Abstract
Rationale: Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and usually lethal fibrotic lung disease characterized by profound changes in epithelial cell phenotype and fibroblast proliferation.
Objectives: To determine changes in expression and role of microRNAs in IPF.
Methods: RNA from 10 control and 10 IPF tissues was hybridized on Agilent microRNA microarrays and results were confirmed by quantitative real-time polymerase chain reaction and in situ hybridization. SMAD3 binding to the let-7d promoter was confirmed by chromatin immunoprecipitation, electrophoretic mobility shift assay, luciferase assays, and reduced expression of let-7d in response to transforming growth factor-β. HMGA2, a let-7d target, was localized by immunohistochemistry. In mice, let-7d was inhibited by intratracheal administration of a let-7d antagomir and its effects were determined by immunohistochemistry, immunofluorescence, quantitative real-time polymerase chain reaction, and morphometry.
Measurements and Main Results: Eighteen microRNAs including let-7d were significantly decreased in IPF. Transforming growth factor-β down-regulated let-7d expression, and SMAD3 binding to the let-7d promoter was demonstrated. Inhibition of let-7d caused increases in mesenchymal markers N-cadherin-2, vimentin, and α-smooth muscle actin (ACTA2) as well as HMGA2 in multiple epithelial cell lines. let-7d was significantly reduced in IPF lungs and the number of epithelial cells expressing let-7d correlated with pulmonary functions. HMGA2 was increased in alveolar epithelial cells of IPF lungs. let-7d inhibition in vivo caused alveolar septal thickening and increases in collagen, ACTA2, and S100A4 expression in SFTPC (pulmonary-associated surfactant protein C) expressing alveolar epithelial cells.
Conclusions: Our results indicate a role for microRNAs in IPF. The down-regulation of let-7d in IPF and the profibrotic effects of this down-regulation in vitro and in vivo suggest a key regulatory role for this microRNA in preventing lung fibrosis.
Clinical trial registered with www.clinicaltrials.gov (NCT 00258544).
Keywords: epithelial–mesenchymal transition, HMGA2 (high-mobility group AT-hook 2), microRNA, transforming growth factor-β
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
The role and regulation of microRNAs in idiopathic pulmonary fibrosis (IPF), a progressive and lethal interstitial lung disease, are unknown.
What This Study Adds to the Field
In this study we determine that let-7d, a microRNA abundantly expressed in epithelial cells in normal lungs, is down-regulated in IPF and its target molecule HMGA2 is overexpressed. The expression of let-7d is inhibited by transforming growth factor-β1. Down-regulation of let-7d expression causes epithelial mesenchymal transition in epithelial cells in vitro and in vivo and increased collagen deposition in mouse lungs in vivo. Taken together, our results suggest that down-regulation of let-7 microRNAs may be important in determining the lung phenotype in IPF.
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive, and usually lethal fibrotic lung disease (1). The disease is characterized by alveolar epithelial cell injury and activation, formation of myofibroblast foci, and exaggerated accumulation of extracellular matrix in the lung parenchyma (1–3). Despite significant progress the etiology and molecular mechanisms underlying the lung phenotype in IPF are largely unknown.
It was suggested that the lung phenotype in IPF is characterized by aberrant recapitulation of developmental programs (4) as evidenced by activation of the WNT/β-catenin pathway (5–8) and evidence of epithelial–mesenchymal transition (EMT) in IPF (9). EMT is the phenomenon in which epithelial cells obtain mesenchymal characteristics, including change in shape, increased motility, and expression of mesenchymal markers such as N-cadherin (CDH2), vimentin (VIM), and α-smooth muscle actin (ACTA2). EMT is implicated in embryonic development, cancer, and kidney fibrosis (10). Transforming growth factor (TGF)-β is considered the major stimulus for EMT through its effects on SNAI1, SNAI2, TWIST, ID2, and their regulator high-mobility group AT-hook 2 (HMGA2) (11, 12). In vitro, alveolar epithelial cells undergo EMT in response to stimulation by TGF-β, and in vivo, cells coexpressing epithelial and mesenchymal markers are found in IPF lungs and in mice after intranasal delivery of TGF-β (9, 13).
Studies suggest a role for microRNAs in EMT (14–17). MicroRNAs are post-transcriptional gene regulators that function by binding to specific sequences, typically in the 3′ untranslated region of the target mRNAs and blocking translation or causing the rapid degradation of the target transcript (18). MicroRNAs are implicated in embryonic development (19), in multiple cancers (20), in nonmalignant diseases such as chronic heart failure (21), and in allergic airway inflammation (22, 23) and are also found in the peripheral blood (24).
Considering that the lung in IPF is characterized by profound changes in the phenotype of lung fibroblasts and epithelial cells, as well as by drastic changes in global patterns of gene expression (4, 25), we studied the expression, regulation, and potential role of microRNAs in IPF lungs. Some of the results have been previously reported in the form of an abstract (26).
METHODS
See the online supplement for details on methods.
IPF Tissues
Ten IPF lung tissue samples were obtained from surgical remnants of biopsies or lungs explanted from patients with IPF who underwent pulmonary transplantation, and 10 control samples were obtained through the University of Pittsburgh Health Sciences Tissue Bank (Pittsburgh, PA) as previously described (27, 28), from samples resected from patients with lung cancer.
Cell Culture
A549 cells (CCL-185; American Type Culture Collection [ATCC], Manassas, VA) and RLE-6TN cells (CRL-2300; ATCC) were grown in F12K medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum at 37°C in a humidified chamber supplemented with 5% CO2. Normal human bronchial epithelial (NHBE) cells (Lonza, Basel, Switzerland) were cultured according to the supplier's protocol. Wherever indicated cells were stimulated with recombinant TGF-β1 (R&D Systems, Minneapolis, MN).
Microarrays
Total RNA from tissues and cells was isolated with an miRNeasy mini kit (Qiagen, Valencia, CA). MicroRNA profiling was performed with an 8 × 15K Agilent human microRNA microarray containing 470 microRNAs (Sanger miRbase release 9.1), in accordance with the protocol described by the manufacturer (Agilent, Santa Clara, CA). The gene expression microarrays have been previously described (29).
Statistical Analysis
MicroRNA microarray data were log2 transformed and normalized to the mean of each array, and a Wilcoxon rank-sum test was used to identify those microRNAs that were differentially expressed (P < 0.05) between IPF and control lungs; each microRNA has three or four unique probes on the array. Only microRNAs with mean expression values, for each probe, greater than 95% of the negative controls under at least one condition were considered for statistical analysis. Data visualization was accomplished with Genomica (30) and Spotfire Decision Site 8.0 (Spotfire Inc., Göteborg, Sweden). For quantitative real-time polymerase chain reaction (qRT-PCR), statistical significance was determined by Student t test, using P < 0.05. In situ hybridizations were analyzed by Mann-Whitney test to compare let-7d–positive alveolar epithelial cells (AECs) per square millimeter between IPF and control lung samples. Tissue and Masson's trichrome quantitation was performed with MetaMorph (Molecular Devices, Sunnyvale, CA).
Quantitative RT-PCR
TaqMan microRNA assays and gene expression assays (Applied Biosystems, Foster City, CA) were used to determine the relative expression levels of microRNAs and mRNAs, respectively. Throughout this article all genes are referred to by their official symbol and their descriptions and gene IDs are included in Table E2 in the online supplement.
MicroRNA Promoter Analysis
SMAD3- and SMAD4-binding site prediction was performed with the Footer algorithm, using default parameters (31).
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) protocol was performed according to the published protocol from the Young laboratory (32).
Electrophoretic Mobility Shift Assay
Cultured A549 cells at 60–70% confluence were treated with TGF-β1 (2 ng/ml) for 1 hour. Nuclear proteins were isolated by a standard rapid micropreparation technique described previously (33).
Luciferase Reporter Assays
pGL4.17 (Promega, Madison, WI) constructs contained the bp −1600 to +87 region of let-7d at the 5′ end of the reporter gene or the 8-bp Smad3 site deletion (GCTGAGTA) plasmid constructed with a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Eighty percent confluent A549 cells were stimulated for 2 hours with TGF-β (10 ng/ml). Reporter DNA was transfected with Lipofectamine 2000 (Invitrogen) at a 1:1 ratio for 4 hours and cell growth was continued for another 16 hours. Luciferase activity was determined with a dual-reporter assay system (Promega).
In Situ Hybridization
The tissue microarrays were previously described (34). Sixty tissue samples consisting of 40 IPF and 20 control tissues derived from the normal part of lungs removed for benign lesions were studied. The Rembrandt Universal RISH and AP detection kit (Invitrogen) was used. The number of let-7d–positive AECs per square millimeter (in five fields per case) was counted.
Immunohistochemistry
Tissue sections were treated as previously described (35). Anti-HMGA2 rabbit polyclonal antibody (4 μg/ml) (Abcam, Cambridge, MA) and anti-ACTA2 (ab21027; Abcam) were used.
Masson's Trichrome Staining
Sections were stained as per the established protocol (36) by the University of Pittsburgh Transplantation Institute Core Facility. The amount of tissue in each section and the blue stain for collagen were quantified with MetaMorph software (Molecular Devices). To avoid any bias we selected images without airways and blood vessels for our morphometric analyses.
Transfection
Cells were transfected with 50 nM hsa-let-7d inhibitor, pre-let-7d, and their corresponding negative controls (Ambion, Austin, TX), using Lipofectamine 2000 according to the manufacturer's instructions.
Immunofluorescence
A549 cells were transfected with 50 nM anti-let-7d for 48 hours and stained for cytokeratin, VIM, CDH2, and ACTA2. Frozen lung tissue into O.C.T. blocks was used for staining and colocalization of S100A4 or ACTA2 with SFTPC. The detailed protocol is included in the online supplement.
Immunoblotting
RLE-6TN cells were transfected with 50 nM anti-let-7d for 72 hours and immunoblotted for cytokeratin, tight junction protein-1 (TJP1), VIM, and ACTA2. The detailed protocol is included in the online supplement.
Animals
Six- to 12-week-old C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed under pathogen-free conditions. The studies were approved by the Animal Care and Use Committee at the University of Pittsburgh. The design of the let-7d antagomir was adapted from Krutzfeldt and colleagues (37). In the shorter protocol, four mice were treated either with let-7d antagomir (10 mg/kg body weight) administered intratracheally in 50 μl or with an equal volume of saline. In the extended protocol, mice were administered the antagomir on Days 1, 2, 3, 8, 9, 10, 15, 16, and 17 and killed on Day 18. Two control mice were administered the same volume of saline. Lungs from bleomycin-treated mice from previous experiments were used for let-7d determination. The detailed protocol is included in the online supplement.
RESULTS
MicroRNAs Are Differentially Expressed in IPF Lungs
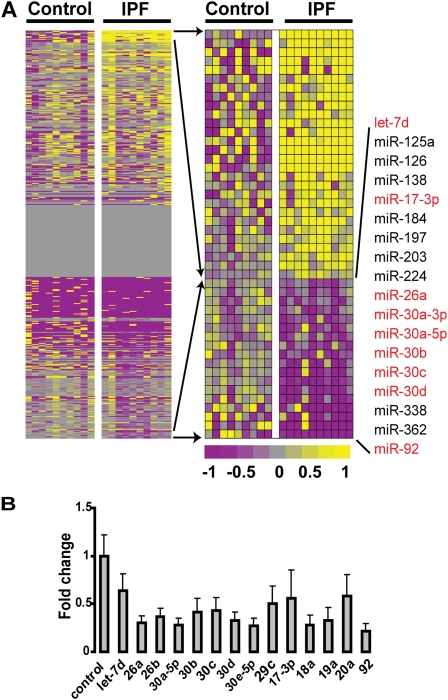
To determine differentially expressed microRNAs in IPF, we analyzed 10 IPF and 10 control lungs on Agilent microRNA microarrays. The complete microRNA microarray data have been deposited in the Gene Expression Omnibus (GSE13316) and is publicly available. Forty-six microRNAs were significantly differentially expressed in IPF lungs. Among the significantly decreased microRNAs in IPF lungs were let-7d, miR-26, and several members of the miR-30 family (Figure 1A), which were also validated by qRT-PCR (Figure 1B). Most of these microRNAs are differentially expressed during development and in various cancers. Analysis of microRNA target databases (TargetScan, miRanda, and PicTar) revealed that down-regulated microRNAs had overlapping targets, some of which were overexpressed in IPF lungs we previously analyzed by gene expression microarrays (29) (Figure E1). The up-regulated microRNAs are listed in Table E1. let-7d expression is also significantly reduced 14 days after bleomycin administration to mice (Figure E2).
Figure 1.
MicroRNAs are differentially expressed in idiopathic pulmonary fibrosis (IPF). (A) The heat map on the left represents global microRNA expression. The heat map on the right represents statistically significant (P < 0.05), differentially expressed microRNAs. Up-regulated microRNAs are shown in progressively brighter shades of yellow, depending on the fold difference, and down-regulated microRNAs are shown in progressively brighter shades of purple. Gray, expression of microRNAs that show no difference between the two groups being compared. The names of the down-regulated microRNAs are provided to the right of the heat map. (B) Quantitative real-time polymerase chain reaction (qRT-PCR) confirmation of the microarray results demonstrates significant changes in all qRT-PCR–measured microRNAs (P < 0.05).
let-7d Expression Is Regulated by SMAD3 Binding to Its Promoter
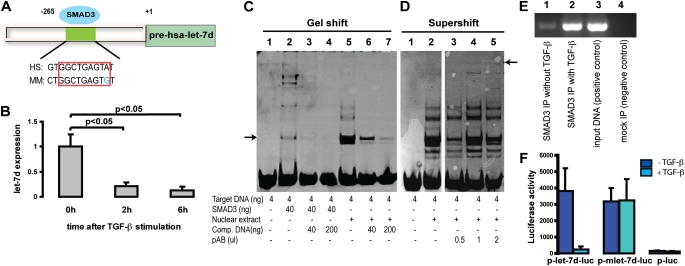
Analysis of the putative promoters of differentially expressed microRNAs by the Footer algorithm (31) revealed a binding site for the TGF-β transcription factor SMAD3 in the region upstream of let-7d (Figure 2A). let-7d expression was significantly suppressed by treatment of A549 cells with recombinant TGF-β1 (P < 0.005, 85% reduction) (Figure 2B). To confirm the computational prediction of SMAD3 binding to the let-7d promoter we performed an electrophoretic mobility shifty assay and ChIP. Incubation of target DNA with either recombinant SMAD3 protein or nuclear extract of A549 cells revealed distinct bands representing the binding of SMAD3 to the let-7d promoter sequence, which diminished in intensity in the presence of increasing concentrations of competitor DNA (Figure 2C). The supershift band representing the DNA–protein–antibody complex was visible with increasing concentrations of SMAD3 antibody (arrow on the right, Figure 2D). SMAD3 ChIP revealed minimal binding of SMAD3 with the let-7d promoter, with a dramatic increase after TGF-β stimulation (Figure 2E). The promoter activity of the 5′ region of let-7d was further analyzed in a luciferase reporter assay (Figure 2F). The 1,687-bp region (positions −1600 to +87) and an 8-bp SMAD3-binding site deletion mutant were amplified by PCR and cloned into the 5′ end of the luciferase gene. The reporter constructs and empty vector controls were transfected into A459 cells with and without TGF-β activation and luciferase activity was measured. The 1,687-bp region increased the average reporter activity by 25-fold. Luciferase activity was reduced to less than 30% in TGF-β–treated cells as compared with untreated controls. This TGF-β–mediated inhibition of luciferase activity was eliminated when we used the 8-bp SMAD3-binding site deletion mutant (Figure 2F). The presented results are representative of at least three experiments. Taken together, our results indicate that TGF-β1 inhibits let-7d expression and that this is mediated through SMAD3 binding to the let-7d promoter.
Figure 2.
let-7d is a transforming growth factor (TGF)-β target molecule. (A) Putative SMAD3 (mothers against decapentaplegic homolog-3)-binding sites identified by the Footer algorithm upstream of hsa-let-7d, and identified to be differentially expressed in idiopathic pulmonary fibrosis (IPF) versus control lung. HS = human sequence; MM = mouse sequence. (B) A549 cells were treated with recombinant TGF-β (3 ng/ml) and let-7d expression was determined 0, 2, and 6 hours after stimulation. Results represent the average expression and SD of triplicate experiments. (C) Electrophoretic mobility shift assay and (D) supershift assay of recombinant SMAD3 protein and nuclear extracts isolated from A549 cells treated with recombinant human TGF-β (2 ng/ml) for 1 hour. Comp. DNA = competitor DNA; pAB = polyclonal antibody. Arrow in C, gelshift band representing the protein-DNA complex; Arrow in D, supershift band representing the DNA-protein-antibody complex. (C) and (D) are from two gels run separately. (E) SMAD3 chromatin immunoprecipitation assay revealed association with let-7d in A549 lung cells. IP = immunoprecipitation. (F) Reporter assays were performed on A549 cells transfected with a recombinant vector containing the base pair (bp) −1600 to +87 let-7d region 5′ to the luciferase gene, referred to as p-let-7d-luc. A similar vector with an 8-bp deletion of the predicted SMAD3-binding site is referred to as p-mlet-7d-luc. The luciferase scale is arbitrary and values represent averages of the triplicate assays. TGF-β stimulation was performed before DNA transfection. The dark blue columns represent the reporter construct without any stimulation, and the light blue columns represent the same with TGF-β stimulation.
let-7d Is Localized to the Alveolar Epithelium in Normal Lungs and Is Significantly Decreased in the Alveolar Epithelium in IPF Lungs
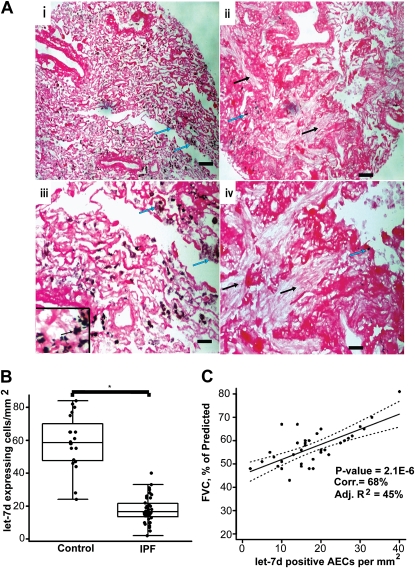
To localize and quantify let-7d expression in the lungs we performed in situ hybridization on tissue microarrays containing 40 IPF tissue and 20 control lung samples previously generated by us (34). Control lungs exhibited abundant expression of let-7d in alveolar epithelial cells (Figure 3A, panels i and iii). In contrast, let-7d expression was almost absent in the alveolar epithelium within areas of fibrotic changes, or adjacent to fibroblastic foci in IPF lungs (Figure 3A, panels ii and iv). let-7d staining was comparable in the bronchial epithelium of both control and IPF lungs. The benefit of tissue arrays is that they allow the analysis of multiple lungs in parallel. We counted let-7d–positive AECs in each tissue core (diameter, 1.5 mm; three in total for each patient). As seen in Figure 3B the number of let-7d–positive cells per square millimeter was significantly lower (P < 0.001) (17.9 ± 7.9) in patients with IPF compared with control lung samples, where 57.2 ± 17.6 AECs per square millimeter were positive for let-7d. Impressively, the number of let-7d–positive cells per square millimeter positively correlated with the FVC, a physiological indicator of disease progression (Figure 3C).
Figure 3.
Tissue microarray analysis reveals that let-7d localizes within normal alveolar epithelium in control lungs and is nearly absent from fibrotic areas in idiopathic pulmonary fibrosis (IPF) lungs. (A) Panels i and iii, let-7d is localized in alveolar epithelial cells of control lungs, as evident by black staining (blue arrows); panels ii and iv, IPF lungs show almost a total absence of let-7d. The black arrows point to areas of dense fibrosis whereas blue arrows point to minimal staining for let-7d in the immediate surrounding areas. Scale bars: panels i and ii, 100 μm; panels iii and iv, 25 μm, in panel iii, 10 μm. (B) Number of let-7d–expressing cells per square millimeter was significantly lower in 40 slides from patients with IPF compared with 20 normal histology controls. *P < 0.001. (C) The forced vital capacity, expressed as a percentage of the predicted value (FVC%), of patients with IPF significantly increased (P = 2.1 × 10−6) with the number of let-7d–expressing cells per square millimeter. AECs = alveolar epithelial cells. Solid line connects the fitted values from a linear regression model. Dotted lines represent 95% confidence intervals on the fitted values.
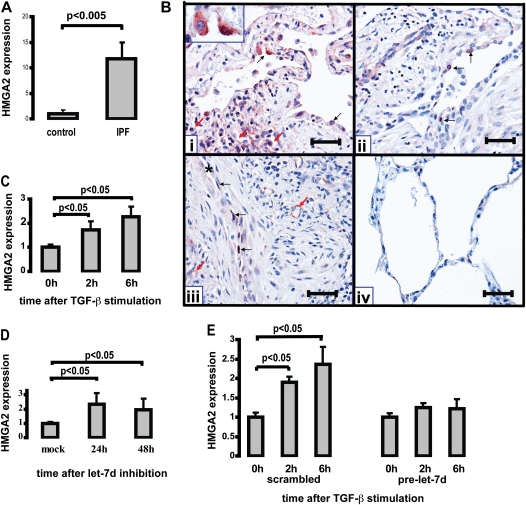
Decreased let-7d Results in Up-Regulation of HMGA2 in Vitro and in Vivo
Because HMGA2 is a known target of let-7 (38) we studied its expression in IPF as a marker of let-7 down-regulation. HMGA2 was significantly overexpressed in IPF lungs compared with controls in our previously published microarray data (27). qRT-PCR performed on the same samples used for microRNA microarrays confirmed the previous microarray data and revealed a 12-fold increase in HMGA2 (P < 0.005) (Figure 4A). Immunohistochemistry revealed expression of HMGA2 in alveolar epithelial cells and in some capillary endothelial cells of IPF lungs but not in control lungs (Figure 4B). To establish a potential relationship between the down-regulation of let-7d and concomitant up-regulation of HMGA2 in IPF lungs we explored whether let-7d was a regulator of TGF-β–induced HMGA2 expression in lung epithelial cell lines. TGF-β stimulation caused a significant increase in HMGA2 (P < 0.005) (Figure 4C). Transfection of A549 cells with a let-7d inhibitor for 24 and 48 hours led to a significant increase in HMGA2 at 24 hours that remained elevated at 48 hours (Figure 4D), an effect that was also observed in RLE-6TN cells. Overexpression of let-7d in RLE-6TN cells by transfection with pre-let-7d prevented TGF-β–mediated induction of HMGA2 (Figure 4E). These results demonstrate that let-7d is a regulator of HMGA2 expression in lung epithelial cell lines and that the increase in HMGA2 after TGF-β stimulation is dependent on inhibition of let-7d by TGF-β. The let-7d inhibitor is specific to the let-7 family of microRNAs (Figures E3A and E3B) but does not affect the levels of unrelated microRNAs (Figure E3C).
Figure 4.
High-mobility group AT-hook 2 (HMGA2) localizes to alveolar epithelial cells in idiopathic pulmonary fibrosis (IPF) lungs and is regulated by transforming growth factor (TGF)-β and let-7 in vitro. (A) HMGA2 mRNA levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR) in 10 control and 10 IPF lungs. (B) Immunolocalization of HMGA2 in IPF lungs (panels i, ii, and iii) and normal lungs (panel iv). Panels i and ii: Two different IPF lungs showing the immunoreactive protein, which was found primarily in alveolar epithelial cells, either in cytoplasm or nuclei (black arrows). The red arrows in panel i point to collapsed airspaces that are lined by HMGA2-positive epithelial cells. Panel iii: The same IPF lung as in panel ii, showing nuclear staining of HMGA2 in elongated epithelial cells (asterisk indicates an alveolar space) and some fibroblast-like cells immersed in a fibroblastic focus. Positive endothelial cells are marked with red arrows. Panel iv: Normal lungs were negative for HMGA2. Scale bars (all panels): 100 μm inset in panel (i) 100x. (C) HMGA2 mRNA levels determined by qRT-PCR in A549 cells at 0, 2, and 6 hours after stimulation with recombinant TGF-β (3 ng/ml). Results represent the average expression and SD of triplicate experiments. (D) HMGA2 mRNA levels determined by qRT-PCR in A549 cells 24 and 48 hours after transfection with 50 nM let-7d inhibitor. (E) HMGA2 mRNA levels in RLE-6TN cells 0, 2, and 6 hours after stimulation with recombinant TGF-β (5 ng/ml) after transfection with pre-let-7d or negative control 24 hours earlier.
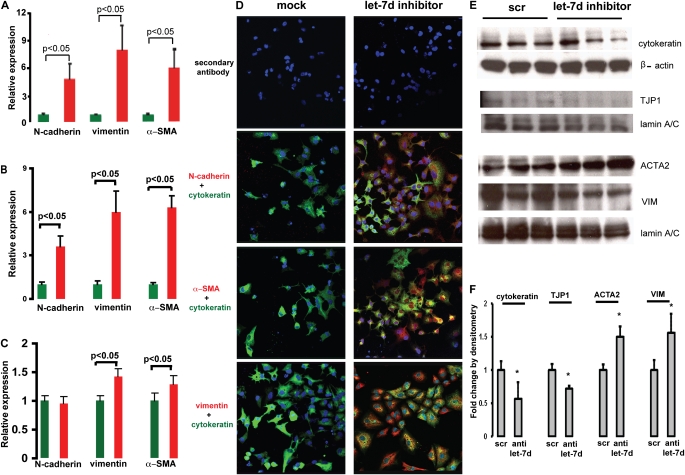
Inhibition of let-7d Causes Expression of Mesenchymal Markers in Lung Epithelial Cell Lines
To determine the potential roles of let-7d inhibition in IPF we studied the effects of let-7d on epithelial cell phenotype. Inhibition of let-7d induced a significant (P < 0.05) increase in expression of the mesenchymal markers CDH2, VIM, and ACTA2 in A549 cells (Figure 5A), RLE-6TN cells (Figure 5B), and primary bronchial epithelial cells (Figure 5C). Inhibition of HMGA2 expression did not fully ablate this effect (Figure E3D), suggesting that it was not completely mediated through HMGA2. Immunofluorescence confirmed the results at the protein level. A549 cells transfected with a let-7d inhibitor stained positive (red) for CDH2, VIM, and ACTA2, 48 hours after transfection, whereas mock-transfected cells did not express positive staining (Figure 5D). RLE-6TN cells transfected with a let-7d inhibitor exhibited increased expression of mesenchymal markers ACTA2 and VIM and decreased expression of the epithelial markers cytokeratin and TJP1 (Figures 5E and 5F) by immunoblotting.
Figure 5.
Inhibition of let-7d results in epithelial–mesenchymal transition changes. N-cadherin (CDH2), vimentin (VIM), and α-smooth muscle actin (ACTA2; α-SMA) mRNA levels were determined by quantitative real-time polymerase chain reaction in (A) A549 cells and (B) RLE-6TN cells 48 hours after transfection and in (C) NHBE cells 24 hours after transfection with 50 nM let-7d inhibitor. (D) Immunofluorescence imaging of A549 cells transfected with 50 nM let-7d inhibitor. Green fluorescence represents cytokeratin, an epithelial marker. Red fluorescence denotes the mesenchymal markers (CDH2, VIM, and ACTA2 [α-SMA]). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Whereas red staining was observed in cells transfected with let-7d inhibitor (right), there was no staining in cells transfected with a control oligonucleotide (left). (E) Western blots of RLE-6TN cells transfected with 50 nM let-7d inhibitor. scr = scrambled sequence; TJP1 = tight junction protein-1. (F) Densitometric analysis of the immunoblots in (E), *P < 0.05. Fold change by densitometry is equivalent to the density of the targeted protein divided by the density of the corresponding housekeeping gene. Results represent averages and SD of triplicate experiments.
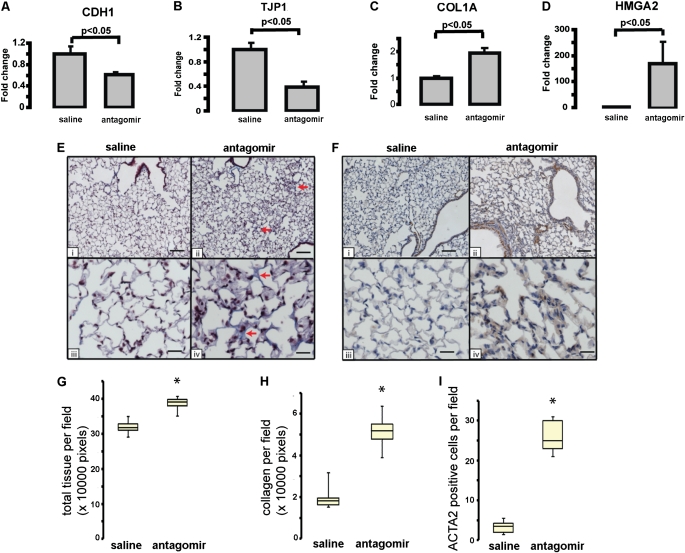
Inhibition of let-7d in Vivo Caused Decreased CDH1 and TJP1 Expression; Increased Collagen, HMGA2, and ACTA2 Expression; and Thickening of Alveolar Septa
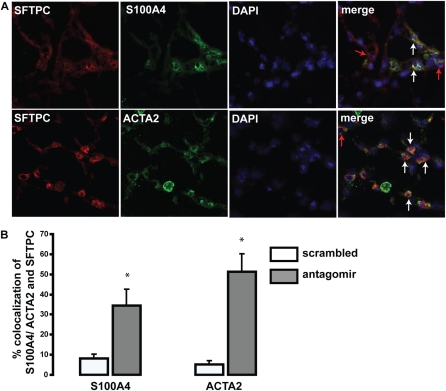
To determine the effects of let-7d inhibition in vivo we administered intratracheally the let-7d antagomir, designed as previously described (37), at a dose of 10 mg/kg body weight into the lungs of four mice on three consecutive days. The mice were killed on the fourth day. Control mice were administered an equal volume of saline. Intratracheal administration of the let-7d antagomir caused a complete knockdown of let-7d in the lungs (Figure E4). let-7d inhibition caused a significant decrease in expression of the epithelial markers CDH1 and TJP1 (Figures 6A and 6B) and a significant increase in COL1A1 and HMGA2 expression in the lungs (Figures 6C and 6D). At this dose the mice appeared sick and the lungs were grossly hemorrhagic, and it was impossible to maintain them longer. To better observe the long-term effects of let-7d inhibition we reduced the dose to 5 mg/kg body weight and treated the animals for 18 days. At this dose we detected a significant decrease in let-7d in the lungs (Figure E4), but the mice did not demonstrate any discomfort. Histological evaluation of formalin-fixed mouse lung demonstrated increased thickening of alveolar septa and Masson trichrome stain revealed increased blue staining indicative of the presence of collagen in let-7d antagomir–treated lungs (Figure 6E). Morphometric analysis confirmed that in the antagomir-treated lungs, the total tissue per field was significantly increased (by 21%) relative to the control sections (Figure 6G). Similarly, quantitation of blue stain revealed a significant (2.6-fold) increase in antagomir-treated lungs (Figure 6H). Immunohistochemical staining for ACTA2 revealed enhanced immunoreactive protein in the alveolar walls of antagomir-treated mouse lungs that was not observed in saline-treated mice (Figure 6F). Manual counting of ACTA2-positive cells revealed a significant increase in ACTA2-expressing cells in the antagomir-treated lungs (Figure 6I). To determine whether the increased ACTA2 was localized in alveolar epithelial cells we colocalized expression of ACTA2, as well the fibroblast-specific marker S100A4, with SFTPC, a marker of alveolar type II cells (Figure 7A). Impressively, colocalization of S100A4 and ACTA2 with SFTPC was observed in, respectively, 38 and 41% of the alveolar epithelial cells in antagomir-treated lungs (Figure 7B). In contrast, control tissues exhibited minimal colocalization (Figure E5). Taken together, these results suggest that in vivo inhibition of let-7 in the lung causes changes consistent with changes in epithelial cell phenotype (increase in ACTA2 and S100A4 expression in SFTPC-expressing cells), as well as a decrease in global lung expression of epithelial markers and increased alveolar septal thickening.
Figure 6.
Effect of let-7 inhibition in vivo by intratracheal antagomir administration. Gene expression levels of (A) E-cadherin (CDH1), (B) tight junction protein-1 (TJP1 [ZO-1]), (C) collagen type 1A (COL1A), and (D) high-mobility group AT-hook 2 (HMGA2) levels in mice (n = 4) treated with intratracheal antagomir (10 mg/kg) or saline for 4 days. In both (E) and (F), panels i and iii are of saline-treated lungs and panels ii and iv are of antagomir-treated lungs. Scale bars: i and ii, 100 μm; iii and iv, 25 μm. (E) Masson trichrome staining after 18 days of saline or antagomir treatment. Arrows in ii and iv point to the blue staining for collagen. Note alveolar septal thickening. (F) Immunolocalization of α-smooth muscle actin (ACTA2). ACTA2 staining (brown) is significantly increased in antagomir-treated mice compared with saline-treated controls. (G) Quantitation of the total tissue in control and antagomir-treated lungs. Ten different fields (original magnification, ×20) from each slide were quantified, *P < 0.05. (H) Quantitation of collagen in control and antagomir-treated lungs. Ten different fields (original magnification, ×20) from each slide were quantified, *P < 0.05. (I) Number of ACTA2-expressing cells stained brown was significantly higher in antagomir-treated lungs in comparison with control lungs, *P < 0.05.
Figure 7.
Colocalization of α-smooth muscle actin (ACTA2) and S100A4 with SFTPC (surfactant, pulmonary-associated protein C). (A) The green fluorescence represents the mesenchymal markers ACTA2 and S100A4. The red fluorescence denotes the alveolar type II cell marker SFTPC. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. The merged image shows colocalization of S100A4 and SFTPC (top) and ACTA2 and SFTPC (bottom). The white arrows point to cells showing colocalization. The red arrows represent cells that express only SFTPC. (B) Relative quantitation of the colocalization of SFTPC (red) and ACTA2/ S100A4 (green) in scrambled control and antagomir-treated lungs. The results represent an average of four different fields per slide and three slides per group.
DISCUSSION
In this study we explored the expression, regulation, and potential role of microRNAs in IPF. Among the significantly decreased microRNAs in IPF lungs were let-7d, miR-26, and several members of the miR-30 family. We focused on let-7d and discovered that it was directly transcriptionally inhibited by the key profibrotic cytokine TGF-β. In vitro inhibition of let-7d induced an increase in mesenchymal markers in lung epithelial cell lines, and in vivo inhibition of let-7d in mouse lungs caused alveolar septal thickening, decreased expression of epithelial markers, and an increase in collagen and expression of ACTA2 and S100A4 in SFTPC-expressing alveolar epithelial cells. In IPF lungs let-7d expression was drastically diminished, whereas HMGA2 expression was increased in alveolar epithelial cells. Increased expression of HMGA2 was induced by let-7d inhibition in vitro and TGF-β–induced expression of HMGA2 was inhibited by let-7d expression, suggesting that the concomitant decrease in let-7d and increase in HMGA2 in human lungs were associated. Taken together, our findings suggest that let-7d inhibition is a key regulatory event in the dramatic phenotypic changes that happen in the alveolar epithelium in IPF (39).
The let-7 family of microRNAs was one of the first discovered (40) and is the most extensively studied; however, this is the first time a member of the let-7 family has been implicated in a nontumor disease. Members of the let-7 family are temporally regulated microRNAs that coordinate developmental timing (40). They are considered tumor suppressors, regulating RAS and many cell cycle genes (41), and their expression is supposed to sustain an “epithelial” gene signature (42). Among the genes suppressed by the let-7 family is HMGA2, a structural transcriptional regulator expressed in early embryonic development, some benign tumors, and lung cancer (38). In the context of IPF it is of interest to note that HMGA2 confers a growth advantage to fibroblasts as evident by the retarded growth in hmga2-deficient mouse embryonic fibroblasts compared with wild-type fibroblasts (43) and is also a mediator of TGF-β–induced EMT (11). HMGA2 facilitates transcription of SNAI1 and TWIST, the transcriptional repressors of adherens, tight, and desmosomal junction components, leading to reduced intercellular adhesions. However, although we found a dramatic decrease in let-7d expression accompanied by an increase in HMGA2 in human IPF lungs, and demonstrated that inhibition of let-7d leads to EMT in vitro and to early fibrotic changes in vivo, we do not have direct evidence that this effect is mediated solely through HMGA2. A more realistic interpretation at this stage would be that in the lungs, down-regulation of let-7d leads to up-regulation of HMGA2 as well as other fibrosis-relevant targets of let-7 such as RAS, insulin-like growth factor-1 (IGF1), and IGF1 receptor (IGF1R) (Figure E1A) and thus leads to the profound and sustained changes in cellular phenotype that are indeed observed in IPF. The fact that let-7 inhibition is sufficient to induce changes consistent with EMT in vitro and expression of mesenchymal markers as well as thickening of alveolar septum in vivo also suggests an effect mediated through modification of the expression of multiple fibrosis-relevant genes and not just a single gene.
Our results add to previous results that implicate a role for inhibition of microRNAs in EMT (14–17). The miR-200 family prevents EMT by targeting the transcription factors ZEB1 and SIPI that repress E-cadherin, an epithelial cell marker (14–17). Similar to let-7d, TGF-β stimulation also decreased expression of the miR-200 family. Overexpression of the miR-200 family could not completely block EMT (15), suggesting that let-7d and the miR-200 family may be acting synergistically, possibly with a few other microRNAs, in preventing EMT. Although the current study does not directly overlap the putative pathway of the miR-200 family, there is no doubt that there may be multiple signaling cascades at work in a complex cellular transition such as EMT. In addition, it is possible that in different organs or during different embryonic stages different microRNAs may have the role of maintaining the epithelial cell phenotype. In this context, it is important to note that although our article focuses on let-7d, the downstream effects of let-7d overexpression or inhibition are probably common to all members of the let-7 family of microRNAs. Although the inhibitory effects we describe do not go through microRNAs that are not members of the let-7 family it is highly unlikely that this effect will be mediated only through let-7d and not through other members of the family.
In this study we demonstrate the influence of TGF-β, the key profibrotic cytokine and a regulator of EMT (44), on the expression of a microRNA through direct effect on its promoter. This result is consistent with the genome-scale analysis of microRNA promoters and evidence for binding by stem cell transcription factors as well as RNA polymerase II, which suggest that the expression of many microRNAs is under dynamic transcriptional regulation by transcription factors (45, 46). So far, changes in let-7 microRNA expression levels have been attributed mostly to their localization to fragile sites (47), to post-transcriptional modification (48), or to epigenetic changes (49). Our discovery that let-7d is directly inhibited by TGF-β is consistent with previous observations that suggested a SMAD3-mediated inhibitory effect on gene expression (50–53). More importantly, it adds a new potential mechanism for regulation of microRNAs and may have important therapeutic implications as inhibitors of TGF-β signaling and activation are currently evaluated for cancer and fibrosis. Furthermore, the suggestion that some of the effects of TGF-β on epithelial cells may be mediated through microRNAs may have significant implications leading to a better understanding of the profound and sustained effects of TGF-β on cell and organ phenotype.
In summary, we demonstrate that IPF lungs are different from control lungs in their microRNA repertoire. We discovered that let-7d, a microRNA significantly down-regulated in IPF, is negatively transcriptionally regulated by TGF-β. Furthermore, inhibition of let-7d alone is sufficient to cause EMT changes in A549, RLE-6TN, and NHBE cells. Concomitant with let-7d down-regulation in IPF lungs, we found significant increases in lung expression of HMGA2, suggesting that let-7 inhibition of HMGA2 is released in IPF lungs. Last, we demonstrate that let-7 inhibition in vivo in the lungs may cause changes in the lung alveolar epithelium with increases in mesenchymal markers, decreases in epithelial markers, and thickening of alveolar septa. The discovery of the down-regulation of let-7d in IPF, its regulation by TGF-β, and its potential role in EMT and fibrosis may have important implications in our understanding of the molecular mechanisms underlying IPF, and may also lead to the development of new therapeutic interventions in this devastating and incurable disease.
Supplementary Material
Acknowledgments
The authors thank Dr. Zohar Yakhini and Dr. Mark Gladwin for useful discussions and comments.
Supported by NIH grants LM009657, HL073745, HL0894932, and RC2HL101715 and by the Dorothy P. and Richard P. Simmons Endowed Chair for Pulmonary Research. M.S. and A.P. were supported by a Universidad Nacional Autónoma de México grant SDI.PTID.05.6. MicroRNA arrays were part of an unrestricted gift from Agilent Technologies.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200911-1698OC on April 15, 2010
Conflict of Interest Statement: K.V.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; D.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; H.Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; M.Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; A.T. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; K.F.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; K.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; S.A.Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; M.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; D.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; T.R. received more than $100,001 from Biogen Idec, Inc. in industry-sponsored grants; M.S. received $5,001–$10,000 from Boehringer in consultancy fees; S.C.W. received more than $100,001 from the NIH in sponsored grants; A.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; A.B.-Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; D.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; O.E. received more than $100,001 from Ergonex Pharma in industry-sponsored research grants; P.R. received more than $100,001 from Centocor, Inc. in industry-sponsored grants; P.V.B. holds up to $1,000 in stocks in Sequenom; N.K. received $1,001–$5,000 from Stromedix and $1,001–$5,000 from Genentech in consultancy fees, holds patents from the University of Pittsburgh for the use of microRNAs in treatment and diagnosis of IPF, peripheral blood biomarkers in IPF, and urinary biomarkers in IPF.
References
- 1.Selman M, King TE, Pardo A. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 2001;134:136–151. [DOI] [PubMed] [Google Scholar]
- 2.American Thoracic Society, European Respiratory Society. Idiopathic pulmonary fibrosis: diagnosis and treatment [international consensus statement]. Am J Respir Crit Care Med 2000;161:646–664. [DOI] [PubMed] [Google Scholar]
- 3.Katzenstein AL, Myers JL. Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med 1998;157:1301–1315. [DOI] [PubMed] [Google Scholar]
- 4.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med 2008;5:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, et al. Aberrant Wnt/β-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol 2003;162:1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konigshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One 2008;3:e2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, et al. Wnt1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest 2009;119:772–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vuga LJ, Ben-Yehudah A, Kovkarova-Naumovski E, Oriss T, Gibson KF, Feghali-Bostwick C, Kaminski N. WNT5A is a regulator of fibroblast proliferation and resistance to apoptosis. Am J Respir Cell Mol Biol 2009;41:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial–mesenchymal transition in alveolar epithelial cells by transforming growth factor-β1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. J Clin Invest 2009;119:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-β employs HMGA2 to elicit epithelial–mesenchymal transition. J Cell Biol 2006;174:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willis BC, Borok Z. TGF-β–induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol 2007;293:L525–L534. [DOI] [PubMed] [Google Scholar]
- 13.Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci USA 2006;103:13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008;9:582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008;283:14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 19.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001;294:858–862. [DOI] [PubMed] [Google Scholar]
- 20.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258–267. [DOI] [PubMed] [Google Scholar]
- 22.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 2009;182:4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA 2009;106:18704–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 2008;3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci USA 2002;99:6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yousef H, Corcoran DL, Handley D, Pandit KV, Ben-Yehudah A, Eickelberg O, Benos PV, Kaminski N. let7d—a microRNA at the junction of TGF-β signaling and epithelial–mesenchymal transition [abstract]. Am J Respir Crit Care Med 2008;177:A303. [Google Scholar]
- 27.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, Lokshin AE, Lindell KO, Cisneros J, Macdonald SD, Pardo A, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med 2008;5:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, Bisceglia M, Gilbert S, Yousem SA, Song JW, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2009;180:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet 2004;36:1090–1098. [DOI] [PubMed] [Google Scholar]
- 31.Corcoran DL, Feingold E, Dominick J, Wright M, Harnaha J, Trucco M, Giannoukakis N, Benos PV. Footer: a quantitative comparative genomics method for efficient recognition of cis-regulatory elements. Genome Res 2005;15:840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 2002;298:799–804. [DOI] [PubMed] [Google Scholar]
- 33.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 1991;19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzouvelekis A, Harokopos V, Paparountas T, Oikonomou N, Chatziioannou A, Vilaras G, Tsiambas E, Karameris A, Bouros D, Aidinis V. Comparative expression profiling in pulmonary fibrosis suggests a role of hypoxia-inducible factor-1α in disease pathogenesis. Am J Respir Crit Care Med 2007;176:1108–1119. [DOI] [PubMed] [Google Scholar]
- 35.Selman M, Ruiz V, Cabrera S, Segura L, Ramirez R, Barrios R, Pardo A. Timp-1, -2, -3, and -4 in idiopathic pulmonary fibrosis: a prevailing nondegradative lung microenvironment? Am J Physiol 2000;279:L562–L574. [DOI] [PubMed] [Google Scholar]
- 36.Gomori G. A rapid one-step trichrome stain. Am J Clin Pathol 1950;20:661–664. [DOI] [PubMed] [Google Scholar]
- 37.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with “antagomirs.” Nature 2005;438:685–689. [DOI] [PubMed] [Google Scholar]
- 38.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007;315:1576–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc 2006;3:364–372. [DOI] [PubMed] [Google Scholar]
- 40.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901–906. [DOI] [PubMed] [Google Scholar]
- 41.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007;67:7713–7722. [DOI] [PubMed] [Google Scholar]
- 42.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 2007;104:11400–11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995;376:771–774. [DOI] [PubMed] [Google Scholar]
- 44.Nawshad A, Lagamba D, Polad A, Hay ED. Transforming growth factor-β signaling during epithelial–mesenchymal transformation: implications for embryogenesis and tumor metastasis. Cells Tissues Organs 2005;179:11–23. [DOI] [PubMed] [Google Scholar]
- 45.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One 2009;4:e5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008;134:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006;20:2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res 2007;67:1419–1423. [DOI] [PubMed] [Google Scholar]
- 50.Lacerte A, Korah J, Roy M, Yang XJ, Lemay S, Lebrun JJ. Transforming growth factor-β inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal 2008;20:50–59. [DOI] [PubMed] [Google Scholar]
- 51.Uchiyama Y, Guttapalli A, Gajghate S, Mochida J, Shapiro IM, Risbud MV. SMAD3 functions as a transcriptional repressor of acid-sensing ion channel 3 (ASIC3) in nucleus pulposus cells of the intervertebral disc. J Bone Miner Res 2008;23:1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall MC, Young DA, Waters JG, Rowan AD, Chantry A, Edwards DR, Clark IM. The comparative role of activator protein 1 and Smad factors in the regulation of Timp-1 and MMP-1 gene expression by transforming growth factor-β1. J Biol Chem 2003;278:10304–10313. [DOI] [PubMed] [Google Scholar]
- 53.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 2002;110:19–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.