromatin accessibility is independent of the developmental stage of the myeloid cells.
The observation that differentiation of promyeloid HL-60 cells to monocytic cells led to the expression of the pro-inflammatory cytokines IL-1β, IL-6 and TNF-α (Fig. 1) is in concordance with the results obtained from human CD34+ progenitors from umbilical cord blood, which have also been enabled to produce IL-1β and IL-6 after differentiation to monocytes.15 Hence, the events detected in primary cells during monopoiesis are applicable to the HL-60 differentiation model.
Analyses of the IL-1β promoter chromatin structure showed that two regions near the transcription start (− 199 to − 17) are significantly more accessible in differentiated HL-60 cells and in primary monocytes compared with undifferentiated promyeloid HL-60 cells. A relatively inaccessible structure has also been described in human B cells and murine B-cell precursors as well as in murine T cells.5,7 These observations on lymphoid cells and the inaccessible promoter structure of promyeloid cells detected in our experiments most likely explain that B cells and undifferentiated HL-60 cells do not produce significant amounts of IL-1β. Liang et al.5 have postulated that the poised promoter architecture is lineage-specific. Based on the detection of IL-1β producing pre-activated cultured daughter cells from haematopoietic stem cells,20 this group assumed the generation of the package of the IL-1β promoter into an accessible structure very early on the hematopoietic precursor cell level. In contrast, our data in concert with recent investigations of CD34+ progenitor cells that do not produce IL-1β after stimulation15 strongly indicate that the highly accessible IL-1β promoter is actively generated during monocytic differentiation and is not a permanent feature in myeloid precursors. In addition, the generation of the accessible IL-1β promoter appears to be an earlier event than induction of the monocyte marker CD14 during monocytic differentiation, as shown by comparing enriched differentiated CD14+ and not enriched but differentiated HL-60 cells (Fig. S1).21
Interestingly, the accessibility of the IL-1β promoter in promyeloid and monocytic differentiated HL-60 cells is not modified upon cellular stimulation (Fig. 2) confirming the unique status of the IL-1β promoter among IL-4, IL-10, TNF-α, interferon-β and interferon-γ promoters,5,7,22 in which chromatin is rapidly reorganized after stimulation.
Surprisingly, we found a significantly higher IL-1β promoter accessibility of region −VIII in primary monocytes in contrast to undifferentiated as well as differentiated HL-60 and THP-1 cells (Fig. 3). Putative transcription-factor-binding motifs of MyoD, GATA-1 and CP2 surrounding the −VIII region were detected, based on results using tfsearch software (available online at http://www.cbrc.jp/research/db/TFSEARCH.html). These transcription factors are involved in myogenesis, normal erythroid, megakaryocytic and progenitor cell maturation.23,24 To what extent these binding motifs are involved in the IL-1β expression of mature monocytes or whether they are a feature of acute myeloid leukaemia cells must be elucidated in future studies.
A key candidate for the induction of IL-1β transcription as well as for the establishment of the poised chromatin structure is the transcription factor PU.1 (Spi-1).25 It has been reported that PU.1 is constitutively bound to the IL-1β promoter, either in association with interferon regulatory factor 8 or C/EBPβ before stimulation or C/EBPβ binding after stimulation.5,7,26 Grondin et al.27 found the proximal promoter bound by PU.1 and C/EBPβ only after monocytic differentiation of the erythro-monocytic cell line TF-1. In context with our results, one can assume that the IL-1β promoter opens during monopoiesis facilitating constitutive binding of mainly PU.1 and C/EBPβ. This triggers the establishment of the poised structure and activating factors post-translationally altered upon stimulation enable the fast secretion of huge amounts of pro-inflammatory IL-1β by mature monocytes. Chromatin remodelling and transcription-factor-binding to the IL-1β promoter may be independent processes controlled within monopoiesis resulting in the poised architecture as a prerequisite for rapid IL-1β expression in mature monocytes.
As LPS stimulation did not modify the chromatin structure within the IL-1β promoter, the impact of methylation on IL-1β gene regulation was examined. Demethylation induced IL-1β expression in promyeloid cells and resulted in enhanced expression in differentiated HL-60 cells (Fig. 4). Kovacs et al.9 found induced IL-1β expression in U937 cells and enhanced IL-1β mRNA levels in THP-1 cells, although in their experiments the presence of calcium ionophores was crucial, because AZA treatment alone could not induce IL-1β expression in unstimulated cells. This might be because of the usage of different cell lines, a different, less sensitive IL-1β detection assay (D10.G4.1 mitogenic assay) and diverse stimulation conditions (48 hr instead of 4 hr). The decrease of responsiveness in this time period might result from remethylation of formerly demethylated sites. Additionally, not responding cells might overgrow AZA-responsive cells during long-term stimulation, because AZA inhibits cell proliferation.
However, we could show that demethylation influences IL-1β expression independent of the developmental status of the cells by detecting a high response to AZA before and after differentiating promyeloid HL-60 into monocytic cells. Many reports showed that AZA treatment led to the reactivation of a variety of genes, but did not result in a non-specific up-regulation of all genes, because glyceraldehyde 3-phosphate dehydrogenase (GAPDH), β-actin (Fig. 4a and data not shown) and IL-1α9 expression remain unchanged. The IL-1β gene28,29 and also other genes without CpG islands can be induced by AZA, suggesting promoter-independent effects of AZA.30,31 This raises the possibility that AZA treatment led to the reactivation of genes involved in IL-1β transcription or to the demethylation of upstream regulators that control IL-1β production or mRNA stability. DNA-incorporated AZA not only binds and inactivates DNA-methyltransferases but can also act as an inhibitor of the methylation of histones.32,33 Therefore, histone methylation cannot be ruled out to affect the establishment of the poised structure during monopoiesis and IL-1β transcription. Experiments using the histone-deacetylase inhibitor TSA revealed that IL-1β transcription is independent of acetylation in differentiated and undifferentiated HL-60 cells. This is in line with data from Liang et al.5 who observed that IL-1β promoter regions −III to −I are not packaged by hyper-acetylated histone 3 in monocytic Mono Mac 6 cells. Sato et al.10 reported an increased IL-1β expression in choriodecidual tissue after TSA treatment, which might be the result of different regulation of pro-inflammatory cytokines in haematological and non-haematological cells.
In summary, the results for the differentiation model can be transferred to the processes taking place during monopoiesis. Our data strongly suggest that the closed, inaccessible form of the IL-1β promoter of promyeloid cells is remodelled into a poised structure during differentiation, enabling differentiated monocytic cells to rapidly secrete IL-1β after stimulation. Furthermore, the poised structure of the IL-1β promoter is cell-specific for monocytes but not for the entire myeloid lineage. Additionally, we found that demethylation resulted in IL-1β expression, probably by activation of IL-1β regulating genes. Methylation in contrast to chromatin remodelling appears to regulate IL-1β expression independently of the differentiation status of the cell.
However, our data indicate that other cells of the myeloid lineage might display distinctive capabilities based on different promoter accessibilities, for example of pro-inflammatory genes. Analysis of promoter accessibilities might offer an additional diagnostic tool for the classification of acute myeloid leukaemia.
Disclosures
The authors declare no conflict of interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Enriched CD14+ VD3differentiated HL-60 cells do not display any differences inIL-1β promoter accessibility in comparison to VD3 differentiated HL-60 cells.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Dinarello CA. Interleukin-1 and interleukin-1 antagonism. Blood. 1991;77:1627–52. [PubMed] [Google Scholar]
- 2.Shirakawa F, Saito K, Bonagura CA, Galson DL, Fenton MJ, Webb AC, Auron PE. The human prointerleukin 1β gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol Cell Biol. 1993;13:1332–44. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krane SM, Conca W, Stephenson ML, Amento EP, Goldring MB. Mechanisms of matrix degradation in rheumatoid arthritis. Ann N Y Acad Sci. 1990;580:340–54. doi: 10.1111/j.1749-6632.1990.tb17943.x. [DOI] [PubMed] [Google Scholar]
- 4.Dinarello CA. IL1 family and inflammatory disease. Clin Exp Rheumatol. 2002;20(Suppl. 27):S1–13. [PubMed] [Google Scholar]
- 5.Liang MD, Zhang Y, McDevit D, Marecki S, Nikolajczyk S. The Interleukin-1β gene is transcribed from a poised promoter architecture in monocytes. J Biol Chem. 2006;281:9227–37. doi: 10.1074/jbc.M510700200. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Hara E, Tenen DG, Hozumi N, Oikawa T. Site-specific DNA methylation of PU.1 and Dnmt3a/b. Oncogene. 2005;25:2477–88. doi: 10.1038/sj.onc.1209272. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Saccani S, Shin H, Nikolajczyk BS. Dynamic protein associations define two phases of IL-1β transcriptional activation. J Immunol. 2008;181:503–12. doi: 10.4049/jimmunol.181.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukada J, Saito K, Waterman WR, Webb AC, Auron PE. Transcription factors NF-IL6 and CREB recognize a common essential site in the human prointerleukin 1β gene. Mol Cell Biol. 1994;14:7285–97. doi: 10.1128/mcb.14.11.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacs EJ, Oppenheim JJ, Carter DB, Young HA. Enhanced Interleukin-1 production by human monocyte cell lines following treatment with 5-Azacytidine. J Leukoc Biol. 1987;41:40–6. doi: 10.1002/jlb.41.1.40. [DOI] [PubMed] [Google Scholar]
- 10.Sato TA, Mitchel MD. Molecular inhibition of histone deacetylation results in major enhancement of the production of IL-1β in response to LPS. Am J Physiol Endocrinol Metab. 2006;290:490–3. doi: 10.1152/ajpendo.00406.2005. [DOI] [PubMed] [Google Scholar]
- 11.Collins SJ. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. J Am Soc Hematol. 1987;70:1233–44. [PubMed] [Google Scholar]
- 12.Tagliafico E, Tenedini E, Bergamaschi A, et al. Gene expression profile of Vitamin D3 treated HL60 cells shows an incomplete molecular phenotypic conversion to monocytes. Cell Death Diff. 2002;9:1185–95. doi: 10.1038/sj.cdd.4401104. [DOI] [PubMed] [Google Scholar]
- 13.Rola-Pleszczynski M, Stankova J. Differentiation-dependent modulation of TNF production by PAF in human HL-60 myeloid leukemia cells. J Leukoc Biol. 1992;51:609–16. doi: 10.1002/jlb.51.6.609. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Wang Y, Gao L, et al. Expression of Toll-like receptors 2 and 4 and CD14 during differentiation of HL-60 cells induced by phorbol 12-myristate 13-acetate and 1α, 25-dihydroxy-vitamin D3. Cell Growth Differ. 2002;13:27–38. [PubMed] [Google Scholar]
- 15.Schröder AK, von der Ohe M, Kolling U, et al. Polymorphonuclear leucocytes selectively produce anti-inflammatory interleukin-1 receptor antagonist and chemokines, but fail to produce pro-inflammatory mediators. Immunology. 2006;119:317–27. doi: 10.1111/j.1365-2567.2006.02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altstaedt J, Kirchner H, Rink L. Cytokine production of neutrophils is limited to interleukin-8. Immunology. 1996;81:563–8. doi: 10.1046/j.1365-2567.1996.d01-784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haase H, Ober-Blöbaum JL, Engelhardt G, Hebel S, Heit A, Heine H, Rink L. Zinc signals are essential for lipopolysaccharide-induced signal transduction in monocytes. Immunology. 2008;181:6491–502. doi: 10.4049/jimmunol.181.9.6491. [DOI] [PubMed] [Google Scholar]
- 18.Friedmann AD, Keefer JR, Kummalue T, Liu H, Wang Q, Cleaves R. Regulation of granulocyte and monocyte differentiation by CCAAT/enhancer binding protein. Blood Cells Mol Dis. 2003;31:338–41. doi: 10.1016/s1079-9796(03)00135-9. [DOI] [PubMed] [Google Scholar]
- 19.Murumägi A, Vähämurto P, Peterson P. Characterization of regulatory elements and methylation pattern of the Autoimmune Regulator (AIRE) Promoter. J Biol Chem. 2003;278:19784–90. doi: 10.1074/jbc.M210437200. [DOI] [PubMed] [Google Scholar]
- 20.Watari K, Mayani H, Lee F, Dragowska W, Lansdorp PM, Schrader JW. Production of interleukin 1β by human hematopoietic progenitor cells. J Clin Invest. 1996;97:1666–74. doi: 10.1172/JCI118593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White SL, Belov L, Barber N, Hodgkin PD, Christopherson RI. Immunophenotypic changes induced on human HL60 leukaemia cells by 1α,25-dihydroxyvitamin D3 and 12-O-tetradecanoylphorbol-13 acetate. Leuk Res. 2005;29:1141–51. doi: 10.1016/j.leukres.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Fields PE, Kim ST, Flavell RA. Changes in histone acetylation at the IL-4 and IFN-γ loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 23.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Bosè F, Fugazza C, Casalgrandi M, et al. Functional interaction of CP2 with GATA-1 in the regulation of erythroid promoters. Mol Cell Biol. 2006;26:3942–54. doi: 10.1128/MCB.26.10.3942-3954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toda Y, Tsukada J, Misago M, Kominato Y, Auron PE, Tanaka Y. Autocrine induction of the human pro-IL-1β gene promoter by IL-1β in monocytes. J Immunol. 2002;168:1984–91. doi: 10.4049/jimmunol.168.4.1984. [DOI] [PubMed] [Google Scholar]
- 26.Unlu S, Kumar A, Waterman WR, Tsukada J, Wang KZQ, Galson DL, Auron PE. Phosphorylation of IRF8 in a pre-associated complex with Spi-1/PU.1 and non-phosphorylated Stat1 is critical for LPS induction of the IL-1β gene. Mol Immunol. 2007;13:3364–79. doi: 10.1016/j.molimm.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grondin B, Lefrancois M, Tremblay M, et al. c-Jun Homodimers can function as a context-specific coactivator. Mol Cell Biol. 2007;27:2919–33. doi: 10.1128/MCB.00936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicklin MJ, Barton JL, Nguyen M, FitzGerald MG, Duff GW, Kornman K. A sequence-based map of the nine genes of the human interleukin-1 cluster. Genomics. 2002;79:718–25. doi: 10.1006/geno.2002.6751. [DOI] [PubMed] [Google Scholar]
- 29.Yakabe S, Soejima H, Yatsuki H, et al. MeCP2 knockdown reveals DNA methylation-independent gene repression of target genes in living cells and a bias in the cellular location of target gene products. Genes Genet Syst. 2008;83:199–208. doi: 10.1266/ggs.83.199. [DOI] [PubMed] [Google Scholar]
- 30.Zhu WG, Dai Z, Ding H, et al. Increased expression of unmethylated CDKN2D by 5-aza-2′-deoxycytidine in human lung cancer cells. Oncogene. 2001;20:7787–96. doi: 10.1038/sj.onc.1204970. [DOI] [PubMed] [Google Scholar]
- 31.Liang G, Gonzalez FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:961–6. [PubMed] [Google Scholar]
- 32.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–15. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JCY, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2-deoxycytidine. Cancer Res. 2002;62:6456–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


