Abstract
Rationale: The NF-E2 related factor 2 (Nrf2)–antioxidant response element (ARE) pathway is essential for protection against oxidative injury and inflammation including hyperoxia-induced acute lung injury. Microarray expression profiling revealed that lung peroxisome proliferator activated receptor γ (PPARγ) induction is suppressed in hyperoxia-susceptible Nrf2-deficient (Nrf2−/−) mice compared with wild-type (Nrf2+/+) mice. PPARγ has pleiotropic beneficial effects including antiinflammation in multiple tissues.
Objectives: We tested the hypothesis that PPARγ is an important determinant of pulmonary responsivity to hyperoxia regulated by Nrf2.
Methods: A computational bioinformatic method was applied to screen potential AREs in the Pparg promoter for Nrf2 binding. The functional role of a potential ARE was investigated by in vitro promoter analysis. A role for PPARγ in hyperoxia-induced acute lung injury was determined by temporal silencing of PPARγ via intranasal delivery of PPARγ-specific interference RNA and by administration of a PPARγ ligand 15-deoxy-Δ12,14-prostaglandin J2 in mice.
Measurements and Main Results: Deletion or site-directed mutagenesis of a potential ARE spanning -784/-764 sequence significantly attenuated hyperoxia-increased Pparg promoter activity in airway epithelial cells overexpressing Nrf2, indicating that the -784/-764 ARE is critical for Nrf2-regulated PPARγ expression. Mice with decreased lung PPARγ by specific interference RNA treatment had significantly augmented hyperoxia-induced pulmonary inflammation and injury. 15 Deoxy-Δ12,14-prostaglandin J2 administration significantly reduced hyperoxia-induced lung inflammation and edema in Nrf2+/+, but not in Nrf2−/− mice.
Conclusions: Results indicate for the first time that Nrf2-driven PPARγ induction has an essential protective role in pulmonary oxidant injury. Our observations provide new insights into the therapeutic potential of PPARγ in airway oxidative inflammatory disorders.
Keywords: antioxidant response element, hyperoxia, inflammation, siRNA, 15d-PGJ2
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Peroxisome proliferator activated receptor γ (PPARγ) is known to act not only on adipogenesis and glucose/lipid homeostasis but also on antiinflammatory responses. Results demonstrate that Nrf2-induced PPARγ plays an essential protective role during the pathogenesis of pulmonary inflammation and oxidative stress in acute lung injury.
What This Study Adds to the Field
Bioinformatic determination of a functional antioxidant response elements and potential antioxidant response elements suggested mutual feedback regulation of Nrf2-PPARγ is a molecular mechanism of cytoprotection in Pparg and suggested mutual feedback regulation of Nrf2-PPARγ will provide keys for molecular mechanisms of cytoprotection.
The lung is at high risk for oxidative stress as it interfaces with various airborne oxidants including air pollutants and high concentrations of oxygen (O2) when used as combined therapy. Reactive oxygen species have been implicated in the pathogenesis of many acute and chronic pulmonary disorders including adult respiratory distress syndrome (ARDS), bronchopulmonary dysplasia, emphysema, idiopathic pulmonary fibrosis, and cancer (1–3). ARDS is a severe form of acute lung injury (ALI) and is a major lung disease affecting millions worldwide. ARDS is characterized by increased permeability and inflammation accompanying abnormal gas exchange and variable late-phase responses including pulmonary fibrosis (4). Hyperoxia (>95% O2) exposure to laboratory rodents induces pulmonary damage that resembles ARDS subphenotypes, and has been widely used to investigate the molecular basis of oxidative lung injury.
Nrf2 is a transcription factor that induces antioxidant and defense gene expression through binding to cis-acting antioxidant response elements (AREs) (5), and is essential in tissue protection from various oxidants and xenobiotics (6). Using mice genetically deficient in Nrf2 (Nrf2−/−), a protective role for the Nrf2-ARE pathway has been demonstrated in oxidant-mediated inflammatory lung injury including hyperoxia toxicity (7–16). To elucidate molecular mechanisms underlying Nrf2-mediated pulmonary protection against O2, we profiled Nrf2-dependent genes regulated during the development of hyperoxia-induced lung injury in Nrf2+/+ and Nrf2−/− mice (17). In addition to the predicted ARE-bearing antioxidant and cytoprotective genes (e.g., Nqo1, Gstp, Txnrd1, Ex, and Cp-2), several novel genes were defined to be Nrf2-dependent (17). They included Pparg (or Nr1C3), which encodes peroxisome proliferator activated receptor γ (PPARγ).
PPARs belong to a nuclear hormone receptor super family, and are classified into isoforms α, β (or δ), and γ. PPARs regulate ligand-specific gene transcription through PPAR response element (PPRE) binding after forming a heterodimeric complex with a retinoid X receptor (RXR) member. Pleiotropic effects of PPARs include lipid and lipoprotein metabolism and adipogenesis, glucose homeostasis, cell cycle regulation, and cellular proliferation and differentiation.
PPARγ has been found in various cells of the immune system, endothelium, epithelial cells, fibroblasts, glial cells, and smooth muscle cells (18–21). Recent in vivo and in vitro studies defined a novel role of PPARγ-RXRα in the pathogenesis of neurologic disorders, cancers, and inflammation and immune diseases (22–25). Use of specific PPARγ ligands or agonists, such as endogenous 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) and exogenous thiazolidinediones (rosiglitazone and pioglitazone), has suggested PPARγ may have a beneficial effect in critical pulmonary disorders including asthma, ALI, chronic obstructive pulmonary disorders, fibrosis, bronchopulmonary dysplasia, and lung cancer (20, 26–28). Interestingly, Nrf2 has also been implicated in animal models of these diseases (7–15).
The present study was designed to characterize the mechanisms through which Nrf2 regulates PPARγ, and to determine the function of PPARγ in the pathogenesis of hyperoxia-induced lung injury in mice. Results of this study demonstrate an essential protective role for Nrf2-driven PPARγ against ALI. Some of the results of this study have been previously reported in an abstract (29).
METHODS
Animals
Nrf2+/+ and Nrf2−/− (ICR background) mice (5) were produced from breeding colonies at the National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health, Research Triangle Park, North Carolina. Mice were provided food (modified AIN-76A) and water ad libitum. Male mice (5–7 wk old) were used for all experiments.
In Vivo PPARγ Specific Interference RNA Treatment
Specific interference RNA (siRNA) sequences for PPARγ were selected based on a previous publication (30). The sense and antisense strands of siRNA (si-PPARγ) were 5′-GAC AUG AAU UCC UUA AUG AUU-3′ and 5′-UCA UUA AGG AAU UCA UGU CUU-3′, respectively, spanning 880–899 bp of mouse Pparg (GI:187960102). si-PPARγ was synthesized in 2′-deprotected, duplex, desalted, and purified form by Dharmacon Research, Inc. (Lafayette, CO). Nrf2+/+ mice were anesthetized with isofluorane and si-PPARγ (2 mg/kg, in 50-μl phosphate-buffered saline for 25 g) or equivalent dose and volume of nonspecific control siRNA duplex (si-NS) (nontargeting siRNA #2, Dharmacon) was administered intranasally (31). Intranasal instillation was done daily for 3 consecutive days (1 d before, day 0, and at 1 d after hyperoxia or air exposure).
Administration of a PPARγ Agonist 15d-PGJ2
Vehicle (phosphate-buffered saline with 4% dimethyl sulfoxide) or 15d-PGJ2 (30 μg/kg, EMD Chemicals, Inc., Gibbstown, NJ) in vehicle was administered intraperitoneally (100 μl) in Nrf2+/+ and Nrf2−/− mice daily starting 1 day before inhalation exposure. The dose was chosen based on previous publications in which PPARγ was efficiently induced in the lung (32, 33). The last injection was done 24 hours before the end of the designated inhalation exposure period.
Inhalation Exposure
Mice were placed individually in a Hazleton M60 battery inside a Hazleton 1000 chamber (Lab Products, Maywood, NJ). After acclimation in the chambers for 24 hours per day for 2–3 days, mice were exposed to more than 95% O2 (UHP grade, Min. purity 99.994% O2 tanks; National Welders, Durham, NC) 24 hours per day for 48 or 72 hours. Water and feed (modified AIN76/A) were provided ad libitum during the exposure. The temperature (72°F ± 3°F) and humidity (50 ± 15%) of the chambers were monitored. Mice for room-air exposure were placed in a Hazleton M60 battery placed on a countertop with food and water provided ad libitum for the exposure duration. The chambers were opened for approximately 30 minutes at the same time each morning to allow animal health checks (morbidity and mortality), water and feed checks, and excreta paper change. Immediately after the end of each exposure, mice were killed by sodium pentobarbital overdose (104 mg/kg). All animal use was approved by the NIEHS Animal Care and Use Committee.
Bronchoalveolar Lavage Analysis
The right lung of each mouse was lavaged in situ four consecutive times with Hanks' balanced salt solution (0.5 ml/25 g body weight) and analyzed for total protein content (a marker of lung permeability) and differential cell count (markers of lung cellular inflammation and epithelial cell exfoliation), as described previously (7).
Lung Histopathology
The left lung from each mouse (n = 3/group) was perfused intratracheally in situ with zinc formalin, removed from the mouse, and fixed at a constant intraairway pressure of 25 cm fixative for 30 minutes The trachea was ligated and the inflated lung lobes were immersed in the same fixative for 1 to 3 days. The fixed left lung lobe was processed to be stained with hematoxylin and eosin (H&E) for histopathologic analysis. Immunohistological staining was done using an anti-PPARγ antibody (sc-7273X; Santa Cruz Biotechnology, Inc., Santa Cruz, CA).
Bioinformatic Analysis of the Pparg Promoter for Putative ARE Screening
Potential ARE sequences containing RTKAYnnnGCR for Nrf2 binding were determined in the 10 kb 5′ upstream region of murine Pparg (GI:83029857) using a position weight matrix (PWM) statistical model that was constructed based on a set of functional ARE sequences curated from published experimental studies (34).
Terminal Deoxynucleotidyl Transferase-mediated dUTP Nick-End Labeling Assay
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was performed on lung tissue sections to determine the cellular DNA fragmentation in situ as a parameter of apoptotic cell death using DeadEnd Colorimetric TUNEL System (Promega, Madison, WI) according to the recommendations of the manufacturer. Briefly, deparaffinized lung sections were fixed in 4% paraformaldehyde, permeabilized with proteinase K, and incubated with biotinylated nucleotide mix (including terminal deoxynucleotidyl transferase) to incorporate biotinylated nucleotide at the fragmented 3′-OH DNA ends. After immunohistochemical procedures, TUNEL-positive nuclei were visualized under a light microscope. Negative control samples that were incubated without terminal deoxynucleotidyl transferase and a positive control sample that was treated with DNase I before incubation with biotinylated nucleotide mix were prepared for references.
Reverse Transcription Polymerase Chain Reaction
Total lung RNA was isolated from homogenates using RNeasy Mini Kit (Qiagen Inc., Valencia, CA). One μg of total lung RNA was reverse transcribed into cDNA in a 50-μl volume (35). For semiquantitative polymerase chain reaction (PCR), cDNA equivalent to 50 ng of RNA was amplified in a 25-μl reaction containing 240 nM of forward and reverse primers specific for mouse HO-1 (7), CD36 (5′-GCTTATTGGGAAGACAATCAA-3′, 5′-GCAAATGTCAGAGGAAAAGAA-3′), Nrf2 (7), NQO1 (7), GST-Ya (7), and 18s rRNA (5′-TACCTGGTTGATCCTGCCAG-3′, 5′-CCGTCGGCATGTATTAGCTC-3′) in a Gene Amp PCR System 9700 (Applied Biosystems, Foster City, CA), and expression levels were quantified with 18s ribosomal RNA as an internal control following the procedures described elsewhere (35).
Lung Protein Isolation
Total lung proteins were prepared from lung homogenates in radioimmunoprecipitation buffer including phenylmethanesulfonyl fluoride (10 μg/ml) and protease/phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) using a polytron homogenizer. Nuclear proteins were prepared from pulverized lung tissues following procedures described previously (36). Proteins were quantified and stored in aliquots at −70°C.
Electrophoretic Mobility Shift Assay
Nuclear DNA binding activities were determined by gel shift analyses of nuclear protein aliquots (3–5 μg) on 3 × 104 cpm (γ32P) ATP end-labeled double-stranded oligonucleotides containing a consensus binding sequence (5′-AGGTCAAAGGTCA-3′) for PPAR/RXR consensus binding sequence (PPRE or direct repeat, sc-2587; Santa Cruz Biotechnology, Santa Cruz, CA), mutant PPRE oligonucleotide (sc-2588; Santa Cruz Biotechnology), mouse Nrf2 promoter-931 PPRE-like sequence (5′-TAATGCTGGTCAAAGGCCAAGGGGT-3′), ARE consensus sequence (35), -784 wild-type (Wt) mouse Pparg-ARE (5′-TCATTGTGACATAGCACTTAT-3′), or -784 mutant (Mt) mouse Pparg-ARE (5′-TCATTGGTACATAGCACTTAT-3′) following the procedure described previously (7). Specific binding activity was determined by preincubation of nuclear proteins with anti-Nrf2 antisera (8) or anti-RXRα (sc-553X, Santa Cruz Biotechnology) for 2 hours in ice followed by electrophoretic mobility shift assay. The gel was autoradiographed using an intensifying screen at −70°C, and bands were scanned using a Bio-Rad Gel Doc 2000 system (Bio-Rad Laboratories, Hercules, CA). Representative band images from independent analyses (n = 3/group) are presented.
Transcription Factor ELISA
To analyze specific binding activity of nuclear p65 nuclear factor kappa B (NF-κB) proteins, transcription factor ELISA was performed using nuclear extracts (2 μg) following the manufacturer's instruction (NF-κB TransAM kit; Active Motif, Carlsbad, CA) and procedures described previously (37).
Western Blot Analyses
Proteins (20–100 μg) were separated on SDS-phage gels and analyzed by Western blotting using specific antibodies against Nrf2 (sc-722X; Santa Cruz Biotechnology); PPARγ (sc-723X, Santa Cruz Biotechnology); RXRα (sc-553X; Santa Cruz Biotechnology); PTEN (sc-7974, Santa Cruz Biotechnology); bax (sc-7480; Santa Cruz Biotechnology); cleaved caspase-8 (#9748; Cell Signaling Technology Inc., Danvers, MA); lamin B (sc-6217, Santa Cruz Biotechnology); or pan actin (sc-1615, Santa Cruz Biotechnology). Representative band images from multiple independent blotting (n = 3/group) are presented. Specific protein bands were scanned and, if applicable, quantified using a Bio-Rad Gel Doc 2000 System.
ELISA for Cytokine Measurement
The concentration of pulmonary proinflammatory cytokine IL-6 was determined in bronchoalveolar lavage (BAL) fluid (50 μl) using mouse-specific ELISA kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.
Plasmid Construction
The 5′-upstream region of Pparg (GI:83029857) was amplified by PCR amplification using oligonucleotide primers bearing restriction enzyme sites (SacI or NheI), 5′-GCGAGCTCCACTGAATTATATTAGGTCA-3′, 5′-GCGAGCTCGTTGCTATTGATAGATAAAC-3′, and 5′-GCGCTAGCGTTTTGTCTATGTCTTGCAA-3′, and Pparg-ARE segment (−1000 to +1), which contains a putative -784 ARE sequence (5′- TCATTGTGACATAGCACTTAT-3′, from -784 to -764), or Pparg-ΔARE segment (-730 to +1), which has deletion of the -784 ARE, were generated. The promoter segments were processed for cloning into SacI and NheI sites of a pGL3-basic vector (Promega) containing a heterologous SV40 promoter and a firefly luciferase reporter gene using a Rapid DNA and Ligation Kit (Roche Applied Science, Indianapolis, IN) to create pPparg-ARE and pPparg-ΔARE. A mammalian overexpression vector for murine Nrf2 was generated by PCR amplification of the DBA/J mouse cDNA using oligonucleotide primers 5′-GGTACCCATGATGGACTTGGGAGTTGCCA-3′ and 5′-TCTAGACTCCATCCTCCCGAACCTAGT-3′. cDNA encoding Nrf2 were then cloned into the KpnI and XbaI sites of p3XFLAG-CMV (Sigma-Aldrich) for subsequent bacterial expression (pFlag-Nrf2). All plasmid sequences were confirmed by sequencing analysis performed in the NIEHS Sequencing Core facility.
Site-Directed Mutagenesis
A point mutation was introduced into the Pparg -784 ARE using a kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions to create mutated AREs (pPparg-AREmt; T-785A) in pPparg-ARE (or pPparg-AREwt). Oligonucleotide primers used for PCR amplification of 1,000 bp upstream containing the mutated ARE were 5′-GAGGGCTCCAATTTTTTCATTGAGACATAGCACTTATCACTTAAA-3′ (forward) and 5′-TTTAAGTGATAAGTGCTATGTCTCAATGAAAAAATTGGAGCCCTC-3′ (reverse). The mutations in the promoter sequences were confirmed after cloning.
Transfection, In Vitro Hyperoxia Exposure, and Reporter Gene Analyses
Transient transfection was performed in BEAS-2B cells (ATCC, Manassas, VA) with 60 to 70% confluency using Effectene Transfection Reagent (Qiagen) following standard procedures. In brief, cells were transfected with 100 ng of each vector, pPparg-ARE or pPparg-ΔARE together with 1 ng of Renilla luciferase plasmid (pRL-TK, Promega). In a separate experiment, pPparg-ARE or pPparg-ΔARE were cotransfected with 100 ng of Nrf2 overexpressing plasmid (pFlag-Nrf2). Immediately after the end of exposure to air or hyperoxia (85 ± 5%; 4, 6, or 12 h), aliquots of cell extracts were assayed for firefly and Renilla luciferase activities using a dual luciferase kit (Promega) with a microplate luminometer (Fluoroskan Ascent FL; Thermo Scientific Inc., Waltham, MA). Background firefly luciferase activity from mock-transfected cell extracts was subtracted from each experimental measurement. Firefly luciferase activity was then normalized to that of Renilla luciferase to correct for variation in transfection efficiency in each cell extract, and the normalized average activity for pPparg-ARE in the air exposure group in the absence of Nrf2 overexpression was assigned a value of 1. For the site-directed mutagenesis assay, pPparg-ARE (pPparg-AREwt) or pPparg-AREmt was transfected into BEAS-2B cells with or without pFlag-Nrf2. All transfections were performed in triplicate, and each experiment was repeated (n = 6–9/group).
Statistics
Data are expressed as the group mean ± SEM. Two-way analysis of variance was used for all the experimental data, and the Student-Newman-Keuls test was used for a posteriori comparisons of means (P < 0.05). All of the statistical analyses were performed using the SigmaStat 3.0 software program (SPSS Science Inc., Chicago, IL).
RESULTS
Differential Expression and Activation of Lung PPARγ in Nrf2+/+ and Nrf2−/− Mice
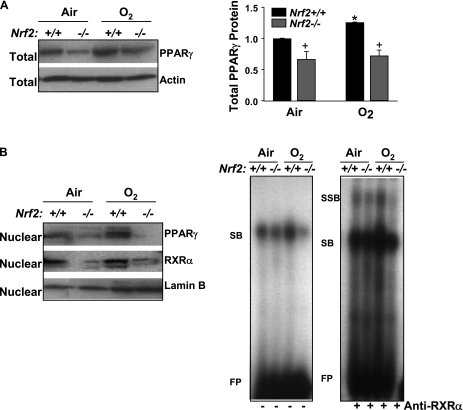
Expression of lung total PPARγ protein was significantly lower in Nrf2−/− mice than in Nrf2+/+ mice at baseline, and PPARγ level was increased (25%) in Nrf2+/+ mice after O2 (48 and 72 h, 48 h data shown), whereas no significant change was found in Nrf2−/− mice (Figure 1A). O2 also enhanced nuclear levels of PPARγ protein in Nrf2+/+ mice but not in Nrf2−/− mice. Nuclear protein level of RXRα that binds to PPRE with PPARγ as a dimer form was also suppressed in Nrf2−/− mice compared with Nrf2+/+ mice after air and O2 exposure (Figure 1B). Gel shift analysis of nuclear proteins determined increased total PPRE binding activity as indicated by shifted bands (SB) in the lungs of Nrf2+/+ mice after O2, whereas no O2-mediated change in the PPRE binding activity was found in Nrf2−/− mice (Figure 1B). Increased specific RXRα-PPRE binding activity was found in the lungs of Nrf2+/+ mice after hyperoxia as indicated by super shifted bands (SSB) (Figure 1B).
Figure 1.
Pulmonary peroxisome proliferator activated receptor γ (PPARγ) level and activity was suppressed in Nrf2-deficient mice. (A) Aliquots of lung total protein were subjected for Western blotting to determine differential protein levels of pulmonary PPARγ in Nrf2+/+ and Nrf2−/− mice after exposure to air and O2 (48 h). Representative images are presented, and group mean ± SEM (n = 3/group) of total PPARγ level normalized to air-exposed Nrf2+/+ mice is depicted in a graph. * = significantly higher than genotype-matched air controls (P < 0.05). + = significantly lower than exposure-matched Nrf2+/+ mice (P < 0.05). (B) Aliquots of lung nuclear extracts isolated from pieces of left lung tissue (n = 3 mice/group) were subjected for Western blotting to determine differential nuclear translocation of pulmonary PPARγ and retinoid X receptor α protein in Nrf2+/+ and Nrf2−/− mice after exposure to air and O2 (48 h). Representative images are presented (n = 3/group). Differential nuclear protein–PPAR response element (PPRE) binding activity in the lungs of Nrf2+/+ and Nrf2−/− mice after exposure to air and O2 (48 h) was determined by gel shift/supershift analysis. Aliquots of nuclear protein were incubated with an end-labeled oligonucleotide probe containing PPRE consensus sequence. Total PPRE binding (shifted bands, left panel) and specific retinoid X receptor α–PPRE binding (super shifted bands, right panel) was determined by gel shift analysis. FP = free probes; RXRα = retinoid X receptor α; SB = shifted bands; SSB = super shifted bands. Representative images from multiple analysis (n = 3/group) are presented.
Pulmonary Localization of PPARγ
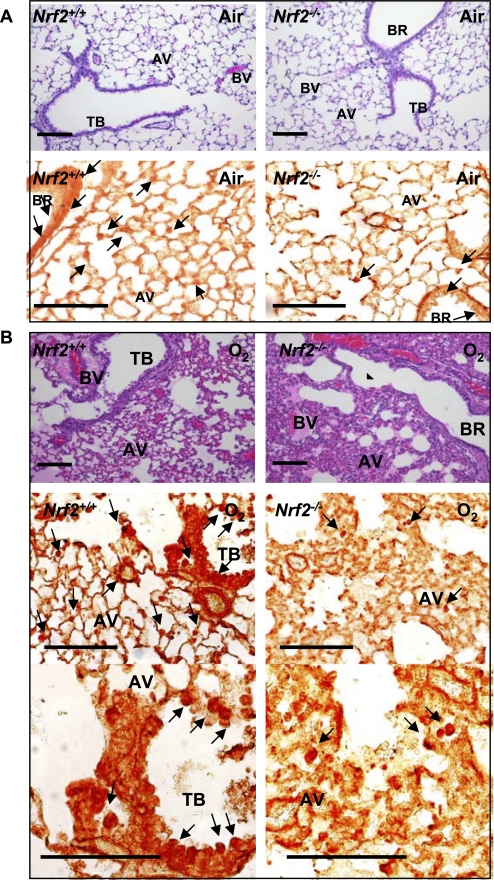
PPARγ was detected largely in alveolar macrophages and in epithelium lining conducting airways in air-exposed mice, and the populations of PPARγ-positive cells were smaller in Nrf2−/− mice than in Nrf2+/+ mice (Figure 2A). After hyperoxia exposure, PPARγ-bearing cells were markedly increased throughout the lungs of Nrf2+/+ mice. PPARγ was detected predominantly in infiltrating inflammatory cells, bronchial epithelial cells, endothelial cells, and smooth muscle cells of injured regions with perivascular and peribronchial edema in (Figure 2B). PPARγ-positive cells were infrequent in the lungs from Nrf2−/− compared with Nrf2+/+ mice after O2 (Figure 2B).
Figure 2.
Pulmonary cellular localization of peroxisome proliferator activated receptor γ (PPARγ). Pulmonary histopathology demonstrated by hematoxylin and eosin staining and PPARγ localization determined by immunohistochemical staining of paraffin-embedded lung sections after 48 hour exposure to air (A) or O2 (B). Hyperoxia-induced protein edema in peribronchiolar and perivascular regions and alveolar air space, epithelial proliferation, and inflammatory cell infiltration shown in hematoxylin and eosin stained sections (A, B, upper panels) was markedly greater in Nrf2−/− mice relative to Nrf2+/+ mice. Cellular PPARγ localized by immunohistochemical staining (A, B, bottom panels) using an anti-PPARγ antibody indicated inflammatory cells and bronchiolar epithelial cells (arrows) as the primary sources of PPARγ in the hyperoxia-injured lung. PPARγ-positive cells were more predominantly enhanced in Nrf2+/+ mice relative to Nrf2−/− mice after hyperoxia. Representative light photomicrographs are shown (n = 3/group). Higher magnification photomicrographs displayed fewer occurrences of PPARγ-bearing cells in Nrf2−/− mice relative to Nrf2+/+ mice after O2. AV = alveoli; BR = bronchi or bronchiole; BV = blood vessel; TB = terminal bronchioles. Bars indicate 100 μm.
Putative AREs Elucidated by Bioinformatic Analysis of the Pparg Promoter
To investigate whether Nrf2 directly or indirectly regulates PPARγ expression, potential ARE sequences for Nrf2 binding in the 5′ upstream region of Pparg were analyzed. We used a PWM statistical model (34) that was constructed based on a set of functional ARE sequences curated from published experimental studies. Theoretical PWM score is proportional to binding energy (38), and as the PWM value increases, so does the likelihood that the ARE is bona fide. Our computational method identified nine putative ARE or ARE-like sequences (Table 1) from 10 kb of the 5′-upstream promoter sequence of the Pparg (GI:83029857). Among nine putative AREs, the most proximal upstream sequence spanning -784 and -764 bp matched the ARE consensus sequence (RTKAYnnnGCR; R = A or G, K = G or T, Y = C or T, n = A, C, G, or T) with the highest PWM score (14.2). This PWM score is compatible with that of validated AREs in pulmonary Nrf2 target genes including Nqo1 (ch16:68318405–68318425, PWM 16.4), Txnrd1 (ch12:103183169–103183189, PWM 17.9), and Gclm (ch1:94087027–94087047, PWM 12.2) (34). This potential ARE (hereafter designated -784) was pursued for further functional analysis.
TABLE 1.
PUTATIVE ANTIOXIDANT RESPONSE ELEMENTS OR ANTIOXIDANT RESPONSE ELEMENT–LIKE SEQUENCES DETERMINED IN THE MOUSE PPARG PROMOTER BY BIOINFORMATIC ANALYSIS
| Location | Sequence | PWM* | MS† | Orientation |
|---|---|---|---|---|
| −9306/-9286 | tcaggGTGAGggaGCTagctt | 7.3 | 0.825 | FW‡ |
| −8688/-8668 | tctcaATAATacaGCAtctga | 9.0 | 0.774 | FW |
| −8211/-8191 | tgtgtTGCttgGTGATggctc | 6.8 | 0.788 | CF§ |
| −7660/-7640 | caaggGGCtatGTCATggtgg | 13.3 | 0.901 | CF |
| −6928/-6908 | tcaaaTTGACagaGAAacatg | 8.8 | 0.788 | FW |
| −6043/-6023 | tttgcTGCtgcATCACagtta | 12.1 | 0.869 | CF |
| −4638/-4618 | aagagAGCtatATCAGggtcc | 7.5 | 0.825 | CF |
| −2150/-2130 | atgaaATGAGtgaGCTacatt | 9.1 | 0.856 | FW |
| −784/-764 | tcattGTGACataGCActtat | 14.2 | 0.91 | FW |
Position weight matrix score.
Matrix similarity score.
Forward.
Complimentary forward.
Antioxidant response element core-like sequences (5′-RTKAYnnnGCR-3′; R = A or G, K = G or T, Y = C or T, n = A, C, G, or T) are underlined, and mismatch sequences relative to the core are italicized.
Functional Analyses of the -784 ARE In Vitro
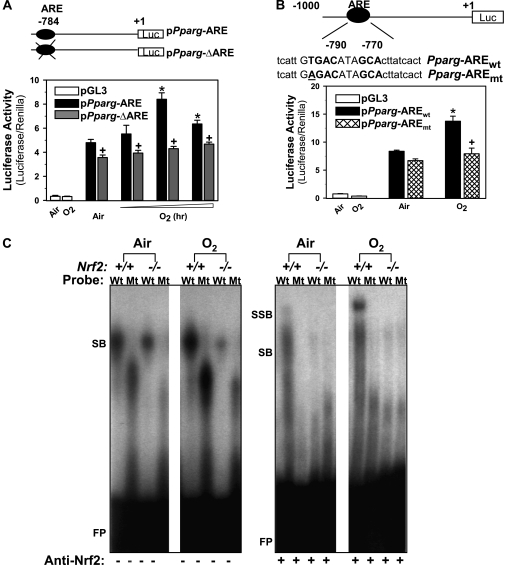
To determine functional relevance of the -784 ARE in regulation of Pparg expression, the 1-kb upstream region of the Pparg promoter bearing the -784 ARE sequence was cloned in a luciferase reporter vector (pPparg-ARE) and the promoter activity was analyzed in airway epithelial cells by transient transfections. A plasmid containing the Pparg promoter constructs with deletion of the -784 ARE region (pPparg-ΔARE) was also generated and used in transfections. In airway epithelial cells overexpressing Nrf2 by transfection with Flag-tagged Nrf2 expression vector (pFlag-Nrf2), hyperoxia exposure significantly enhanced the luciferase activity (at 6 or 12 h) after transfection with pPparg-ARE (Figure 3A). Promoter activity of cells transfected with pPparg-ΔARE was not significantly changed by O2, and was significantly lower after either air or O2 exposure relative to the pPparg-ARE transfection (Figure 3A). Further, site-directed mutagenesis analysis determined significantly lower luciferase activity in cells transfected with the vector containing a point mutation in the ARE (GAGACATAGC, pPparg-AREmt) than in cells transfected with the wild type ARE (GTGACATAGC, pPparg-AREwt) after O2 exposure (Figure 3B). Together, these results consistently demonstrated that Nrf2 binding on Pparg -784 ARE is critical for hyperoxia-induced PPARγ expression.
Figure 3.
Functional assessment of a putative antioxidant response elements (ARE) on -784 region of Pparg promoter. (A) Transactivating activity of -784 ARE sequence was determined by promoter deletion analysis. Nrf2 overexpressing airway epithelial cells (BEAS-2B) were prepared by transfection with murine Nrf2 expression vector (pFlag-Nrf2). The cells were then transfected by Pparg promoter-luciferase reporter constructs with (pPparg-ARE) or without (pPparg-ΔARE) -784 ARE region, and exposed to either air or hyperoxia (4–12 h). Mean ± SEM. presented (n = 6–9/group). * = significantly higher than air exposure under same transfection condition (P < 0.05). + = significantly lower than exposure-matched pPparg-ARE transfection (P < 0.05). (B) Transactivating activity of -784 ARE sequence was further determined by mutation analysis. Briefly, BEAS-2B cells overexpressing Nrf2 were transfected with a vector containing wild-type Pparg promoter bearing -784 ARE (pPparg-AREwt) or a vector containing the promoter with a mutation in -784 (pPparg-AREmt) incorporated by site-directed mutagenesis. The cells were then exposed to either air or hyperoxia (12 h). * = significantly higher than air exposure under the same transfection condition (P < 0.05). + = significantly lower than exposure-matched pPparg-AREwt (P < 0.05). (C) Gel shift analysis was performed on an aliquot of lung nuclear proteins from Nrf2+/+ and Nrf2−/− mice exposed to air or O2 (48 h) using γP32-end labeled wild-type (Wt; tcattGTGACataGCActtatcact) or mutated (Mt; tcattGGTACataGCActtatcact) -784 ARE-like probe. Hyperoxia increased Wt probe binding activity of nuclear proteins from Nrf2+/+, but not those from Nrf2−/− mice. Hyperoxia-induced increase of total DNA binding and specific Nrf2 binding (+ anti-Nrf2 antisera) was detected only on Wt probe by nuclear proteins from Nrf2+/+ mice. FP = free probes; SB = shifted bands; SSB = super shifted bands.
Functional Analyses of the -784 ARE In Vivo
Nrf2 binding to the -784 ARE was further analyzed by gel shift assays using lung nuclear proteins from Nrf2+/+ and Nrf2−/− mice. Total DNA binding activity (SB) of nuclear proteins on the Wt -784 ARE probe was higher at baseline (air) in Nrf2+/+ mice than in Nrf2−/− mice (Figure 3C). The Wt -784 ARE binding activity was increased by O2 in Nrf2+/+ mice but not in Nrf2−/− mice (Figure 3C). Total DNA binding activity of nuclear proteins on a mutated -784 ARE probe (Mt) demonstrated loss of DNA binding activity in Nrf2+/+ and Nrf2−/− mice exposed to air and O2 (Figure 3C). Specific Nrf2 binding activity (SSB) on the Wt probe as determined by addition of anti-Nrf2 antisera (8) was detected only in Nrf2+/+ mice, and was greatly increased by O2 (Figure 3C). No specific Nrf2 binding activity was found in any of the experimental groups when Mt -784 ARE probe was used for the assay (Figure 3C).
Effect of In Vivo RNA Interference on Pulmonary PPARγ Silencing
Intranasal administration of si-PPARγ suppressed total (40%) expression of lung PPARγ proteins relative to si-NS treatment in Nrf2+/+ mice exposed to air (Figure 4A). This reduction level is similar to the degree of PPARγ suppression in various tissues of Pparg+/− mice (39, 40). After 72 hours O2, significant elevation of total PPARγ levels (45%) was found in si-NS–treated mouse lungs (Figure 4A). Total PPARγ level in mice treated with si-PPARγ was not significantly changed after hyperoxia (Figure 4A). The nuclear level of PPARγ did not differ significantly between treatments in air control mice (Figure 4A). Nuclear translocation of lung PPARγ was significantly increased by O2 in mice treated with si-NS, whereas it remained unchanged in mice treated with si-PPARγ (Figure 4A). No hyperoxia-enhanced PPRE binding was found in mice treated with si-PPARγ (Figure 4B).
Figure 4.
In vivo peroxisome proliferator activated receptor γ (PPARγ) specific interference RNA (siRNA) treatment inhibited pulmonary PPARγ expression and activity. (A) Aliquots of lung total and nuclear proteins were used for Western blotting to determine differential protein levels of pulmonary PPARγ in mice treated with nonspecific siRNA (si-NS) or PPARγ siRNA (si-PPARγ) followed by air and O2 exposure (72 h). Representative images from multiple analyses are presented, and group mean ± SEM (n = 3/group) of total and nuclear PPARγ normalized to air-exposed NS-treated mice are depicted in graphs. * = significantly higher than treatment-matched air controls (P < 0.05). + = significantly lower than exposure-matched NS-treated mice (P < 0.05). (B) Differential nuclear protein PPAR response element (PPRE) binding activity in the lungs of mice treated with si-NS or si-PPARγ after exposure to air and O2 (72 h). Aliquots of nuclear protein isolated from pieces of left lung tissue (n = 3 mice/group) were incubated with an end-labeled oligonucleotide probe containing PPRE consensus sequence. Total PPRE binding was determined by gel shift analysis. FP = free probes; SB = shifted bands of total bindings (PPRE motif-protein complex). Representative images from multiple analysis (n = 3) are presented.
Effect of PPARγ Inhibition on Hyperoxia-induced Lung Inflammation
Relative to si-NS controls, si-PPARγ treatment significantly exacerbated (10-fold) hyperoxia-induced pulmonary neutrophilic infiltration in Nrf2+/+ mice (Figure 5A). si-PPARγ also caused a slight, but statistically significant, increase in mean numbers of BAL neutrophils after air exposure. Significant hyperoxia-induced increases in the number of BAL lymphocytes (Figure 5A) and eosinophils (Figure 5A) were observed only in mice treated with si-PPARγ. In addition to the cellular inflammation, O2 caused significantly greater increase in pulmonary NF-κB p65 activity (Figure 5B) and BAL IL-6 concentrations (Figure 5C) in mice treated with si-PPARγ than in mice treated with si-NS.
Figure 5.
In vivo peroxisome proliferator activated receptor γ (PPARγ) specific interference RNA (siRNA) treatment augmented pulmonary inflammation after hyperoxia. (A) Bronchopulmonary lavage (BAL) analysis determined the number of lung neutrophils, lymphocytes, and eosinophils in mice treated with nonspecific siRNA (si-NS) or PPARγ siRNA (si-PPARγ) followed by air and O2 exposure (72 h). Data are presented as group mean ± SEM (n = 3–6/group). (B) Transcription factor ELISA quantified specific DNA binding activity of nuclear p65 NF-κB. Group mean ± SEM (n = 3/group) normalized to air-exposed mice treated with si-NS are presented. (C) BAL levels of a proinflammatory cytokine IL-6 were determined by ELISA. Data presented as group mean ± SEM (n = 4–6/group). * = significantly higher than treatment-matched air controls (P < 0.05). + = significantly higher than O2-exposed si-NS-treated mice (P < 0.05).
Effect of PPARγ Inhibition on Pulmonary Apoptosis after Hyperoxia
TUNEL-positive cells were detected mainly in epithelium lining bronchi, bronchioles, and terminal bronchioles in all air-exposed mice (Figure 6A). Hyperoxia exposure enhanced TUNEL staining of the airway epithelium, and caused marked apoptosis in inflammatory cells (macrophages and neutrophils), alveolar and bronchial/bronchiolar endothelial cells, and endothelium and smooth muscles of perivascular and peribronchial regions (Figure 6A). Hyperoxia-induced increases in TUNEL-positive cells were more obvious in mice treated with si-NS relative to mice treated with si-PPARγ (Figure 6A). Furthermore, O2 caused greater protein levels of signal transducers known to be associated with PPARγ-induced cellular apoptosis, phosphatase and tensin homolog deleted on chromosome 10 (PTEN), caspase-8 (active form), and bax in the lungs of mice treated with si-NS relative to the si-PPARγ–treated lungs (Figure 6B).
Figure 6.
In vivo peroxisome proliferator activated receptor γ (PPARγ) specific interference RNA (siRNA) treatment suppressed pulmonary apoptosis markers after hyperoxia. (A) Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay determined cells undergoing apoptosis in lung tissue sections after air or hyperoxia exposure (72 h). Hyperoxia caused cellular apoptosis predominantly in inflammatory, endothelial, and epithelial cells in all mice. Intensity and abundance of TUNEL-positive cells increased by hyperoxia were greater in mice treated with nonspecific siRNA (si-NS) compared with mice treated with PPARγ-specific siRNA (si-PPARγ). Higher magnification photomicrographs displayed differentially less abundant apoptotic macrophages, epithelial cells, and endothelial cells in hyperoxia-exposed lungs treated with si-PPARγ than in lungs treated with si-NS. AV = alveoli; BR = bronchi or bronchiole; BV = blood vessel; TB = terminal bronchioles. Arrows indicate TUNEL-stained nuclei. Bars indicate 100 μm. Representative light photomicrographs are shown (n = 3/group). (B) O2-enhanced lung levels of PPARγ-mediated apoptosis signaling proteins, PTEN, caspase-8, and bax determined by Western blot analysis in aliquots of total lung proteins were greater in mice treated with si-NS than in mice treated with si-PPARγ (72 h). Representative band images are shown (n = 3/group).
Effect of PPARγ Inhibition on Cytoprotective Genes and Nrf2
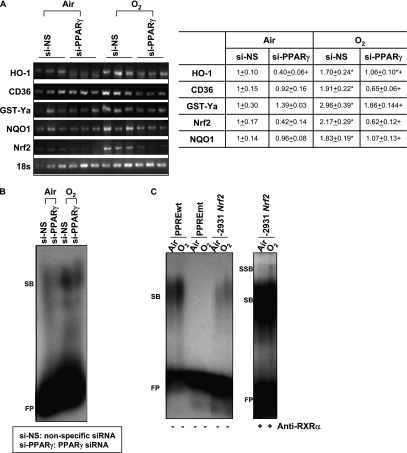
Recent studies indicate that PPARγ may control the expression of cytoprotective genes including GST-α, HO-1, and CD36 directly through their PPRE or indirectly through Nrf2 (41–45). To determine whether PPARγ modulates these cytoprotective genes and Nrf2 in the lung, their expression was compared in wild type mice treated with si-PPARγ or si-NS (Figure 7A). Constitutive mRNA expression of HO-1 was significantly suppressed in the lungs of Nrf2+/+ mice after si-PPARγ treatment. Significant up-regulation of HO-1 mRNA by O2 was inhibited in mice treated with si-PPARγ. Message level of CD36 was significantly elevated in si-NS–treated mice, but not in si-PPARγ–treated mice after O2. Similarly, O2 significantly induced mRNA expression of Nrf2, NQO1, and GST-Ya in si-NS–treated mice, but their expression levels were not significantly affected by O2 in si-PPARγ–treated mice. Gel shift analysis determined that total ARE binding activity of pulmonary nuclear proteins (SB) was suppressed at baseline and after hyperoxia (72 h) in mice treated with si-PPARγ compared with mice treated with si-NS (Figure 7B). We applied the computational PWM statistical method that has been used to search AREs, and identified two potential sequences for PPAR binding (PPRE) in the 5′ upstream region of murine Nrf2 (Table 2). Gel shift analysis of lung nuclear proteins on the Nrf2 promoter -2931 PPRE-like sequence identified enhanced binding activity (SB) after O2 (72 h) with a similar pattern of band shift as shown on PPRE consensus sequence (Figure 7C). Addition of anti-RXRα antibody identified elevated specific PPRE-PPARγ-RXRα binding activity (SSB) after O2. These findings suggested involvement of the potential PPRE in transcriptional activation of Nrf2, and warrant further functional study. Overall, attenuated expression of Nrf2 and ARE- or PPRE-bearing genes and suppressed nuclear ARE binding activity in the lung concurrently with suppressed PPARγ level suggests that PPARγ could serve not only as a Nrf2 downstream effector but may also modulate Nrf2-ARE pathways in the pathogenesis of hyperoxia.
Figure 7.
In vivo peroxisome proliferator activated receptor γ (PPARγ) specific interference RNA (siRNA) treatment suppressed cytoprotective gene expression and antioxidant response elements (ARE) binding activity after hyperoxia. (A) Message levels of cytoprotective genes HO-1, CD36, GST-Ya, and NQO1 and Nrf2 were detected by semiquantitative reverse transcriptase–polymerase chain reaction using total lung RNA isolated from mice treated with nonspecific siRNA (si-NS) or PPARγ-specific siRNA (si-PPARγ) after air and O2 exposure (72 h). cDNA band images for each gene and quantified relative intensities to air-exposed NS-treated mice of digitized cDNA bands normalized to the intensity of each 18s band are shown. Data are presented as group mean ± SEM (n = 3/group). * = significantly higher than treatment-matched air controls (P < 0.05). + = significantly lower than exposure-matched si-NS-treated mice (P < 0.05). (B) Differential nuclear protein-ARE binding activity in the lungs of mice treated with si-NS or si-PPARγ after exposure to air and O2 (72 h). Aliquots of nuclear protein isolated from pooled pieces of left lung tissues (n = 3 mice/group) were incubated with an end-labeled oligonucleotide probe containing ARE consensus sequence. Total ARE binding was determined by gel shift analysis. FP = free ARE probes; SB = shifted bands of total bindings (ARE motif-protein complex). Representative images from multiple analysis (n = 2) are presented. (C) Increased total nuclear protein binding activity (SB) and specific PPARγ-retinoic acid X receptor (RXRa) binding activity (SSB) on an Nrf2 PPAR response element (PPRE)–like sequence in the lungs of mice after exposure to O2 (72 h). Aliquots of nuclear protein isolated from pooled pieces of left lung tissue exposed to either air or O2 (n = 3 mice/group) were incubated with an end-labeled oligonucleotide probe containing -2931 Nrf2 PPRE-like sequence (-2931 Nrf2). PPRE consensus sequence (PPREwt) was used as a positive control for band shift, and mutant PPRE (PPREmt) as a no binding control. FP = free DNA probes; SB = shifted bands of total bindings (PPRE motif-protein complex); SSB = super shifted bands of specific RXRα bindings (PPRE motif-protein–anti-RXRα antibody complex). Representative images from multiple analysis (n = 2) are presented.
TABLE 2.
PUTATIVE PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR DNA BINDING SEQUENCES DETERMINED IN THE MURINE NRF2 PROMOTER BY BIOINFORMATIC ANALYSIS
| Location on ch2 (Map on Nrf2 Promoter) | Sequence | PWM* |
|---|---|---|
| 75545629/75545641 (-2931) | TGGTCAAAGGCCA | 13.1 |
| 75547419/75547431 (-4721) | AGGACAAAGGAGG | 9.2 |
Position weight matrix score.
Mismatch sequences relative to putative peroxisome proliferator activated receptor DNA binding sequences core (5′-AGGTCAAAGGTCA -3′) are italicized.
Effect of a PPARγ Ligand on Hyperoxia-induced Pulmonary Injury
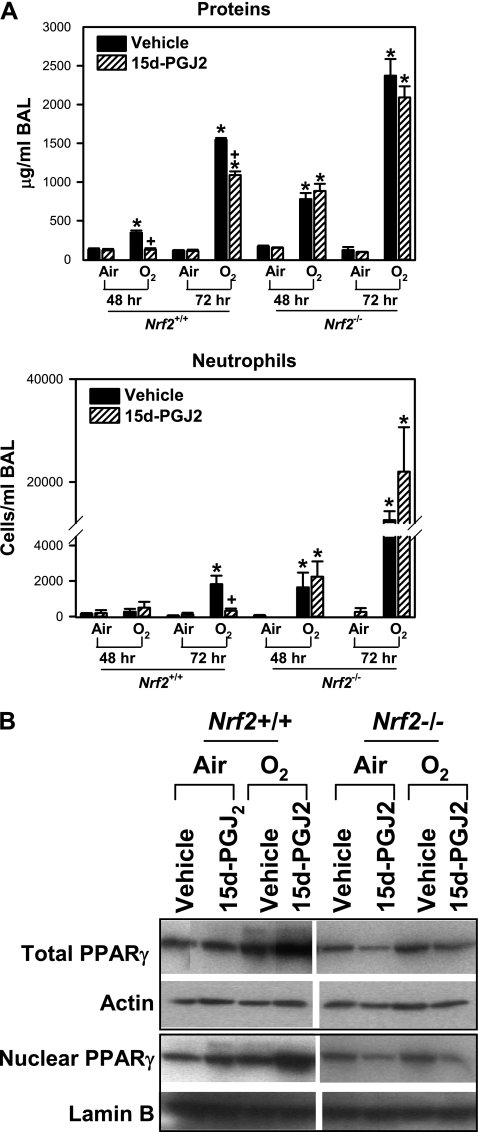
A functional role of PPARγ in the pulmonary pathogenesis response to hyperoxia was further determined by administration of a widely used ligand, 15d-PGJ2. Relative to vehicle treatment, 15d-PGJ2 significantly decreased hyperoxia-induced increase in the number of neutrophils (at 72 h) and the concentration of total BAL proteins (at 48 and 72 h) in Nrf2+/+ mice (Figure 8A). No significant differences in the O2-increased numbers of macrophages, lymphocytes, and epithelial cells were found between vehicle and 15d-PGJ2 treatments in Nrf2+/+ mice (data not shown). In Nrf2−/− mice, hyperoxia caused markedly greater lung injury determined by BAL cell numbers and total protein concentration relative to Nrf2+/+ mice as we demonstrated previously (7). However, a significant effect of 15d-PGJ2 administration was not found on the lung injury phenotypes in Nrf2−/− mice (Figure 8A). Administration of 15d-PGJ2 increased basal levels of total and nuclear PPARγ in the lungs of Nrf2+/+ mice (Figure 8B). Hyperoxia-induced increase in total and nuclear PPARγ level was higher in the lungs of Nrf2+/+ mice treated with 15d-PGJ2 relative to these mice treated with vehicle (Figure 8B). 15d-PGJ2-mediated increase of pulmonary PPARγ protein level was not found in Nrf2−/− mice (Figure 8B).
Figure 8.
Treatment with 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) enhanced lung peroxisome proliferator activated receptor γ (PPARγ) expression and suppressed pulmonary injury caused by hyperoxia in Nrf2+/+ mice. (A) Bronchopulmonary lavage (BAL) analysis determined the concentration of total proteins and the number of lung neutrophils in Nrf2+/+ and Nrf2−/− mice pretreated with either vehicle or 15d-PGJ2 followed by air and O2 exposure (48 and 72 h). Data are presented as group mean ± SEM (n = 3–7/group). * = significantly higher than genotype- and treatment-matched air controls (P < 0.05). + = significantly lower than genotype- and exposure-matched vehicle-treated mice (P < 0.05). (B) Aliquots of lung total and nuclear proteins were subjected for Western blotting to determine PPARγ levels in Nrf2+/+ and Nrf2−/− mice treated with vehicle or 15d-PGJ2 after air or O2 exposure (72 h). Representative images from multiple analysis (n = 3 for total proteins, n = 2 for nuclear proteins) are presented.
DISCUSSION
Results of the present study suggested an antiinflammatory role for PPARγ in the setting of hyperoxia-induced lung injury. We determined that the PPARγ ligand 15d-PGJ2 significantly reduced hyperoxia-induced inflammation, whereas in vivo partial silencing of PPARγ by siRNA treatment significantly exacerbated O2-induced pulmonary inflammation. The results are consistent with recent observations that PPARγ functions not only in adipogenesis and glucose/lipid metabolism but also exerts antiinflammatory effects. Moreover, our results using Nrf2−/− and Nrf2+/+ mice suggest that the antiinflammatory effects of PPARγ in the lung seems to be at least in part Nrf2-dependent.
The antiinflammatory role of PPARγ was first demonstrated in macrophage or monocyte activation and inflammatory cytokine production, which resulted in altered native and acquired immune responses (46). Subsequently, studies on experimental models of inflammatory bowel diseases, renal disorders, periodontitis, and various cancers have supported PPARγ as a potential therapeutic target for tissue inflammation and carcinogenesis (47–51). PPARγ is ubiquitously found in most cell types and tissues including immune system (18–21). The amount of PPARγ in monocytes is relatively low (52) but increases during differentiation into macrophages (46, 53), which corresponds to the role for PPARγ agonists in monocyte-macrophage differentiation (52, 54). Other inflammatory and immune cells including neutrophils, dendritic cells, B and T lymphocytes, eosinophils, natural killer cells, and mast cells are also identified as sources for PPARγ (18, 19, 55–59). Augmented PPARγ expression has been detected in airway epithelium, bronchial submucosa, and smooth muscle of asthmatics (60). The up-regulation of PPARγ in lung tissues was coincident with markers of apoptosis (caspase-3), airway remodeling (Ki67), and collagen deposition in these asthma patients (60). Supporting the implication for PPARγ in asthma pathogenesis, a PPARG polymorphism haplotype (Pro12Ala, C1431T) was found to be associated with increased risk for asthma exacerbation (61).
The mechanism underlying antiinflammatory effects of PPARγ in multiple tissues has been under investigation. It is known that PPARγ antagonizes signal transducing kinases or transcriptional regulators, such as activator protein-1, nuclear factor of activated T cell, and NF-κB (62). PPARγ also transrepresses inflammatory mediators, such as tumor necrosis factor-α; inducible nitric oxide synthase; IL-1β; chemokine receptors (e.g., chemokine receptors-7); adhesion molecules; and matrix metalloproteinase-9 (63, 64). Moreover, PPARγ can potentiate inflammatory cell apoptosis (65, 70, 71) and transactivate antiinflammatory mediators including IL-10 (68). Enhanced pulmonary p65 NF-κB and IL-6 and reduced inflammatory cell apoptosis in mice with suppressed pulmonary PPARγ determined in the current study supports the antiinflammatory mechanisms through PPARγ in ALI.
Less well-known are the regulatory mechanisms of PPARγ expression. We tested the hypothesis that Nrf2 regulates PPARγ through binding to its promoter ARE sites, and this interaction is important in protecting the lung against hyperoxia-induced injury. We initially found that lung PPARγ was more highly expressed and activated in Nrf2+/+ mice than in Nrf2−/− mice basally and after hyperoxia exposure, which suggested that PPARγ expression was at least partially dependent on Nrf2. To address the molecular mechanism through which Nrf2 regulated PPARγ, we initially used bioinformatic tools (34). This analysis identified a number of potentially important promoter AREs in Pparg that could bind Nrf2, and we focused our efforts on the -784 ARE because it had the highest PWM score with the greatest similarity to the consensus ARE sequence of all of the Pparg AREs. Site-directed mutation and deletion experiments confirmed a significant role for the -784 ARE in Nrf2-mediated pulmonary PPARγ expression after hyperoxia. Although our analysis suggest the -784 site is a functionally important ARE, potential AREs further upstream in the promoter need to be tested.
Because conventional deletion of Pparg resulted in embryonic lethality (69, 70), animal studies on PPARγ have been done primarily using agonists/antagonists to determine its role in airway inflammation. Asthma and allergy models are under intense investigation, and thiazolidinediones (e.g., rosiglitazone, ciglitazone, and pioglitazone) reduced eosinophilic inflammation and allergic symptoms by various allergens in mice (26, 58, 71–73). Studies using the agonists or adenovirus carrying Pparg cDNA in mice also determined that enhanced PTEN and suppressed phosphorylated Akt/phosphoinositol 3 kinase and NF-κB was involved in the antiasthmatic effect of PPARγ on eosinophilia and airway hyperresponsiveness (66). Similar protective effects of PPARγ were observed in ALI models including carrageenin-induced pleurisy (32) and lipopolysaccharide-induced neutrophilia and airway hyperresponsiveness (67, 74, 75). PPARγ ligands also have antifibrogenic potential in murine airways. For example, administration of thiazolidinediones or 15d-PGJ2 inhibited bleomycin-induced pulmonary injury and fibrosis in mice (33, 76). PPARγ agonists are postulated to act through cell cycle arrest and TGF-β1 inhibition for myofibroblast differentiation and collagen deposition (76, 77). Enhanced PPARγ was protective against respiratory syncytial virus infection in lung airway epithelial cell lines, and reduced ICAM-1 expression and NF-κB activation was associated with this effect (78). Overall, these observations strongly indicate that PPARγ is an important down-regulator of airway inflammation and injury.
Maternal Pparg deletion in mice (conditional knockout mice) caused inflammatory milk production because of repression of 12-lipoxygenase and elevation of lipid oxidation enzymes in lactating mammary gland, which resulted in skin inflammation and hair loss of nursing offspring (79). This observation suggested that PPARγ-mediated tissue protection may be associated with antioxidant defense mechanisms. In airways, PPARγ agonists modulated nicotinamide adenine dinucleotide phosphate hydrogen oxidase and reduced reactive oxygen species production of asthmatic mice (60, 80). Because murine Nrf2 is known to contribute to the pulmonary protection from allergy, sepsis, emphysema, ALI-ARDS, viral infection, or fibrosis, and PPARγ has shown defensive roles in these disease models (7–16), it is hypothesized that PPARγ- and Nrf2-mediated molecular events are closely associated in airway protection against oxidants. Supporting this concept, ALI induced by carrageenin was exacerbated in Nrf2−/− mice relative to Nrf2+/+ mice, and administration of 15d-PGJ2 reversed the augmented airway inflammation and protein hyperpermeability in Nrf2+/+ mice caused by a cyclooxygenase-2 inhibitor, which led to depletion of lung 15d-PGJ2 and Nrf2-dependent antioxidants (81). Rosiglitazone and 15d-PGJ2 also reduced pulmonary injury caused by bleomycin in mice (28, 33).
Although we found that Nrf2 modulates pulmonary PPARγ through its ARE, we also found that Nrf2 expression was attenuated in the lung with decreased PPARγ levels by siRNA silencing. This indicates that PPARγ could act on upstream signal pathways for Nrf2 activation. In support of this function, accumulating evidence has indicated that PPARγ agonists induce antioxidant and defense genes encoding GST-α, HO-1, and CD36 through regulation of Nrf2 (43–45). The current study also supported this concept, first by showing suppressed Nrf2 expression and ARE binding activity after PPARγ silencing, and then by elucidating potential PPAR binding motifs in the Nrf2 promoter. Although further investigations are desirable to determine whether PPARγ regulates Nrf2 expression directly or indirectly, it is likely that these two redox sensitive transcriptional factors regulate each other through autoregulatory mechanisms for pulmonary protection against oxidative stress.
Conclusion
Our results provide the first evidence that PPARγ is an ARE-driven Nrf2 effector molecule that contributes significantly to protection against oxidative lung injury. Our observations provide new insights into the therapeutic potential of PPARγ or its agonist in airway oxidative and inflammatory disorders including ALI-ARDS and bronchopulmonary dysplasia.
Acknowledgments
Hyperoxia exposures were conducted at the NIEHS Inhalation Facility under contract to Alion Science and Technology, Inc. The authors thank Dr. Daniel Morgan and Mr. Herman Price for coordinating the exposures. Drs. Michael Resnick, Daniel Menendez, and Mike Fessler at the NIEHS provided excellent critical review of the manuscript.
Supported by the Intramural Research program of the National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, and in part by grant HL66109 (S.P.R.).
Originally Published in Press as DOI: 10.1164/rccm.200907-1047OC on March 11, 2010
Conflict of Interest Statement: H-Y.C. is an employee of the Intramural Research program of the National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, Research Triangle Park, NC (NIEHS-NIH). W.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. X.W. is an employee of the NIEHS-NIH. B.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.B. is an employee of the NIEHS-NIH. S.P.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.R.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Bowler RP, Barnes PJ, Crapo JD. The role of oxidative stress in chronic obstructive pulmonary disease. COPD 2004;1:255–277. [DOI] [PubMed] [Google Scholar]
- 2.Chow CW, Herrera Abreu MT, Suzuki T, Downey GP. Oxidative stress and acute lung injury. Am J Respir Cell Mol Biol 2003;29:427–431. [DOI] [PubMed] [Google Scholar]
- 3.Paz-Elizur T, Krupsky M, Elinger D, Schechtman E, Livneh Z. Repair of the oxidative DNA damage 8-oxoguanine as a biomarker for lung cancer risk. Cancer Biomark 2005;1:201–205. [DOI] [PubMed] [Google Scholar]
- 4.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 5.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small MAF heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997;236:313–322. [DOI] [PubMed] [Google Scholar]
- 6.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116. [DOI] [PubMed] [Google Scholar]
- 7.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of Nrf2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol 2002;26:175–182. [DOI] [PubMed] [Google Scholar]
- 8.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor Nrf2 protects against pulmonary fibrosis. FASEB J 2004;18:1258–1260. [DOI] [PubMed] [Google Scholar]
- 9.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol 2001;173:154–160. [DOI] [PubMed] [Google Scholar]
- 10.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci U S A 1999;96:12731–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii Y, Itoh K, Morishima Y, Kimura T, Kiwamoto T, Iizuka T, Hegab AE, Hosoya T, Nomura A, Sakamoto T, et al. Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J Immunol 2005;175:6968–6975. [DOI] [PubMed] [Google Scholar]
- 12.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest 2004;114:1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med 2005;202:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 2006;116:984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy NM, Kleeberger SR, Cho HY, Yamamoto M, Kensler TW, Biswal S, Reddy SP. Deficiency in Nrf2-GSH signaling impairs type II cell growth and enhances sensitivity to oxidants. Am J Respir Cell Mol Biol 2007;37:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho HY, Imani F, Miller-DeGraff L, Walters D, Melendi GA, Yamamoto M, Polack FP, Kleeberger SR. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am J Respir Crit Care Med 2009;179:138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of Nrf2-mediated protection against oxidative injury. Free Radic Biol Med 2005;38:325–343. [DOI] [PubMed] [Google Scholar]
- 18.Standiford TJ, Keshamouni VG, Reddy RC. Peroxisome proliferator-activated receptor-{gamma} as a regulator of lung inflammation and repair. Proc Am Thorac Soc 2005;2:226–231. [DOI] [PubMed] [Google Scholar]
- 19.Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, Teissier E, Minnich A, Jaye M, Duverger N, et al. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the abca1 pathway. Nat Med 2001;7:53–58. [DOI] [PubMed] [Google Scholar]
- 20.Spears M, McSharry C, Thomson NC. Peroxisome proliferator-activated receptor-gamma agonists as potential anti-inflammatory agents in asthma and chronic obstructive pulmonary disease. Clin Exp Allergy 2006;36:1494–1504. [DOI] [PubMed] [Google Scholar]
- 21.Patel HJ, Belvisi MG, Bishop-Bailey D, Yacoub MH, Mitchell JA. Activation of peroxisome proliferator-activated receptors in human airway smooth muscle cells has a superior anti-inflammatory profile to corticosteroids: relevance for chronic obstructive pulmonary disease therapy. J Immunol 2003;170:2663–2669. [DOI] [PubMed] [Google Scholar]
- 22.Kapadia R, Yi JH, Vemuganti R. Mechanisms of anti-inflammatory and neuroprotective actions of PPAR-γagonists. Front Biosci 2008;13:1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnan A, Nair SA, Pillai MR. Biology of PPAR gamma in cancer: a critical review on existing lacunae. Curr Mol Med 2007;7:532–540. [DOI] [PubMed] [Google Scholar]
- 24.Debril MB, Renaud JP, Fajas L, Auwerx J. The pleiotropic functions of peroxisome proliferator-activated receptor gamma. J Mol Med 2001;79:30–47. [DOI] [PubMed] [Google Scholar]
- 25.Fajas L, Debril MB, Auwerx J. Peroxisome proliferator-activated receptor-gamma: from adipogenesis to carcinogenesis. J Mol Endocrinol 2001;27:1–9. [DOI] [PubMed] [Google Scholar]
- 26.Ward JE, Fernandes DJ, Taylor CC, Bonacci JV, Quan L, Stewart AG. The PPARgamma ligand, rosiglitazone, reduces airways hyperresponsiveness in a murine model of allergen-induced inflammation. Pulm Pharmacol Ther 2006;19:39–46. [DOI] [PubMed] [Google Scholar]
- 27.Rehan VK, Wang Y, Patel S, Santos J, Torday JS. Rosiglitazone, a peroxisome proliferator-activated receptor-gamma agonist, prevents hyperoxia-induced neonatal rat lung injury in vivo. Pediatr Pulmonol 2006;41:558–569. [DOI] [PubMed] [Google Scholar]
- 28.Lakatos HF, Thatcher TH, Kottmann RM, Garcia TM, Phipps RP, Sime PJ. The role of PPARS in lung fibrosis. PPAR Res 2007;2007:71323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho H-Y, Wang X, Bell DA, Reddy SP, Yamamoto M, Kleeberger SR. Peroxisome proliferator-activated receptor gamma inhibits hyperoxia-induced pulmonary inflammation and injury in mice. Proc Amer Thor Soc 2006;3:A898. [Google Scholar]
- 30.Bilban M, Bach FH, Otterbein SL, Ifedigbo E, de Costa d'Avila J, Esterbauer H, Chin BY, Usheva A, Robson SC, Wagner O, et al. Carbon monoxide orchestrates a protective response through PPARgamma. Immunity 2006;24:601–610. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Shan P, Jiang D, Noble PW, Abraham NG, Kappas A, Lee PJ. Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. J Biol Chem 2004;279:10677–10684. [DOI] [PubMed] [Google Scholar]
- 32.Cuzzocrea S, Wayman NS, Mazzon E, Dugo L, Di Paola R, Serraino I, Britti D, Chatterjee PK, Caputi AP, Thiemermann C. The cyclopentenone prostaglandin 15-deoxy-delta(12,14)-prostaglandin j(2) attenuates the development of acute and chronic inflammation. Mol Pharmacol 2002;61:997–1007. [DOI] [PubMed] [Google Scholar]
- 33.Genovese T, Cuzzocrea S, Di Paola R, Mazzon E, Mastruzzo C, Catalano P, Sortino M, Crimi N, Caputi AP, Thiemermann C, et al. Effect of rosiglitazone and 15-deoxy-delta12,14-prostaglandin j2 on bleomycin-induced lung injury. Eur Respir J 2005;25:225–234. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet 2007;16:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho HY, Jedlicka AE, Reddy SP, Zhang LY, Kensler TW, Kleeberger SR. Linkage analysis of susceptibility to hyperoxia. Nrf2 is a candidate gene. Am J Respir Cell Mol Biol 2002;26:42–51. [DOI] [PubMed] [Google Scholar]
- 36.Cho HY, Morgan DL, Bauer AK, Kleeberger SR. Signal transduction pathways of tumor necrosis factor–mediated lung injury induced by ozone in mice. Am J Respir Crit Care Med 2007;175:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho HY, Jedlicka AE, Clarke R, Kleeberger SR. Role of toll-like receptor-4 in genetic susceptibility to lung injury induced by residual oil fly ash. Physiol Genomics 2005;22:108–117. [DOI] [PubMed] [Google Scholar]
- 38.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics 2000;16:16–23. [DOI] [PubMed] [Google Scholar]
- 39.Rieusset J, Seydoux J, Anghel SI, Escher P, Michalik L, Soon Tan N, Metzger D, Chambon P, Wahli W, Desvergne B. Altered growth in male peroxisome proliferator-activated receptor gamma (PPARgamma) heterozygous mice: involvement of PPARgamma in a negative feedback regulation of growth hormone action. Mol Endocrinol 2004;18:2363–2377. [DOI] [PubMed] [Google Scholar]
- 40.Lu J, Imamura K, Nomura S, Mafune K, Nakajima A, Kadowaki T, Kubota N, Terauchi Y, Ishii G, Ochiai A, et al. Chemopreventive effect of peroxisome proliferator-activated receptor gamma on gastric carcinogenesis in mice. Cancer Res 2005;65:4769–4774. [DOI] [PubMed] [Google Scholar]
- 41.Kronke G, Kadl A, Ikonomu E, Bluml S, Furnkranz A, Sarembock IJ, Bochkov VN, Exner M, Binder BR, Leitinger N. Expression of heme oxygenase-1 in human vascular cells is regulated by peroxisome proliferator-activated receptors. Arterioscler Thromb Vasc Biol 2007;27:1276–1282. [DOI] [PubMed] [Google Scholar]
- 42.Sato O, Kuriki C, Fukui Y, Motojima K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J Biol Chem 2002;277:15703–15711. [DOI] [PubMed] [Google Scholar]
- 43.Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, Mann GE. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res 2004;94:609–616. [DOI] [PubMed] [Google Scholar]
- 44.Kim EH, Surh YJ. 15-deoxy-delta12,14-prostaglandin j2 as a potential endogenous regulator of redox-sensitive transcription factors. Biochem Pharmacol 2006;72:1516–1528. [DOI] [PubMed] [Google Scholar]
- 45.Park EY, Cho IJ, Kim SG. Transactivation of the PPAR-responsive enhancer module in chemopreventive glutathione S-transferase gene by the peroxisome proliferator-activated receptor-gamma and retinoid x receptor heterodimer. Cancer Res 2004;64:3701–3713. [DOI] [PubMed] [Google Scholar]
- 46.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature 1998;391:79–82. [DOI] [PubMed] [Google Scholar]
- 47.Ramakers JD, Verstege MI, Thuijls G, Te Velde AA, Mensink RP, Plat J. The PPARgamma agonist rosiglitazone impairs colonic inflammation in mice with experimental colitis. J Clin Immunol 2007;27:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takaki K, Mitsuyama K, Tsuruta O, Toyonaga A, Sata M. Attenuation of experimental colonic injury by thiazolidinedione agents. Inflamm Res 2006;55:10–15. [DOI] [PubMed] [Google Scholar]
- 49.Lee S, Kim W, Moon SO, Sung MJ, Kim DH, Kang KP, Jang YB, Lee JE, Jang KY, Park SK. Rosiglitazone ameliorates cisplatin-induced renal injury in mice. Nephrol Dial Transplant 2006;21:2096–2105. [DOI] [PubMed] [Google Scholar]
- 50.Katayama K, Wada K, Nakajima A, Mizuguchi H, Hayakawa T, Nakagawa S, Kadowaki T, Nagai R, Kamisaki Y, Blumberg RS, et al. A novel PPAR gamma gene therapy to control inflammation associated with inflammatory bowel disease in a murine model. Gastroenterology 2003;124:1315–1324. [DOI] [PubMed] [Google Scholar]
- 51.Di Paola R, Mazzon E, Maiere D, Zito D, Britti D, De Majo M, Genovese T, Cuzzocrea S. Rosiglitazone reduces the evolution of experimental periodontitis in the rat. J Dent Res 2006;85:156–161. [DOI] [PubMed] [Google Scholar]
- 52.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 1998;93:241–252. [DOI] [PubMed] [Google Scholar]
- 53.Chinetti G, Griglio S, Antonucci M, Torra IP, Delerive P, Majd Z, Fruchart JC, Chapman J, Najib J, Staels B. Activation of proliferator-activated receptors alpha and gamma induces apoptosis of human monocyte-derived macrophages. J Biol Chem 1998;273:25573–25580. [DOI] [PubMed] [Google Scholar]
- 54.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 1998;93:229–240. [DOI] [PubMed] [Google Scholar]
- 55.Nencioni A, Grunebach F, Zobywlaski A, Denzlinger C, Brugger W, Brossart P. Dendritic cell immunogenicity is regulated by peroxisome proliferator-activated receptor gamma. J Immunol 2002;169:1228–1235. [DOI] [PubMed] [Google Scholar]
- 56.Padilla J, Kaur K, Cao HJ, Smith TJ, Phipps RP. Peroxisome proliferator activator receptor-gamma agonists and 15-deoxy-Delta(12,14)(12,14)-pgj(2) induce apoptosis in normal and malignant B-lineage cells. J Immunol 2000;165:6941–6948. [DOI] [PubMed] [Google Scholar]
- 57.Clark RB, Bishop-Bailey D, Estrada-Hernandez T, Hla T, Puddington L, Padula SJ. The nuclear receptor PPAR gamma and immunoregulation: PPAR gamma mediates inhibition of helper t cell responses. J Immunol 2000;164:1364–1371. [DOI] [PubMed] [Google Scholar]
- 58.Woerly G, Honda K, Loyens M, Papin JP, Auwerx J, Staels B, Capron M, Dombrowicz D. Peroxisome proliferator-activated receptors alpha and gamma down-regulate allergic inflammation and eosinophil activation. J Exp Med 2003;198:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Rodriguez-Galan MC, Subleski JJ, Ortaldo JR, Hodge DL, Wang JM, Shimozato O, Reynolds DA, Young HA. Peroxisome proliferator-activated receptor-gamma and its ligands attenuate biologic functions of human natural killer cells. Blood 2004;104:3276–3284. [DOI] [PubMed] [Google Scholar]
- 60.Benayoun L, Letuve S, Druilhe A, Boczkowski J, Dombret MC, Mechighel P, Megret J, Leseche G, Aubier M, Pretolani M. Regulation of peroxisome proliferator-activated receptor gamma expression in human asthmatic airways: relationship with proliferation, apoptosis, and airway remodeling. Am J Respir Crit Care Med 2001;164:1487–1494. [DOI] [PubMed] [Google Scholar]
- 61.Palmer CN, Doney AS, Ismail T, Lee SP, Murrie I, Macgregor DF, Mukhopadhyay S. PPARG locus haplotype variation and exacerbations in asthma. Clin Pharmacol Ther 2007;81:713–718. [DOI] [PubMed] [Google Scholar]
- 62.Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, Keilbaugh SA, Flanigan A, Murthy S, Lazar MA, Wu GD. A novel therapy for colitis utilizing PPAR-gamma ligands to inhibit the epithelial inflammatory response. J Clin Invest 1999;104:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crosby MB, Svenson J, Gilkeson GS, Nowling TK. A novel PPAR response element in the murine INOS promoter. Mol Immunol 2005;42:1303–1310. [DOI] [PubMed] [Google Scholar]
- 64.Dubuquoy L, Bourdon C, Peuchmaur M, Leibowitz MD, Nutten S, Colombel JF, Auwerx J, Desreumaux P. Peroxisome proliferator-activated receptor (PPAR) gamma: a new target for the treatment of inflammatory bowel disease. Gastroenterol Clin Biol 2000;24:719–724. [PubMed] [Google Scholar]
- 65.Moller DE, Berger JP. Role of PPARS in the regulation of obesity-related insulin sensitivity and inflammation. Int J Obes Relat Metab Disord 2003;27:S17–S21. [DOI] [PubMed] [Google Scholar]
- 66.Lee KS, Park SJ, Hwang PH, Yi HK, Song CH, Chai OH, Kim JS, Lee MK, Lee YC. PPAR-gamma modulates allergic inflammation through up-regulation of PTEN. FASEB J 2005;19:1033–1035. [DOI] [PubMed] [Google Scholar]
- 67.Birrell MA, Patel HJ, McCluskie K, Wong S, Leonard T, Yacoub MH, Belvisi MG. PPAR-gamma agonists as therapy for diseases involving airway neutrophilia. Eur Respir J 2004;24:18–23. [DOI] [PubMed] [Google Scholar]
- 68.Becker J, Delayre-Orthez C, Frossard N, Pons F. Regulation of inflammation by PPARS: a future approach to treat lung inflammatory diseases? Fundam Clin Pharmacol 2006;20:429–447. [DOI] [PubMed] [Google Scholar]
- 69.Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell 1999;4:597–609. [DOI] [PubMed] [Google Scholar]
- 70.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 1999;4:585–595. [DOI] [PubMed] [Google Scholar]
- 71.Ueki S, Matsuwaki Y, Kayaba H, Oyamada H, Kanda A, Usami A, Saito N, Chihara J. Peroxisome proliferator-activated receptor gamma regulates eosinophil functions: a new therapeutic target for allergic airway inflammation. Int Arch Allergy Immunol 2004;134:30–36. [DOI] [PubMed] [Google Scholar]
- 72.Cuzzocrea S. Peroxisome proliferator-activated receptors and acute lung injury. Curr Opin Pharmacol 2006;6:263–270. [DOI] [PubMed] [Google Scholar]
- 73.Hammad H, de Heer HJ, Soullie T, Angeli V, Trottein F, Hoogsteden HC, Lambrecht BN. Activation of peroxisome proliferator-activated receptor-gamma in dendritic cells inhibits the development of eosinophilic airway inflammation in a mouse model of asthma. Am J Pathol 2004;164:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu D, Zeng BX, Zhang SH, Yao SL. Rosiglitazone, an agonist of peroxisome proliferator-activated receptor gamma, reduces pulmonary inflammatory response in a rat model of endotoxemia. Inflamm Res 2005;54:464–470. [DOI] [PubMed] [Google Scholar]
- 75.Kaplan JM, Cook JA, Hake PW, O'Connor M, Burroughs TJ, Zingarelli B. 15-deoxy-delta(12,14)-prostaglandin j(2) (15d-pgj(2)), a peroxisome proliferator activated receptor gamma ligand, reduces tissue leukosequestration and mortality in endotoxic shock. Shock 2005;24:59–65. [DOI] [PubMed] [Google Scholar]
- 76.Milam JE, Keshamouni VG, Phan SH, Hu B, Gangireddy SR, Hogaboam CM, Standiford TJ, Thannickal VJ, Reddy RC. PPAR-{gamma} agonists inhibit pro-fibrotic phenotypes in human lung fibroblasts and bleomycin-induced pulmonary fibrosis. Am J Physiol 2008;294:L891–L901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burgess HA, Daugherty LE, Thatcher TH, Lakatos HF, Ray DM, Redonnet M, Phipps RP, Sime PJ. Ppargamma agonists inhibit TGF-beta induced pulmonary myofibroblast differentiation and collagen production: implications for therapy of lung fibrosis. Am J Physiol 2005;288:L1146–L1153. [DOI] [PubMed] [Google Scholar]
- 78.Arnold R, Neumann M, Konig W. Peroxisome proliferator-activated receptor-gamma agonists inhibit respiratory syncytial virus-induced expression of intercellular adhesion molecule-1 in human lung epithelial cells. Immunology 2007;121:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wan Y, Saghatelian A, Chong LW, Zhang CL, Cravatt BF, Evans RM. Maternal PPAR gamma protects nursing neonates by suppressing the production of inflammatory milk. Genes Dev 2007;21:1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee KS, Kim SR, Park SJ, Park HS, Min KH, Jin SM, Lee MK, Kim UH, Lee YC. Peroxisome proliferator activated receptor-gamma modulates reactive oxygen species generation and activation of nuclear factor-kappaB and hypoxia-inducible factor 1alpha in allergic airway disease of mice. J Allergy Clin Immunol 2006;118:120–127. [DOI] [PubMed] [Google Scholar]
- 81.Mochizuki M, Ishii Y, Itoh K, Iizuka T, Morishima Y, Kimura T, Kiwamoto T, Matsuno Y, Hegab AE, Nomura A, et al. Role of 15-deoxy delta(12,14) prostaglandin j2 and Nrf2 pathways in protection against acute lung injury. Am J Respir Crit Care Med 2005;171:1260–1266. [DOI] [PubMed] [Google Scholar]