Abstract
The Galacto-, Homoserine-, Mevalonate-, Phosphomevalonate-kinase (GHMP) superfamily encompases a wide-range of protein function. Three members of the family (mevalonate kinase, phosphomevalonate kinase and diphosphomevalonate decarboxylase) comprise the mevalonate pathway found in S. pneumoniae and other organisms. We have determined the 1.9 Å crystal structure of phosphomevalonate kinase (PMK) from S. pneumoniae in complex with phosphomevalonate and AMPPNP·Mg2+. Comparison of the apo and ternary PMK structures suggests that ligand binding reverses the side-chain orientations of two anti-parallel lysines residues (100 and 101) with the result that lys101 is “switched” into a position in which its ammonium ion is in direct contact with the β,γ-bridging atom of the nucleotide, where it is expected to stabilize both the ground and transition states of the reaction. Analysis of all available GHMP kinase ternary-complex structures reveals that while their Cα-scaffolds are highly conserved, their substrates bind in one of two conformations, which appear to be either reactive or non-reactive. The active site of PMK seems spacious enough to accommodate interconversion of the reactive and nonreactive conformers. A substantial fraction of the PMK active site is occupied by ordered water, which clusters near the charged regions of substrate. Notably, a water pentamer that interacts extensively with the reactive groups of both substrates was discovered at the active site.
Keywords: phosphomevalonate kinase, mevalonate pathway, isoprenoid, structure
The isoprenoid family of compounds contains roughly 25,000 distinct natural products (1, 2), and the classes within the family are often chemically sufficiently diverse to provide the extensive functional overlap often seen in robust metabolic systems. Ubiquinones, the electron carriers of redox chains, are one such class (3); the side-chains that functionalize heme rings are another (4); the isoprenoid tethers that control localization and function of proteins (5, 6) and tRNA (7-9) are a third; carotenoids (10), cholesterol (11), steroid hormones (12)), bile acids (13) - all of these and more fall within the family.
Eukaryotes, archea and a small number of bacteria convert mevalonate to isopentenyl diphosphate, the “building block” of isoprenoids, via the mevalonate pathway (Figure 1) (2, 14), which consists of three enzymes - mevalonate kinase (MK), phosphomevalonate kinase (PMK), and diphosphomevalonate decarboxylase (DPM-DC). Each of these enzymes is a member of the GHMP kinase superfamily (15), the N- and C-terminal sequence-signatures of which appear in ∼ 0.75 % of the sequences in the UniProt database (16).
Figure 1. The Mevalonate Pathway.
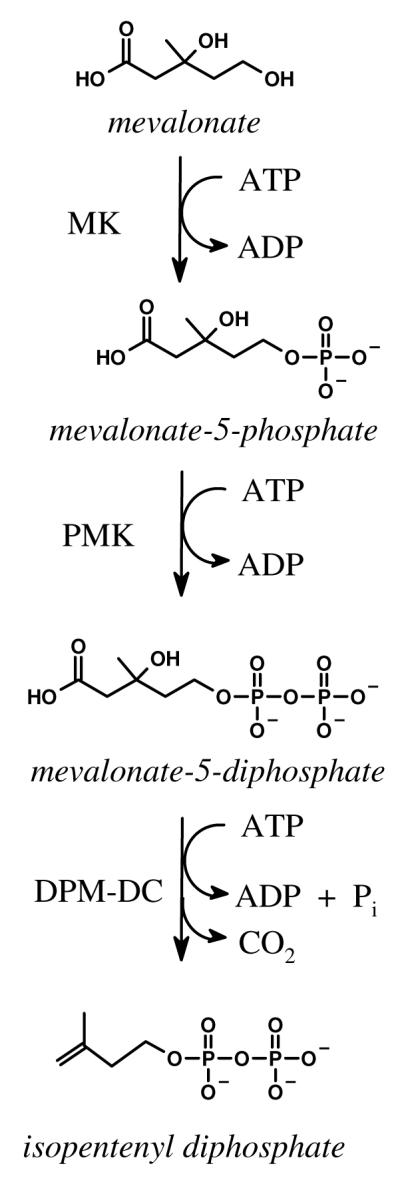
The acronyms: MK, mevalonate kinase (ATP: (R)-mevalonate phosphotransferase, 2.7.1.36), PMK, phosphomevalonate kinase (ATP: (R)-phosphomevalonate kinase, 2.7.4.2), and DPM-DC, diphosphomevalonate decarboxylase (ATP:(R)-5-diphosphomevalonate carboxy-lyase, 4.1.1.33).
Given the pervasive role of isoprenoids in metabolism, it is not surprising that imbalances in isoprenoid synthesis and utilization can lead to cellular dysfunction and disease. Controlling flux through these pathways is used to prevent and cure human disease — statins, which decrease cellular levels of mevalonate by inhibiting HMG-CoA reductase (17, 18), are used widely to control cholesterol levels in humans (19), and may prove useful in the treatment of other diseases (20-24).
The mevalonate pathway is essential for the survival of Streptococcus pneumoniae in lung and serum (25, 26). PMK produces an allostere, diphosphomevalonate, that feedback-inhibits mevalonate kinase. This regulation occurs in S. pneumoniae, but not in humans (27), and is it hoped that careful study of the mevalonate pathway enzymes will provide the opportunity to intervene therapeutically in novel ways to prevent the devastating effect of this organism, which is estimated to kill ∼ 3700 people daily, worldwide (28). Here we present the 1.9 Å X-ray structure of PMK in a ternary complex with phosphomevalonate and AMPPNP and compare it to the structures of the non-liganded form of the enzyme and other members of the family to assess its mechanism of action and structural traits of the family.
Materials and Methods
AMPPNP, MES, MgCl2, polyethylene glycol 4000, were purchased from Sigma-Aldrich. Phosphomevalonate was synthesized enzymatically and purified as described previously (29). The expression, in E. coli, and purification of recombinant phosphomevalonate kinase from S. pneumoniae has been described in detail (30).
Crystals of PMK in complex with Pmev and AMPPNP were prepared under silicon oil using the sitting drop method by mixing 2 μl of 12 mg/ml PMK in 25 mM Hepes/K+, pH 7.5, 0.25 mM Pmev, 8 mM AMPPNP, 10 mM MgCl2, with 2 μl of 36 % (w/v) polyethylene glycol 4000, 100 mM (MES/Na+), pH 6.0. The crystals were briefly transferred to a solution containing 36% polyethylene glycol 4000, 100 mM MES, pH 6.0 and 20% ethylene glycol prior to cooling to 125K. The crystals belonged to space group P21212 (a = 69.0, b = 115.3, c = 40.0 Å) and diffracted x-rays to 1.9 Å resolution using an in-house Rigaku R-Axis IV++ image plate detector and RU-H3R rotating anode x-ray generator equipped with Osmic Blue optics and operating at 50 kV and 100 mA. The data were reduced with HKL (31).
The structure was solved by molecular replacement with EPMR (32), using as the search probe a single subunit of apoenzyme S. pneumoniae PMK (1K47). Model building and atomic parameter refinements were carried out with ARP/wARP (33, 34), O (35) and CNS (36), employing thermal factor restraints. The Ramachandran plot of main chain torsion angle pairs places Tyr 16 in the disallowed region and Asp 297 in the generously allowed region (37, 38).
Results and Discussion
Overall Structure of PMK
The crystal structure of PMK was determined to 1.9 Å resolution in ternary complex with the substrate Pmev, the non-hydrolyzable nucleotide analog AMPPNP and Mg2+ (Fig. 2A, 3). Its overall conformation is very similar to that of the apoenzyme from the same source with the exception of several conformational changes that can be directly attributed to ligand binding. Unlike the apoenzyme, which crystallized in a hexameric arrangement, the enzyme studied here crystallizes as a monomer, in agreement with gel filtration results (data not shown).
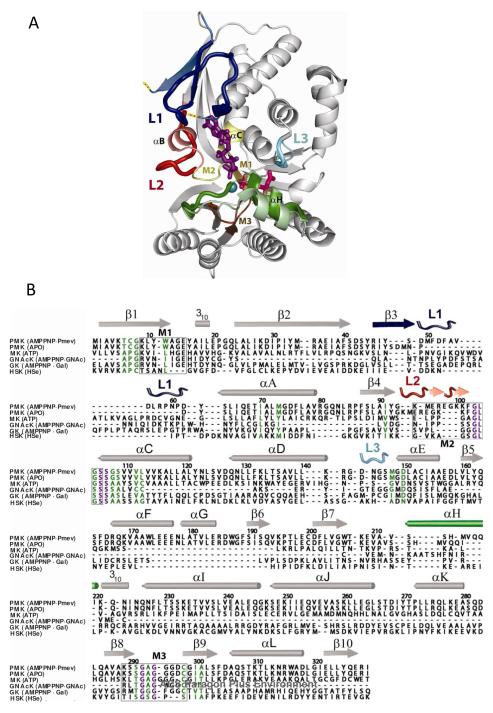
Figure 2. Structure and sequence alignments of Sp PMK.
(A) Superposition of the apo and ternary complex structures reveals the changes that occur upon binding of ligands. Regions insensitive to ligand are gray, and the four responsive elements (L1, L2 and L3, αH) are colored in blue, red, cyan, and green, respectively — the more intense colors are associated with the ternary complex. The active site contains Pmev (magenta), AMPPNP (purple), and Mg2+ (turquoise). The structures of the three conserved sequence motifs originally used to define the GHMP superfamily are highlighted in light brown (M1), pale yellow (M2), and dark brown (M3). Yellow dots indicate where the missing/disordered segment of Loop 1 attaches to the apoenzyme. (B) Structure-based sequence alignment of ternary/apo Sp PMK and representative members of the GHMP kinase superfamily. From top to bottom, the structures include the ternary (2pg9) and apo (1k47) forms of PMK from Streptococcus pneumoniae, ATP-bound mevalonate kinase from Rattus norvegicus (1kvk), the ternary complexes of n-acetygalactose kinase (2a2d) and galactose kinase (1wuu) from Homo sapiens, and homoserine-bound homoserine kinase from Methanococcus jannaschii (1h72). Invariant residues are purple and conserved residues (>60% similarity) are green. The nomenclature for secondary structural elements of ternary PMK, which are colored according to the scheme in (A), is identical to that assigned to apo-PMK (53). Boxes surround motifs M1-3. Panels A and B were produced using Pymol (54) and Alscript (55), respectively.
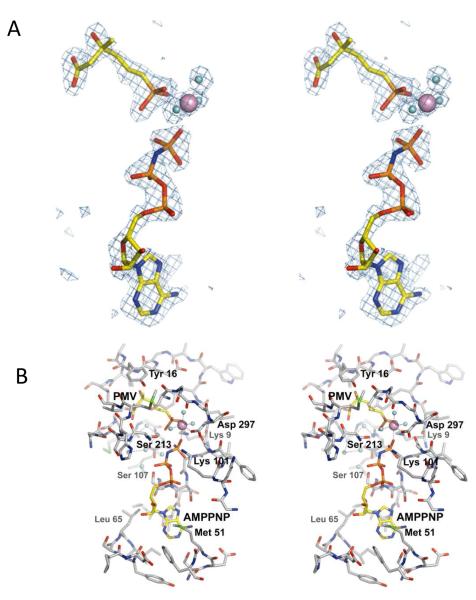
Figure 3. Conformational Changes of SpPMK.
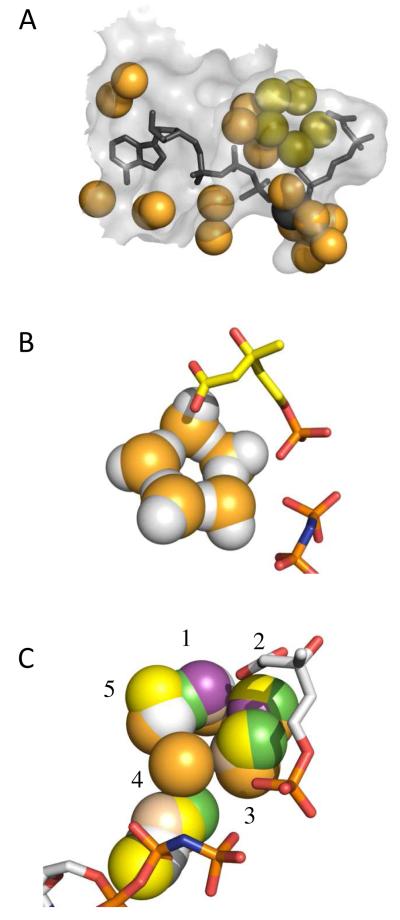
(A) Stereoview of the omit Fo-Fc electron density (3.0 sigma level) surrounding ligands at the active site. (B) Stereo representation of the active site of SpPMK in complex with Pmev, AMPPNP and Mg2+. The dotted line segments depict the octahedral coordination of the Mg2+ ion as well as the interactions between the pentamer of water molecules that comprise a portion of the PMK binding site.
Substrate-Binding Induced Active-Site Conformational Changes
A comparison of the ternary complex and apo structures of PMK reveals the conformational changes that occur upon binding of ligands. Loops L1-3 and helix αH change noticeably. The Pmev phosphoryl-group is hydrogen bonded to both Lys 9 and the main-chain amide of Ala 293, while its carboxyl group is hydrogen bonded Ser 147. The isopentenyl body forms extensive hydrophobic interactions with Tyr 11, Tyr 16, Ile 18 and Leu 19 (Fig. 3B). Initial-rate kinetic experiments indicate that ground-state interaction between ligand and Lys 9, a highly conserved residue in the GHMP kinase family, is focused nearly exclusively on the phosphoryl-moiety of Pmev; this residue is also involved in other, as yet undefined, kcat-related steps (30). Consistent with these initial-rate findings, the structure shows the side chain of Lys 9 interacting with a phosphoryl-oxygen of Pmev (2.8 Å) and the γ-phosphoryl group of AMPPNP. Upon formation of the ternary complex, αH, the so-called “mobile” helix (30), shifts to enclose Pmev, resulting in hydrophobic contacts with Pmev (Val 217), and hydrogen bonding (Ser 213) with the terminal phosphoryl oxygens of both substrates. A 2.8-Å displacement of the nearby L3 loop appears to stabilize the position of αH through interactions of the L3, Asn 145 side chain with Ser 214, Val 217 and Gln 218 of αH.
AMPPNP interacts with both stationary and mobile regions of the PMK scaffold. The stationary region includes Ile 65 and Leu 66 as well as the glycine/serine-rich region (F102GLGSSGLV110) of motif II. The mobile regions include loops L1 and L2, which reorganize considerably to accommodate the nucleotide. L1, which is disordered over a 9-residue stretch in the apoenzyme, assumes an extended conformation in the ternary complex that places Ser 48 in hydrogen-bonding contact with adenine N1, and transfers Pro 61 and Asp 62 from a position that blocks adenine binding to one that promotes a solvent-mediated interaction between the Asp 62 carbonyl oxygen and both adenine N3 and ribose O2. Interestingly, the adenine binding site is similarly disordered in the diphosphomevalonate-bound structure of mevalonate kinase, in which the nucleotide site is empty and the acceptor site is filled. Thus, acceptor binding appears to be insufficient to reorganize the ATP-binding pocket, which is consistent with the lack of synergy in the steady-state affinities of Pmev and ATP (29). Notably, other GHMP superfamily members (galactokinase (2cz9), 4-cytidine 5′-diphospho-2C-metyl-D-erythritol kinase (1uek), and homoserine kinase (1h72)) present an open, solvent-filled framework that remodels upon ternary complex formation (39-43). It should also be noted that the presence of a crystal packing interface comprising N- and C-terminal segments of the apo-PMK L1 loop tempers predictions regarding the extent of L1 disorder under solution conditions.
Ligand Binding Repositions Lys 101
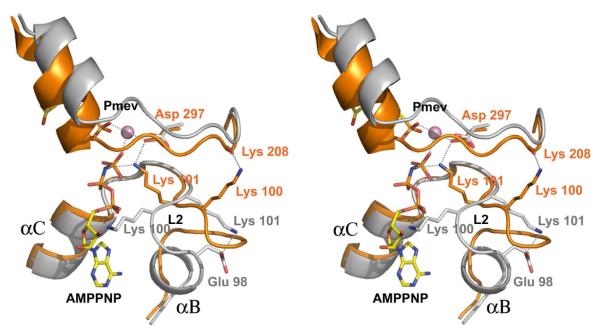
Ligand binding fosters an L2 conformational change that delivers the primary ammonium ion of Lys 101 into Van der Waals contact with the α,β-bridging atom of the nucleotide tripolyphosphate chain where it can electrostatically stabilize both nucleotide binding and the development of negative charge that is expected to occur at the bridging atom in the presumed dissociative transition state (44) (Fig. 4). In the apoenzyme, Lys 100 and Lys 101 reside on a small stretch of random coil that connects the αC helix to the αB helical-turn. The lysine side chains adopt extended conformations and point away from one another, as if to maximize separation of their ammonium groups (13.9 Å). In the apo form Lys 101 is predicted to obstruct nucleotide binding, and is tethered by a salt-bridge to the carboxylate of Glu 98. Upon binding of ligand, the salt bridge is broken, the αB helix unravels, and the lysine side chains “swing” past one another into a conformation that is roughly the opposite of that seen in the apo form - Lys 101 now projects into the binding pocket and is in direct contact with the β,γ-bridging atom of AMPPNP, and Lys100 points away from the pocket and appears to be stabilized by an interaction with the backbone carbonyl oxygen of Lys 208. Thus, ligand binding is linked to the switching of Lys 101, a highly conserved residue among bacterial phosphomevalonate kinases, between what appear to be catalytic and non-catalytic positions.
Figure 4. The Active Site of PMK.
Ribbon-and-stick stereo superposition of the PMK active site corresponding to apoenzyme (gray) and the complex with ligands (orange). Figures were produced using PyMol (54).
A non-reactive ternary complex?
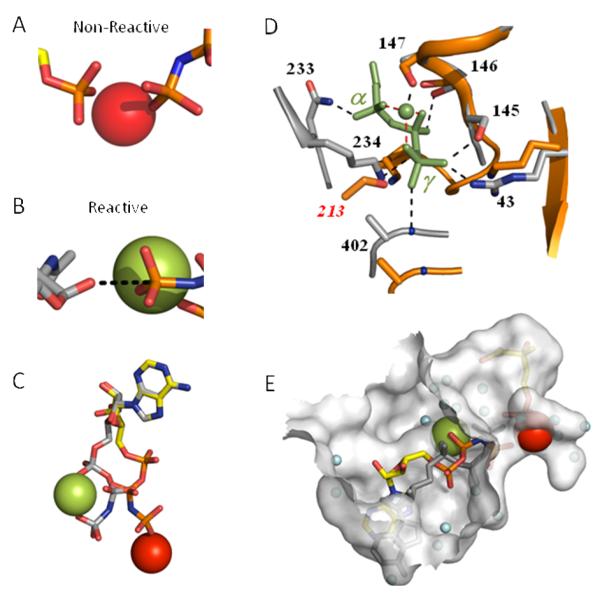
The juxtaposition of the γ-phosphoryl group of AMPPNP and non-bridging phosphoryl-oxygens of Pmev in the ternary complex suggests that these groups must reorient to undergo in-line displacement chemistry, Fig. 5A. It is possible to achieve a reactive geometry by adjusting P-O-P torsion angles; however, doing so appears to require that both reactive groups be displaced from the first-coordination sphere of the metal ion. While plausible, a comparison of the available ternary-complex GHMP kinase structures offers an alternative interpretation. The PDB lists a total of six ternary-complex structures in addition to that of PMK. Three of these exhibit the “non-reactive” configuration seen in PMK (Type I, Fig. 5A); in the remaining three, the reactants are well positioned to accomplish in-line displacement (Type II, Fig. 5B). Notably, erythritol kinase (1uek) exhibits both Type I and II configurations. The fact that either or both of the Type I and II conformers are found distributed roughly evenly across these seven highly structurally conserved active sites suggests that the ability to bind either Type I or II complexes is an intrinsic property of the family.
Figure 5. Type I and II ligand conformations in GHMP kinase ternary structures.
(A) The Type I conformation. The positioning of the γ- and Pmev-phosphoryl groups in the PMK ternary complex (shown on the left and right-hand sides of the panel, respectively) suggests that they are non-reactive. Similiar conformations are found in three of the seven GHMP-kinase ternary complex structures. (B) The Type II conformation. Positioning of the reactive moieties in the ternary complex of Gal-NAc kinase — a GHMP kinase. The Gal-NAc nucleophillic oxygen and the γ-phosphoryl group of AMPPNP appear to be positioned well to accomplish in-line displacement. This orientation is also found in three of the seven GHMP kinase ternary complex structures. (C) Alignment of the Type I and II M2+·nucleotides. The adenine bases of the Type I and II nucleotides were aligned. The PMK nucleotide color coding: carbon (yellow), phosphorus (orange), divalent cation (red); the Gal-NAc kinase nucleotide color coding: carbon and phosphorus (grey), divalent cation (green). (D) Comparison of the Type I and II active-sites. Resides (grey) within 3 Å of the PPPi·Mg2+ moiety (green) of Glc-NAc kinase complex (Type II) are linked to the moiety by blank dotted lines. Analogous residues in the PMK complex (Type I) are shown in orange. Dashed red lines connect the divalent cation to the α, β and γ-phosphoryl groups to which it is coordinated. (E) Positioning the Type II nucleotide·M2+ in the Type I active site. The Type I and II active-sites were superposed using Cα-alignments of conserved regions of PMK and Gal-NAc kinase. The GalNac nucleotide (Type II) is shown in grey and its associated cation (Mn2+) is red. The transition from Type I to II requires the cation to migrate across the active-site cavity. Blue spheres represent ordered water. Figures were produed using PyMol (54) and the surface, shown in E, was created with Hollow (56).
To assess the potential of PMK to accommodate the Type II conformer, the structure of the PMK active site was compared to that of the human GalNAc kinase (45) - an example of a Type II ternary complex. Consistent with the conservation of the protein regions with which they interact, the adenosine moieties of the Type I and II nucleotides exhibit similar conformations; the conformations of the tripolyphosphate·M2+ moieties, however, are very different (see Fig. 5C). When the adenine bases are superposed, it is clear how the C4′-pucker and tripolyphosphate dihedral angles must reorient to align the nucleotide, and that such an alignment causes the divalent cation to migrate (perhaps as part of dihedral rotation) “across” the nucleotide, ultimately to embed in the opposing wall of the active site (Fig. 5E). Conservation of the Type II residues in contact with PPPi-Mg2+ in the Type I PMK structure was evaluated by aligning the Gal-NAc kinase contact residues with the corresponding resides in PMK (Fig. 5D). The seven Gal-NAc kinase residues (grey) containing atoms within 3 Å of the PPPi-Mg2+ moiety (green) are linked to it by dotted lines. Of these seven, five are spatio-chemically conserved in PMK. Of the remaining two, Ser145, which forms a hydrogen bond with the γ-P, is not conserved spatially (Gly 105 occupies this position in PMK) but its function can be provided by PMK Ser213, which, although it approaches the γ-P from the “opposite” side, is also well positioned to H-bond to it. The PMK structure does not appear to provide a good substitute for Asn233 of GK. Thus, on balance, the capacity to bind the Type II conformer appears to have been well preserved in PMK.
It is particularly interesting that the ammonium ion of PMK Lys 101 (which is switched into contact with the β,γ-bridge as a consequence of ligand binding) sits nearly exactly in the position occupied by Lys234 in the Gal-NAc kinase structure (i.e., in β,γ-bridge contact (45)); yet, these residues reside in very different regions of their respective structures. Moreover, Lys 234 is reported to adopt two conformations that place it either in or out of the active site (45). Thus, the ability to switch lysine (101 or 234) between what appear to catalytic and non-catalytic positions has been maintained; yet, the lys has migrated significantly within the GHMP framework.
Interconversion of the Type I and Type II conformations can occur either on the surface of the enzyme, in which case the conformers could represent discrete stages of binding, or via departure and rebinding of ligands, in which case the Type I represents a nonproductive complex that competes with Type II for the enzyme. Whether the PMK active site might accommodate the Type II nucleotide·Mn2+ complex of Gal-NAc kinase was assessed by positioning the nucleotide (via Cα-alignment) at the active site of the PMK ternary complex (Fig. 5E). The figure demonstrates that the cation shifts from “wall-to-wall” as a consequence of the interconversion, and that the active site seems spacious enough to accommodate a surface-interconversion of the complexes.
Active-site solvent
Twenty-one ordered water molecules fill the interstices between the Van der Waals surfaces of the PMK active site and its ligands (Fig. 6, panel A). The majority of this water is organized into clusters that contact charged regions of the ligands. In addition to ordered water, the cavity exhibits significant voids (also primarily near charged regions) that are presumed to contain disordered or rapidly exchanging solvent and/or its contents. The percentage of the surface area of the charged moieties (the tripolyphosphate of ATP and the phosphate and carbolylate of Pmev) in Van der Waals contact with the protein, as opposed to water, voids, or Mg2+, is calculated at ∼ 50% (46). Thus, a significant fraction of these reactive regions are coated in a shell of water, raising the issue of how the PMK active site manages to prevent β,γ-bond hydrolysis during its catalytic cycle. It appears that the active site of PMK, and likely, to varying degrees, members of its family, has evolved to bind charged ligand-regions indirectly, through clusters of ordered water.
Figure 6. Ordered water at the active site of PMK.
(A) Ordered water molecules in the active site of the PMK ternary complex. Water oxygen atoms are either orange or olive. The olive atoms form a pentamer of water. Ligands and their coordinated Mg2+ ion are slate gray. (B) The ternary-complex water pentamer. The olive pentamer oxygens (Panel A) are replaced with water molecules. The proton geometry was adjusted to conform to that predicted for the pentamer and to reflect the likely hydrogen bonding between the radial protons of the pentamer and the charged substrate moieties (carboxyl and phosphoryl groups of Pmev, and the β- and γ-phosphoryl groups of AMPPNP). (C) Pentamer in the absence in the absence of ligand. The Cα-backbone of each of the six subunits of the apo-structure asymmetric unit was aligned with that of the ternary complex, and the ordered water in the vicinity of the ternary-complex pentamer was compared. Oxygen in the ternary complex pentamer is orange; oxygen in the apo structure is color coded according to subunit (A-F) [green (A), grey (B), purple (C), yellow (D), beige (E), white (F)].
An active-site water pentamer
Of particular interest is the water pentamer, shown in olive in Figure 6A. The first coordination sphere of oxygen in liquid water is defined typically by four protons arranged in the tetrahedral geometry defined by the sp3 hybridization state of the oxygen (47, 48). These tetrahedral building blocks assemble into closed polygons (H2O)n, and extensive experimental and computational work predict that liquid water is composed largely of five- and six-membered rings that organize into cage-like structures (48-50). In the pentamer, each oxygen, or vertex, donates one of its protons to a nearest neighbor and accepts one from the other; the second proton of each oxygen radiates outward from the pentamer, like a spoke (48, 50). The radial protons alternately lie slightly above or below the ring - their interconversion is extremely rapid and occurs via quantum tunneling (50). The pentamer geometry, which is slightly puckered, is unique among small water-clusters in that it allows perfectly linear hydrogen bonds between donor and acceptor oxygens, leading to minimum inter-oxygen distances and atypically high-energy, ring-proton hydrogen bonds (48, 50-52), which, in turn, is expected to acidify the radial protons, enabling them to act as general acids.
To begin to understand how a water pentamer might interact with the PMK ligands, the pentamer oxygens were replaced with water molecules (Fig. 6B). The protons were arranged in accordance with the predicted pentamer bonding geometry (50), and oriented as if hydrogen bonded to the ligands. While the proton positioning is likely to be inaccurate in detail, the Van der Waals proximity of the radial aspect of the pentamer to the anionic oxygens of the substrate (the carboxyl- and phosphoryl-oxygens of Pmev and the β- and γ-phosphoryl oxygens of AMPPNP) supports hydrogen bonding and suggests that such structures participate directly in the binding of ligand.
To assess whether the active-site pentamer forms in the absence of ligands, the six subunits of the apo-PMK asymmetric unit were queried for its presence by aligning the Cα backbones of each subunit with that of the ternary complex and inspecting the water structure in the vicinity of the ternary-complex pentamer. Subsets of the pentamer were observed in each of the apo-subunits (Fig. 6C) with the weakest occupancy observed at position four. The alignments support formation of a pentamer-like structure even in the absence of ligand, and argue that this structure forms incompletely/transiently in preparation for the receipt of ligands that, once docked, stabilize the water scaffold on which they bind.
Conclusions
Comparison of the structure of the PMK ternary complex with that of apo-PMK and the ternary complexes of other GHMP kinases offers several insights into the structural basis of function in the enzyme and its family. While the Cα-architecture is conserved well across the family, the active site ligands of the ternary complexes crystallize within that architecture in one of two forms (I and II) - one of which appears poised for catalysis (Type II), while the other seems unlikely to react (Type I). The PMK active site houses twenty-one ordered water molecules and numerous voids, and appears sufficiently spacious to allow the isomers to interconvert on the surface of the enzyme, which calls into question whether the non-reactive isomer represents a binding intermediate. A pentamer of water with its edge positioned to hydrogen bond to the reactive regions of both ATP and Pmev is observed in the PMK structure - its presence strongly supports a role in ligand-binding and perhaps catalysis. Notably, a partially formed pentamer appears in the apo-enzyme, indicating that the pentamer is not “delivered” to the active site by substrate, but rather that it is largely present prior to binding and is completed, or stabilized, by the substrates as they add to, and depart from the active site.
Finally, Lys 101 appears to be switched into what appears to be a catalytic position by the binding of ligand. In the resting state of the enzyme, Lys 101 is approximately antiparallel with Lys 100, which sits in the ATP-binding pocket. Upon binding of ligand, the residues “swap” positions with the result that the ammonium ion of 101 is placed in direct contact with what would be the β,γ-bridging oxgen of ATP, where it is expected to strongly stabilize the ground and transition states of the reaction. Remarkably, a Lys similar to that of 101 (Lys 234) is delivered from a very different region of the Cα-scaffold of Gal-NAc kinase. Thus, the Lys switch has been conserved and yet allowed to migrate over evolutionary time around the edge of the active-site of the GHMP-kinase Cα-scaffold.
Table 1.
Data Measurement and Refinement Statisticsa
| Space group | P21212 |
| Unit cell parameters (Å) | a = 69.0, b = 115.3, c = 40.0 |
| Data Measurement and Phasing | |
| x-ray wavelength (Å) | 1.5418 (CuK )a |
| resolution (Å) | 23.4 – 1.90 |
| no. of observed reflections | 97,975 |
| no. of unique reflections | 23,685 |
| I > 3Σ(I) (%) | 80.5 (48.8) |
| Rmerge (%) | 4.5 (14.5) |
| completeness (%) | 91.3 (81.4) |
| Atoms | |
| no. of protein atoms | 2559 |
| no. of Pmev atoms | 14 |
| no. of ANPPNP atoms | 31 |
| no. of magnesium ions | 1 |
| no. of solvent molecules | 241 |
| Average thermal factor (Å2) | |
| protein atoms | 17.4 |
| Pmev atoms | 15.0 |
| ANPPNP atoms | 25.9 |
| magnesium ions | 20.3 |
| solvent molecules | 28.4 |
| RMS deviation from ideality | |
| bond lengths (Å) | 0.006 |
| bond angles (°) | 1.4 |
| Rfactor/Rfree (%) | 18.5 / 20.7 |
| (18.8 / 19.2) |
Rmerge (%) = Σ | Ii - <I> |/Σ | Ii | × 100. Rfactor (%) = Σ |Fo - Fc|/Σ |Fo| × 100 for all available data, but excluding data reserved for the calculation of Rfree. Rfree (%) = Σ |Fo - Fc|/Σ |Fo| × 100 for a 5% subset of x-ray diffraction data omitted from refinement calculations. Values in parentheses refer to the corresponding statistic calculated for data in the highest resolution bin.
Abbreviations
- AMPPNP
adenosine 5′-[β,γ-imido]triphosphate
- GHMP
Galacto kinase, Homoserine kinase, Mevalonate kinase, Phosphomevalonate kinase
- Hepes
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- MES
2-(N-morpholino)ethanesulfonic acid.
Footnotes
Supported by the National Institutes of Health Grant AI068989
References
- 1.Eisenreich W, Schwartz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A. The deoxyxylulose phosphate pathway of terpenoid biosyntesis in plants and microorganisms. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 2.Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci U S A. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meganathan R. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol Lett. 2001;203:131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- 4.Chemler JA, Yan Y, Koffas AG. Biosynthesis of Isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae. Microbial Cell Factories. 2006;5:20. doi: 10.1186/1475-2859-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 6.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 7.Xie W, Zhou C, Huang RH. Structure of tRNA dimethylallyltransferase: RNA modification through a channel. J Mol Biol. 2007;367:872–881. doi: 10.1016/j.jmb.2007.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA. Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol. 2008;26:139–145. doi: 10.1016/j.tibtech.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Moebius FF, Fitzky BU, Glossmann H. Genetic defects in postsqualene cholesterol biosynthesis. Trends Endocrinol Metab. 2000;11:106–114. doi: 10.1016/s1043-2760(00)00235-6. [DOI] [PubMed] [Google Scholar]
- 12.Sanderson JT. The steroid hormone biosynthesis pathway as a target for endocrine-disrupting chemicals. Toxicol Sci. 2006;94:3–21. doi: 10.1093/toxsci/kfl051. [DOI] [PubMed] [Google Scholar]
- 13.Javitt NB. Bile acid synthesis from cholesterol: regulatory and auxiliary pathways. Faseb J. 1994;8:1308–1311. doi: 10.1096/fasebj.8.15.8001744. [DOI] [PubMed] [Google Scholar]
- 14.Smit A, Mushegian A. Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 2000;10:1468–1484. doi: 10.1101/gr.145600. [DOI] [PubMed] [Google Scholar]
- 15.Tsay YH, Robinson GW. Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase. Mol Cell Biol. 1991;11:620–631. doi: 10.1128/mcb.11.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium U. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolman KG. The liver and lovastatin. Am J Cardiol. 2002;89:1374–1380. doi: 10.1016/s0002-9149(02)02355-x. [DOI] [PubMed] [Google Scholar]
- 18.Bruckert E. New advances in lipid-modifying therapies for reducing cardiovascular risk. Cardiology. 2002;97:59–66. doi: 10.1159/000057673. [DOI] [PubMed] [Google Scholar]
- 19.Spatz ES, Canavan ME, Desai MM. From here to Jupiter: Identifying new patients for statin therapy using data from the 1999 2004 national health and nutrition survey. Circ Cardiovasc Qual Outcomes. 2009;2:41–48. doi: 10.1161/CIRCOUTCOMES.108.832592. [DOI] [PubMed] [Google Scholar]
- 20.Wang CY, Liu PY, Liao JK. Pleiotropic effects of statin therapy: molecular mechanisms and clinical results. Trends Mol Med. 2008;14:37–44. doi: 10.1016/j.molmed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo H, Elson CE. Studies of the isoprenoid-mediated inhibition of mevalonate synthesis applied to cancer chemotherapy and chemoprevention. Exp Biol Med (Maywood) 2004;229:567–585. doi: 10.1177/153537020422900701. [DOI] [PubMed] [Google Scholar]
- 22.Cole SL, Vassar R. Isoprenoids and Alzheimer’s disease: a complex relationship. Neurobiol Dis. 2006;22:209–222. doi: 10.1016/j.nbd.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Capeans C, Pineiro A, Pardo M, Carneiro C, Blanco MJ, Vinuela JM, Salorio M. Sanchez, Dominguez F. Role of inhibitors of isoprenylation in proliferation, phenotype and apoptosis of human retinal pigment epithelium. Graefes Arch Clin Exp Ophthalmol. 2001;239:188–198. doi: 10.1007/s004170000250. [DOI] [PubMed] [Google Scholar]
- 24.Bliznakov EG. Diabetes and the role of isoprenoid biosynthesis. FEBS Lett. 2002;525:169–170. doi: 10.1016/s0014-5793(02)03062-4. [DOI] [PubMed] [Google Scholar]
- 25.Popjak G, Boehm G, Parker TS, Edmond J, Edwards PA, Fogelman AM. Determination of mevalonate in blood plasma in man and rat. Mevalonate “tolerance” tests in man. J Lipid Res. 1979;20:716–728. [PubMed] [Google Scholar]
- 26.Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J Bacteriol. 2000;182:4319–4327. doi: 10.1128/jb.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreassi JL, 2nd, Dabovic K, Leyh TS. Streptococcus pneumoniae isoprenoid biosynthesis is downregulated by diphosphomevalonate: an antimicrobial target. Biochemistry. 2004;43:16461–16466. doi: 10.1021/bi048075t. [DOI] [PubMed] [Google Scholar]
- 28.World HO. Pneumococcal vaccines. Weekly Epidemiological Record. 1999;74:177–184. [Google Scholar]
- 29.Pilloff D, Dabovic K, Romanowski MJ, Bonanno JB, Doherty M, Burley SK, Leyh TS. The kinetic mechanism of phosphomevalonate kinase. J Biol Chem. 2003;278:4510–4515. doi: 10.1074/jbc.M210551200. [DOI] [PubMed] [Google Scholar]
- 30.Andreassi JL, 2nd, Leyh TS. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry. 2004;43:14594–14601. doi: 10.1021/bi048963o. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Kissinger CR, Gehlhaar DK, Smith BA, Bouzida D. Molecular replacement by evolutionary search. Acta Crystallogr D Biol Crystallogr. 2001;57:1474–1479. doi: 10.1107/s0907444901012458. [DOI] [PubMed] [Google Scholar]
- 33.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods in Enzymology. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 34.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 39.Zhou T, Daugherty M, Grishin NV, Osterman AL, Zhang H. Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily. Structure. 2000;8:1247–1257. doi: 10.1016/s0969-2126(00)00533-5. [DOI] [PubMed] [Google Scholar]
- 40.Andreassi JL, 2nd, Bilder PW, Vetting MW, Roderick SL, Leyh TS. Crystal structure of the Streptococcus pneumoniae mevalonate kinase in complex with diphosphomevalonate. Protein Sci. 2007;16:983–989. doi: 10.1110/ps.072755707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miallau L, Alphey MS, Kemp LE, Leonard GA, McSweeney SM, Hecht S, Bacher A, Eisenreich W, Rohdich F, Hunter WN. Biosynthesis of isoprenoids: crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritol kinase. Proc Natl Acad Sci U S A. 2003;100:9173–9178. doi: 10.1073/pnas.1533425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishna SS, Zhou T, Daugherty M, Osterman A, Zhang H. Structural basis for the catalysis and substrate specificity of homoserine kinase. Biochemistry. 2001;40:10810–10818. doi: 10.1021/bi010851z. [DOI] [PubMed] [Google Scholar]
- 43.Wada T, Kuzuyama T, Satoh S, Kuramitsu S, Yokoyama S, Unzai S, Tame JR, Park SY. Crystal structure of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis. J Biol Chem. 2003;278:30022–30027. doi: 10.1074/jbc.M304339200. [DOI] [PubMed] [Google Scholar]
- 44.Mildvan AS. Mechanisms of signaling and related enzymes. Proteins. 1997;29:401–416. [PubMed] [Google Scholar]
- 45.Thoden JB, Holden HM. The molecular architecture of human N-acetylgalactosamine kinase. J Biol Chem. 2005;280:32784–32791. doi: 10.1074/jbc.M505730200. [DOI] [PubMed] [Google Scholar]
- 46.Sanner MF, Spehner J-C, A.J. O. Reduced surfaces:an efficient way to compute mlecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Smith JD, Cappa CD, Wilson KR, Messer BM, Cohen RC, Saykally RJ. Energetics of hydrogen bond network rearrangements in liquid water. Science. 2004;306:851–853. doi: 10.1126/science.1102560. [DOI] [PubMed] [Google Scholar]
- 48.Keutsch FN, Saykally RJ. Water clusters: untangling the mysteries of the liquid, one molecule at a time. Proc Natl Acad Sci U S A. 2001;98:10533–10540. doi: 10.1073/pnas.191266498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinhold F. Quantum cluster equilibrium theory of liquids: Illustrative application to water. Journal of Chemical Physics. 1998;109:373–384. [Google Scholar]
- 50.Harker HA, Viant MR, Keutsch FN, Michael EA, McLaughlin RP, Saykally RJ. Water pentamer: characterization of the torsional-puckering manifold by terahertz VRT spectroscopy. J Phys Chem A. 2005;109:6483–6497. doi: 10.1021/jp051504s. [DOI] [PubMed] [Google Scholar]
- 51.Xantheas SS, Burnham CJ, Harrison RJ. Development of transferable interaction potentials for water: II. Accurate energetics of the first few water custers from first principles. J. Chem. Phys. 2002;116:1493–1499. [Google Scholar]
- 52.Cruzan JD, Brown MG, Liu K, Braly LB, Sakally RJ. The far-infrared vibration-rotation-tunneling spectrum of the water tetramer-d8. J. Chem. Phys. 1996;105:6634–6644. [Google Scholar]
- 53.Romanowski MJ, Bonanno JB, Burley SK. Crystal structure of the Streptococcus pneumoniae phosphomevalonate kinase, a member of the GHMP kinase superfamily. Proteins. 2002;47:568–571. doi: 10.1002/prot.10118. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 56.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]