Abstract
Joints are essential for skeletal form and function, yet their development remains poorly understood. In zebrafish fins, joints form between the bony fin ray segments providing essentially unlimited opportunities to evaluate joint morphogenesis. Mutations in cx43 cause the short segment phenotype of short fin (sof b123) mutants, suggesting that direct cell-cell communication may regulate joint location. Interestingly, increased cx43 expression in the another long fin (alf dty86) mutant appears to cause joint failure typical of that mutant. Indeed, knockdown of cx43 in alf dty86mutant fins rescues joint formation. Together, these data reveal a correlation between the level of Cx43 expression in the fin ray mesenchyme and the location of joints. Cx43 was also observed laterally in cells associated with developing joints. Confocal microscopy revealed that the Cx43 protein initially surrounds the membranes of ZNS5-positive joint cells, but at later stages becomes polarized toward the underlying Cx43-positive mesenchymal cells. One possibility is that communication between the Cx43-positive mesenchyme and the overlying ZNS5-positive cells regulates joint location, and up-regulation of Cx43 in joint-forming cells contributes to joint morphogenesis.
Introduction
Joint development is critical for skeletal form and function, but little is understood about the process. Synovial joints, which articulate previously uninterrupted cartilaginous templates, are the most studied type of joint due to their clinical significance. Defects affecting joints include overuse malfunction, osteoarthritis, and congenital defects (Pacifici et al., 2006; Raz et al., 2008). The development of synovial joints is a multistep process that begins with condensation of a discrete band of mesenchymal cells at the presumptive joint location (Pacifici et al., 2006). This condensation is referred to as the interzone, which appears first as a thin layer of closely associated elongated cells in mammalian joints (Archer et al., 2003). The interzone is required for later steps of joint morphogenesis (Koyama et al., 2007; Pacifici et al., 2006), and may serve as a signaling center to the surrounding cells (Archer et al., 2003). In spite of their obvious significance, the molecular mechanisms underlying joint morphogenesis remain largely unclear.
The zebrafish caudal fin is highly amenable to the study of developing joints for several reasons. First, the fin is a rich source of joints. The caudal fin is comprised of 16-18 fin rays, each comprised of multiple bony segments separated by fibrous joints (Borday et al., 2001). Second, the fin provides essentially unlimited opportunities to evaluate joint morphogenesis. Fin growth occurs throughout the lifetime of the fish, with each segment addition (distally) also resulting in a new joint. Third, differences in joint maturity may be evaluated by comparing distal, or young joints, with the more proximal and older joints. Finally, joint formation may be monitored during fin regeneration, which proceeds rapidly following amputation on the researcher's time schedule.
Fin regeneration proceeds through several stages. Wound healing occurs 12-24 hours post amputation (hpa), followed by the establishment of a specialized structure called a blastema in the distal mesenchyme (Poss et al., 2000). By 72 hpa the blastema becomes organized into a distal compartment of non-dividing cells (10-50 μm) and a more proximal compartment of rapidly proliferating cells (100-200 μm) (Nechiporuk and Keating, 2002). Once the blastema is organized, outgrowth proceeds by coordinated cell proliferation and differentiation to replace lost tissue. It is during this stage where growth and segmentation occurs.
In addition to the advantages mentioned above, fin length mutants have been identified that may affect joint morphogenesis. Such mutants provide further opportunities to evaluate the molecular mechanisms underlying joint formation. For example, the fin overgrowth mutant another long fin (alf dty86) fails to produce regularly-spaced joints (van Eeden et al., 1996). The result is an occasional normal-sized segment and frequent long segments. Fractures of the fin ray are also frequent, perhaps due to the lack of flexibility in the fin (which is also too long). A second potential fin length mutant affecting joint formation is short fin (sof b123). The sof mutant exhibits short segments (or premature joints) due to defects in the gap junction gene connexin43 (cx43) (Iovine et al., 2005). Gap junctions are required for the exchange of small molecules (≤ 1000 Da) among neighboring cells. It is not clear how mutations in gap junctional coupling lead to defects in bone size and shape. However, defects in mammalian CX43 also cause skeletal malformations (Paznekas et al., 2003), suggesting that direct cell-cell communication contributes to a general mechanism regulating bone growth and/or length.
During fin regeneration cx43 mRNA is expressed in two locations: in the population of dividing cells in the blastmal mesenchyme, and in cells flanking the joints between recently separated segments (i.e. in the most proximal and most distal cells in completed segments, Iovine et al., 2005). Targeted gene knockdown of cx43 in the blastema leads to defects in both cell proliferation and segment length, indicating that Cx43 function in dividing cells contributes both to the level of cell proliferation and to segment size (Hoptak-Solga et al., 2008). In heterologous assays, wild-type Cx43 forms functional gap junctions but does not form functional hemichannels (Hoptak-Solga et al., 2007). In contrast, missense alleles of Cx43 exhibit aberrant gap junctional communication in heterologous assays and reduced cell proliferation in vivo. Therefore, it is believed that direct cell-cell communication via Cx43 gap junctions is required for normal levels of cell proliferation and segment length (Hoptak-Solga et al., 2007; Hoptak-Solga et al., 2008).
In this study we examine the relationship between Cx43 expression in the mesenchyme and in joint cells during joint morphogenesis. It may seem obvious that the requirement for Cx43 in cell proliferation itself is responsible for regulating segment length (i.e. less proliferation, shorter segments; greater proliferation, longer segments). However, it has been shown that manipulation of fin growth rates is not sufficient to alter segment length. Indeed, it is possible to reduce the rate of fin growth by raising zebrafish in a crowded environment. Comparison of young/rapidly growing fish with old/slowly growing fish reveals that segment length is the same in similarly sized fins, suggesting that the rate of cell proliferation does not determine segment size (Iovine and Johnson, 2000). Further, in treatments causing reduced cell proliferation by blocking either shh or Fgfr1, segment length is not affected or reduced (Lee et al., 2005; Quint et al., 2002). We suggest instead that Cx43 coordinates signals regulating cell division and joint formation by directing communication between the Cx43-positive mesenchymal cells and cells surrounding newly forming joints.
Materials and Methods
Fish rearing
Zebrafish were raised at constant temperature of 25°C with 14 light: 10 dark photoperiod (Westerfield, 1993). Wild-type (C32), sof b123 (Iovine and Johnson, 2000), and alf dty86 (van Eeden et al., 1996; available from the Zebrafish International Resource Center) fish stocks were all used in this study.
ZNS5 detection
Fins were harvested 5 days post amputation (dpa) at the 50% level and fixed overnight in 4% paraformaldehyde/PBS at 4°C. Fins were stored in 100% methanol at -20°C. Fins were rehydrated using successive 5 min washes in decreasing methanol/PBS solutions. After washing in blocking solution (2% BSA/PBS) fins were treated with ZNS5 (at 1:200, Zebrafish International Resource Center) in block at 4° C overnight. Following three 5 min washes in block solution fins were treated with 2° antibody (Alexa 488 at 0.01 mg/ml, Molecular Probes) in block overnight at 4° C. After three 10 min washes in block solution fins were mounted in Vectashield or glycerol on Superfrost microscope slides. Imaging was completed using either the Zeiss LSM 510 Meta confocal microscope (below) or using a Nikon Eclipse E80 compound microscope. For detection using horse-radish peroxidase fins were treated similarly until treatment with the Alexa 488 conjugated secondary antibody. Instead, fins were treated with goat anti-mouse antibody (Sigma, 0.01 mg/ml) for 2 hours at room temperature, washed using three 5 min washes in block, and treated with mouse peroxidase anti-peroxidase (mouse PAP is from Sigma, 0.03 mg/ml) overnight. Following several washes in PBS, fins were transferred to 0.03 % diaminobenzidine (DAB, Polysciences) in 0.01 M phosphate buffer and hydrogen peroxide was added to a final concentration of 0.01 %. Development proceeded for 10-15 minutes at room temperature before stopping in PBS, mounting in 50 % glycerol, and visualization using a Nikon Eclipse E80 microscope.
ZNS5 and Cx43 double immunofluorescence and confocal microscopy
The Cx43 antibody was described previously and shown to be specific for Cx43 by competition experiments followed by quantitative immunoblots (Hoptak-Solga et al., 2008). Since the Cx43 antigen is sensitive to fixation conditions, a modified protocol was established for double labeling. Five day regenerating fins were fixed in 2% (wt/vol) paraformaldehyde in 25 mM phosphate buffer (PB) for 30 min at room temperature. Following three 10 min. washes in 25mM PB, fins were trypsinized (Trypsin/EDTA, Gibco) for 10 min on ice, washed in 25mM PB, and then incubated in blocking solution (1M Tris-HCl, 5M NaCl + 0.3% Triton X-100 + 4% goat serum) for 30 min. The fins were then incubated with rabbit anti-Cx43 polyclonal antibody (Hoptak-Solga et al., 2008) and with ZNS5 monoclonal antibody (both at 1:200 in blocking solution) overnight at 4°C. After 5 washes in 25 mM PB, the fins were incubated for 2 hr at room temperature with Alexa Fluor 488 goat anti-mouse IgG (at 0.01 mg/ml in blocking solution) and Alexa Fluor 546 goat anti-rabbit (at 0.01 mg/ml in blocking solution) (Molecular Probes). Fins were washed 3 times in PBS and mounted in glycerol.
Laser scanning confocal imaging was performed (Piehl and Cassimeris, 2003) on whole fins double-labeld for ZNS5 and Cx43. Images were acquired using a 40× N.A. 1.4 PlanApo DIC objective on an inverted microscope (Axiovert 200M, Carl Zeiss, Jena, Germany) equipped with an LSM510META scan head (Carl Zeiss, Jena, Germany). Argon ion (488) and 543 HeNe lasers were used to generate the excitation lines, and multitrack sequential excitation was utilized to avoid bleed-through between fluorophores. Z-stacks were collected lateral to medial by focusing first on the primarily green ZNS5-positive cells and moving in Z towards the primarily red mesenchyme. Two color 512 × 512 images were acquired using two-line mean averaging in a Z-series typically containing X - Y overlapping sections of 0.61 μm depth. Files were exported as TIFF files.
Cryosectioning
Following double-staining with ZNS5 and Cx43, fins were rinsed in 1xPBS (3×10 min) and embedded in 1.5% agarose/5% sucrose blocks, and submerged in 30% sucrose overnight at 4 °C. Blocks were frozen on dry ice and mounted using O.C.T. Compound (Tissue Tek®, Sakura, the Netherlands), and 10μm sections were cut using a cryostat (Leica 2800 Frigocut E; Cambridge Instruments, Germany). Sections were collected on Superfrost Plus slides.
Calcein staining
Calcein staining was completed as described (Du et al., 2001). Briefly, fish were allowed to swim in 0.2 % calcein (pH 7.0) for 10 min at room temperature, followed by 10 min in fresh fish water. Fins were harvested and mounted in 50 % glycerol for immediate visualization using a Nikon Eclipse E80 microscope.
In situ hybridization
Antisense probe for cx43 was generated as described (Iovine et al., 2005). Tissue was fixed overnight with 4% paraformaldehyde in PBS and stored in 100% methanol at -20°C. Gradual aqueous washes were completed in methanol/PBST. Tissue was treated with 5 μg/ml proteinase K (5 min for embryos; 45 min for fins) and re-fixed for 20 min. Prehybridization (50 % formamide, 5× SSC, 10 mM citric acid, 0.1 % Tween20) occurred for 1 hour at 65°C, and hybridization in the presence of digoxigenin-labeled antisense probes was completed overnight. Gradual washes into 0.2× SSC were followed by gradual washes into PBST. Anti-digoxigenin Fab fragments (pre-absorbed against zebrafish tissue) were used at 1:5000 overnight. Following extensive washes in PBST followed by three short washes in staining buffer (100 mM Tris, 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1 % Tween20, pH 9.0). Tissue was next transferred to staining solution (staining buffer plus 0.22 mg/ml NBT and 0.175 mg/ml BCIP) and development proceeded until purple color was observed.
qRT-PCR analysis
Trizol reagent (Gibco) was used to isolate mRNA from 5 dpa regenerating fins (5-10 fins were pooled) and first strand cDNA was prepared using oligodT(12-15) and reverse transcriptase. Dilutions of template cDNA were prepared (1:5, 1:50, 1:500, 1:5000). Oligos flanking introns were designed for cx43 (F-TCGCGTACTTGGATTTGGTGA; R-CCTTGTCAAGAAGCCTTCCCA) and keratin4 (F-TCATCGACAAAGTGCGCTTC; R-TCGATGTTGGAACGTGTGGT) using Primer Express software. The cx43 and keratin4 amplicons were amplified independently using the Power SYBR green PCR master mix (Applied Biosystems). Samples were run in triplicate on the ABI7300 Real Time PCR system and the average cycle number (CT) was determined for each amplicon. Delta CT (ΔCT) values represent normalized cx43 levels with respect to keratin4, the internal control. Delta Delta CT (ΔΔCT) values represent cx43 levels of the test sample (sof or alf) minus the cx43 levels of the calibrator sample (wild-type). The fold-change was calculated using the double ΔCT method (i.e. using the equation 2 −ΔΔCT). Four independent experiments were completed.
Segment length analysis and measurements of distance in fins
The third fin ray from the ventral side (V+3) of the caudal fin was examined for all measurements (i.e. distance of joints in ZNS5 fins, distance of physical breaks in calcein treated fins, segment length measurements in alf and wild-type fins). A minimum of 10 fins were examined. Images were collected using ImagePro software. Measurements were also completed using ImagePro.
Morpholino Injection and Electroporation
Microinjection and electroporation methods were used to conduct gene knockdowns in both wild type and alf as described (Hoptak-Solga et al., 2008). Briefly, ten wild type fins and ten alf fins were amputated and allowed to regenerate for three days. The first three fin rays on the dorsal side of each fin were injected with ∼50 nl of either the targeting (cx43-MO2) or control (5mmcx43-MO2) morpholino (n = 5 for each treatment). Morpholinos were purchased from Gene Tools, LLC and have been described (Hoptak-Solga et al., 2008). Immediately after injection, both the dorsal and ventral lobes of the fins were electorporated. The fins were then allowed to grow for four days post injection and electroporation (4 dpe) before staining for ZNS5 to facilitate identification of joints (i.e. and to distinguish joints from fin ray breaks that are typical in alf dty86 fin rays). Segment length was measured using ImagePro software.
Results
Fin ray joints progress through different stages of maturation
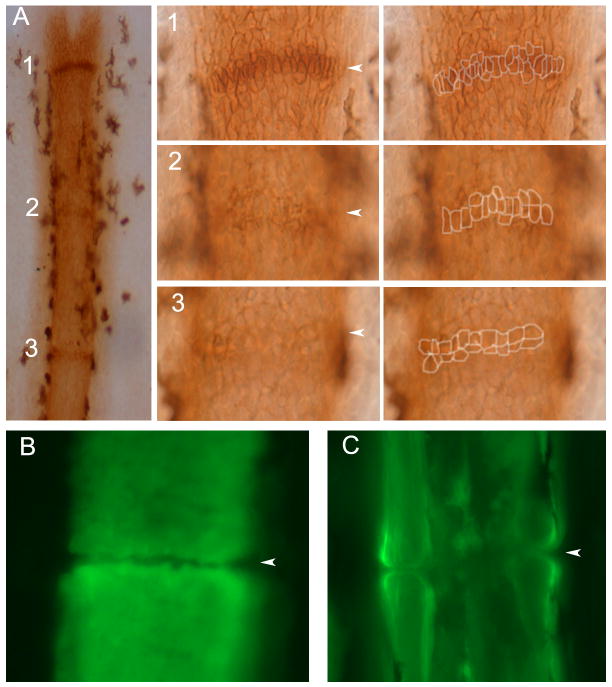
During fin regeneration, new segments and joints are continually added to the distal end of the fin ray. Segments are comprised of two hemirays of bone matrix surrounding a central mesenchymal compartment. Thus, osteoblasts are found in lateral compartments, where they secrete bone matrix directly (Santamaria et al., 1992). Previously, the monoclonal antibody ZNS5 has been used to visualize osteoblasts surrounding newly deposited bone matrix (Johnson et al., 1995; Poss et al., 2002; Smith et al., 2006; Smith et al., 2008). Here we show that when observed in whole mount staining, it is possible to detect cells surrounding joints as well. Indeed, fins stained with ZNS5 show condensations of ZNS5-positive cells along the proximal-distal axis at distances consistent with the locations of joints (Figure 1A). Note that the morphologies of the ZNS5-positive joint cells differ depending on the proximal-distal location of each condensation. Less mature joints (such as joint 1) exhibit elongated ZNS5-positive cells in a single row, while the most-mature joints (similar to joint 3) exhibit two rows of cells appearing more squat and rounded. The first appearance of ZNS5 condenstions as in joint 1 occurs at approximately 300 μm from the end of the fin, proximal to the dividing cells of the blastema and proximal to the distal-most ZNS5-positive cells. The distance between ZNS5 condensations is relatively constant (Δ1,2 and Δ2,3), while the distance of each joint to the end of the fin ray is more variable (Table 1). This is consistent with previous findings that segment length within a fin ray is constant (Iovine and Johnson, 2000), and that the elongated ZNS5-positive condensations characteristic of joint 1 represent an actual or imminent joint in the fin ray.
Figure 1.
Joint formation in zebrafish fin rays. (A) Fins were stained with the osteoblast marker ZNS5, and detected using HRP (brown). At this level joints are detected as condensations of ZNS5-positive cells, which appear with differing maturities along the proximal-distal axis. Joint 1 is the most-distal joint and the least mature. At higher magnification distinct morphologies of ZNS5-positive cells can be observed (and outlines of the ZNS5-positive cells are shown to the right). (B) The morphology of the bone matrix in the newest joint is observed using calcein. (C) The morphology of the bone matrix in a fully mature joint is observed using calcein. Arrowheads point to joints.
Table 1.
Measurements of condensed ZNS5-positive cells and joints to the end of the fin
| Structure | distance to end of fin (μm) |
|---|---|
| joint 1 | 301 +/- 87 |
| joint 2 | 607 +/- 99 |
| joint 3 | 925 +/- 99 |
| Δ1,2 | 305 +/- 27 |
| Δ2,3 | 317 +/- 19 |
| physical separation | 422 +/- 76 |
Fins were stained for ZNS5 to detect cells surrounding joints or using calcein to detect bone matrix. Distances were measured in the V + 3 fin ray from a minimum of 10 fins (n = 12 for ZNS5-cell measurements; n = 17 for calcein measurements).
To determine when the physical separation of bony plates first occurs, fins were stained with calcein to detect calcified bone matrix (Du et al., 2001). The first appearance of a physical joint is found at approximately 422 μm (Table 1), proximal to the first appearance of elongated ZNS5 condensations. In joints at this distance, the break appears as a simple separation between two flat plates of bone (Figure 1B). In more mature joints, where the bone is much thicker, the joints appear as rounded bones facing one another (Figure 1C). In both cases, the mesenchymal compartment remains continuous and the physical joint occurs only in the bone matrix. This is consistent with the previous characterization of fin ray joints as ‘fibrous’ (Borday et al., 2001), defined as joints connected by connective tissue, and with other published longintudinal sections of joints (Iovine et al., 2005). The finding that the most distal physical separation of bone matrix occurs proximal to the position of joint 1 suggests two important conclusions regarding joint morphogenesis. The first is that the most distal ZNS5 condensations at the level of joint 1 represent presumptive joints rather than true joints, while the ZNS5 condensations at the level of joint 2 are actual joints. The second is that the fin ray joint matures by articulating previously deposited bone matrix coincident with characteristic changes in cell morphology of the ZNS5-positive condensations.
Cx43 localization during joint morphogenesis
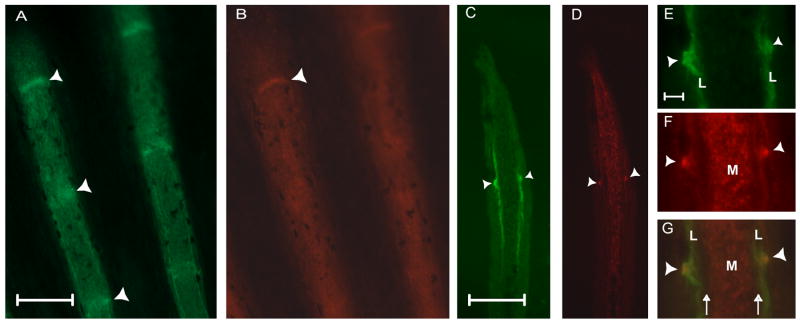
In contrast to ZNS5 staining, the Cx43 antibody has been shown to recognize the centrally located mesenchymal cells in addition to the cells surrounding the most distal one to two joints (Hoptak-Solga et al., 2008 and Figure 2 B). In other words, the Cx43 antibody can recognize joints in positions 1 and 2, but has not been detected in the third fin ray joint. That Cx43 is not readily detectable in the more proximal joints suggests that Cx43 protein associates preferentially with presumptive and newly formed joints. Fins co-stained with ZNS5 and Cx43 were sectioned to visualize the relative locations of immunostaining (Figure 2 C,D). As described, ZNS5-positive cells are located laterally in association with newly forming bone matrix, while Cx43 is found medially in the mesenchyme. Identification of presumptive joints was also possible as ZNS5 and Cx43 positive cells on the lateral surface of the osteoblast compartment (Figure 2 E,F,G).
Figure 2.
ZNS5 and Cx43 detect different populations of cells in the fin ray, and are both expressed in newly developing joints. (A) Whole mount staining of ZNS5. (B) Whole mount staining of Cx43. (C) Longitudinal cryosection showing lateral ZNS5 staining and detection of one joint (arrowhead). (D) Longitudinal cryosection showing mesenchymal Cx43 expression and detection of one joint (arrowhead). (E,F,G) Higher magnification of the joint observed in C and D shown for ZNS5, Cx43, and the overlap. Arrowheads point to joints; arrows indicate the location of bone matrix. L, lateral; M, medial. Scale bars for A-D, 100 μm. Scale bar for E-G, 10 μm.
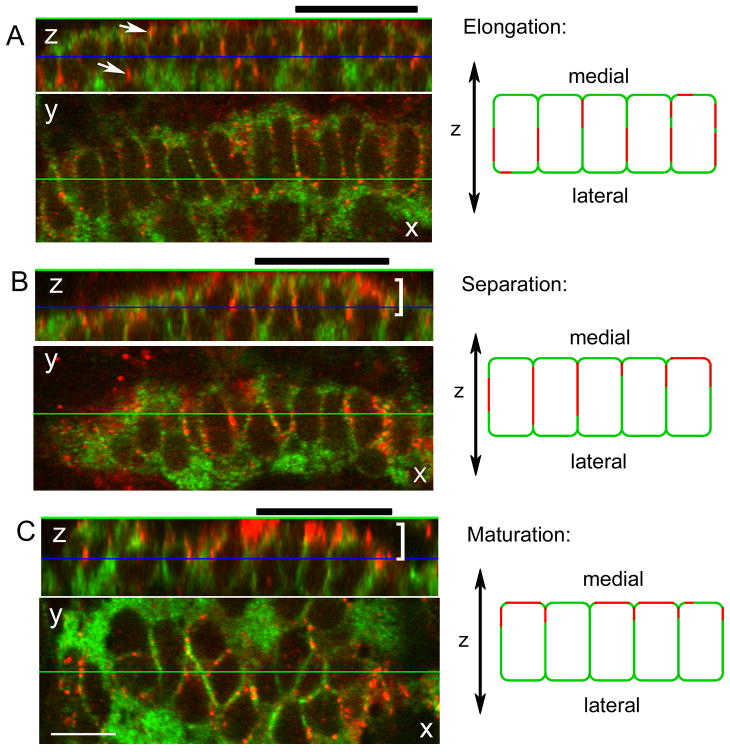
To begin to evaluate normal joint morphogenesis we monitored ZNS5 and Cx43 positive cells in joints at positions 1 and 2 by whole mount confocal microscopy. Joint cells in these two locations were found to display a continuum of morphologies similar to what was observed in Figure 1. For reference, we identified presumptive joints characterized by different cellular morphologies based on the appearance of ZNS5-positive cells: elongated, separating, and maturating (Figure 3, X-Y images). ‘Elongated’ joints exhibit highly elongated ZNS5-positive cells similar to joint 1 from Figure 1 (n = 19, Figure 3A). ‘Separating’ joint cells were less elongated and some of the cells also appeared pinched at one end or were becoming rounded (n = 26, Figure 3B). In fact, ‘separating’ joints in this analysis likely represent a stage in between the joint 1 and joint 2 morphologies observed in Figure 1. Cells of separating joints appear to be moving away from each other in the process of forming two rows of ZNS5-positive cells. In ‘maturing’ joints, most of the cells were rounded and beginning to arrange into two rows of cells, similar to joints 2 and 3 from Figure 1 (n = 10, Figure 3C). This stage most closely resembles the appearance of fully mature joints, where two rows of ZNS5-positive cells flank the physical separation between segments.
Figure 3.
Cx43 localization during joint morphogenesis. A single image from a Z-stack collected by confocal microscopy is shown in the X-Y plane. The display on top represents a slice through the Z-stack along the horizontal green line (i.e., the X-Z plane). The blue line in this display indicates the location of the front image within the Z-stack. During elongation (A), Cx43 (arrows) is found surrounding the ZNS5 positive cells. During separation (B) and maturation (C), Cx43 appears polarized (brackets) toward the medial face of the ZNS5-positive cells. A group of five cells from the X-Z planes are represented by cartoons to the right of each image. Thick black bars over each X-Z plane identify the cells represented by cartoons at the right. Scale bar, 10 μm.
As part of this analysis we noted that the cellular localization of Cx43 changes with maturity of the developing joints. To evaluate Cx43 localization with respect to joint formation, Z-stacks were collected by confocal microscopy. The localization of Cx43 may be observed as increased Cx43 staining in more medial sections of the Z-stack (not shown), and by use of the “orthogonal” tool which displays the depth of the Z-stack in a 2-dimensional image (Figure 3, X-Z images). Here, single images from Z-stacks of elongating, separating, and maturing joints are shown. The displays on top represent a slice through the Z-stack in the plane indicated by the horizontal green line (i.e. the X-Z plane). In joints of the elongated morphology, Cx43 was found in a punctate pattern all around the membranes of ZNS5-positive cells (Figure 3A and cartoon). This is most clearly observed by the appearance of red Cx43 signal surrounding the green ZNS5 signal in the X-Z plane with no apparent bias of Cx43 staining on either the medial or lateral face of the ZNS5-positive cells. In contrast, in joints with the separating and maturing morphologies, Cx43 localization did appear polarized toward the medial-facing membranes of the ZNS5 positive cells (Figure 3B,C and cartoon). This is observed by the localization of the red Cx43 signal primarily on one side of the green ZNS5-positive cells in the X-Z plane. Cx43 polarization was observed in 70 % of joints characterized as separating and mature joints (18/26 separating joints and 7/10 maturing joints), but in only 15 % of joints characterized as elongating (3/19 initiating joints). It is interesting that Cx43 becomes localized towards the mesenchymal cells since Cx43 is also found on the membranes of those cells (Figure 2 D). One possibility is that Cx43 establishes communication between the ZNS5-positive joint cells and the underlying mesenchymal cells, and this communication determines the directionality of the developing joint (i.e. from lateral to medial).
Expression levels of cx43 are correlated with joint specification
We next evaluated joint formation in two mutants that may perturb the process, sof b123 and alf dty86. In sof b123 mutants, cx43 mRNA and protein levels are reduced but not absent (Hoptak-Solga et al., 2008; Iovine et al., 2005). We found no difference in joint morphologies in sof b123 fins when examined by ZNS5 (data not shown). Since Cx43 protein levels are reduced in sof b123, we did not attempt to evaluate Cx43 localization.
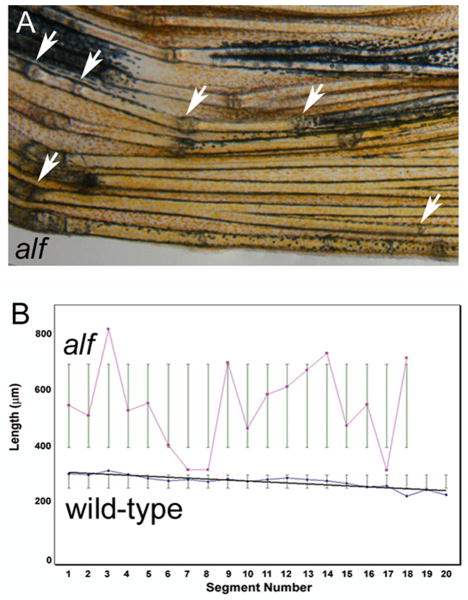
The alf dty86 mutant was reported to make irregular segments (van Eeden et al., 1996 and Figure 4A). Indeed, we found that joint formation is inconsistent in alf dty86 fins. We first evaluated the frequency of joint formation by measuring segment length across a single fin ray (n=10 fin rays in each wild-type and alf dty86). Segment length in wild-type fins exhibits a pattern of progressively decreasing segment length in a proximal to distal fashion (also observed in Iovine and Johnson, 2000). In contrast, segment length appears as a somewhat random distribution in alf dty86 fins (Figure 4B). Joint formation is not completely random, however, since segments smaller than normal (i.e. less than 250 μm) were not observed. Wild-type segments averaged 290.7 ± 32.7 μm across the fin ray while the alf dty86 segments averaged 609.5 ± 318.9 μm. Thus, alf dty86 segments were on average twice as long as wild type segments while also exhibiting a very broad deviation from the mean (Figure 4B). These results indicate a failure to conform to a standard segment length rather than a simple segment overgrowth phenotype. Together, these data suggest a stochastic disturbance in the initiation or placement of a joint.
Figure 4.
Segment length is irregular in alf dty86 fins. (A) Bright-field image of an alf dty86 fin. Arrows point to joints. (B) Segment length in wild-type and alf dty86 fins. Segment length was measured along the V + 3 fin ray in each of 10 fins. Segment length gradually decreases along the proximal-distal axis in wild-type fins, but appears random in alf dty86 fins.
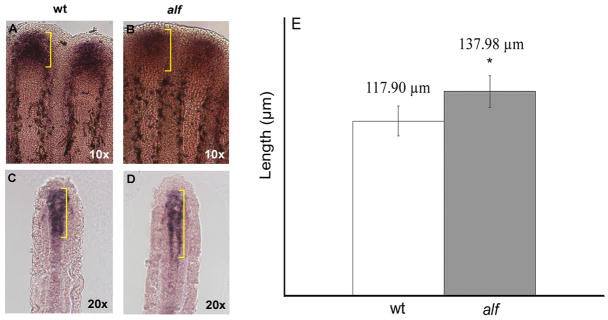
Since sof b123 mutant fins exhibit reduced cx43 expression and what appears to be premature joint formation, we wondered if up-regulation of cx43 underlies irregular joint formation in alf dty86 fins. The molecular lesion causing the alf dty86 phenotype is not known, and in fact is not due to a gain-of-function mutation in the cx43 gene (data not shown). Still, it remained possible that overexpression of cx43 might contribute to the alf dty86 phenotype. To test this hypothesis wild type and alf dty86 fins were examined for cx43 expression by in situ hybridization (Figure 5). We found that mesenchymal cx43 mRNA expression was expanded in alf dty86 fins (117.90 ± 10.14 μm in wild-type and 137.98 ± 10.75 μm in alf dty86, p=0.0001).
Figure 5.
The cx43 mRNA is overexpressed in alf dty86 fins. (A,B) Whole mount in situ hybridization shows that the distal expression domain of cx43 is expanded in alf dty86 fins. (C,D) The expansion of the mesenchymal cx43 expression domain is also apparent following cryosectioning of stained fins. (E) The length of the cx43 expression domain was measured from the V + 3 fin ray in each of ten fins, and is statistically different between alf dty86 and wild-type fins.
To examine this difference more carefully, qRT-PCR was completed on wild-type, sof b123, and alf dty86 regenerating fins from four independent tissue samples (Table 2). Delta CT (ΔCT) values represent the normalized cycle number when the cx43 amplicon becomes detectable from each sample (i.e. lower ΔCT levels indicate higher mRNA levels in the sample). Delta CTs were converted to fold-difference using the double delta CT method (ΔΔCT = ΔCT test sample − ΔCT wild-type). First, we found that cx43 expression is 15-20-fold lower in sof b123 fins, consistent with our previous report showing that Cx43 protein levels are reduced in sof b123 animals (Hoptak-Solga et al., 2008). When examining alf dty86 fins we found that cx43 levels are consistently 1.5-2-fold higher than wild-type. This difference is moderate but significant, and consistent with the expanded expression domain observed by in situ hybridization. Therefore, decreased cx43 expression of sof b123 mutants is correlated with premature joint formation while increased cx43 expression of alf dty86 mutants is correlated with the failure to form joints at regular intervals.
Table 2.
Expression levels of cx43 in sof b123 and alf dty86 fins.
| ΔΔCT values | fold-change | |
|---|---|---|
| sof b123 | 3.948 +/- 0.49 | -15.43 |
| 4.085 +/- 0.66 | -16.97 | |
| 4.330 +/- 0.10 | -20.11 | |
| 3.880 +/- 0.41 | -14.72 | |
| alf dty86 | -1.310 +/- 0.46 | 2.48 |
| -1.420 +/- 0.03 | 2.68 | |
| -0.640 +/- 0.10 | 1.56 | |
| -0.863 +/- 0.33 | 1.82 | |
Cx43 levels in the blastema contributes to joint specification
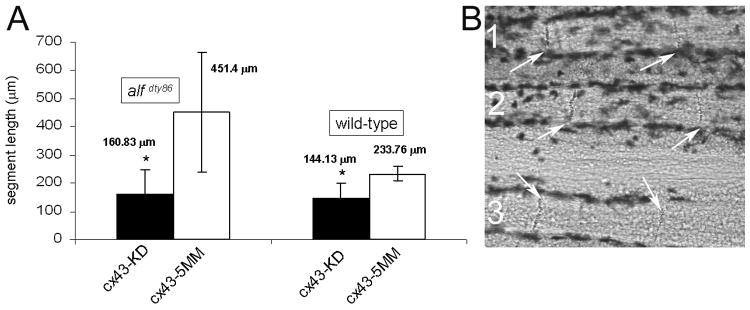
Based on the proposed correlation between cx43 expression levels and relative joint formation, we predicted that reducing cx43 expression in alf dty86 fins would restore the regularity of joint location. Recently, we described results of cx43 gene knockdown in wild-type fins and documented the efficacy of two cx43 morpholinos (Hoptak-Solga et al., 2008). This method permits knockdown of the target gene in the cells of the regeneration blastema (Thummel et al., 2006). Targeted gene knockdown was accomplished similarly here using the cx43-MO2 and the non-targeting mismatch morpholino, 5mmcx43-MO2. Indeed, cx43 gene knockdown in alf dty86 fins restored regular joint formation as predicted (Figure 6). The average segment length for fin rays treated with cx43-MO2 was 160.83 ± 63.46 μm compared with 451.40 ± 169.58 μm for alf dty86 fin rays treated with 5mmcx43-MO2 (p < 0.0001). Thus, segment length is reduced and the standard deviation is also much tighter in the cx43 knockdown fin rays. Unexpectedly, the average segment length for treated alf dty86 fin rays was not significantly different than the average segment length for cx43 knockdown in wild-type fin rays (145.62 ± 49.65 μm, p > 0.5). This is consistent with our findings that cx43 is only moderately overexpressed in alf dty86 fins, and further suggests a threshold level of Cx43 required for wild-type segment length. It is interesting to note that cx43 knockdown in the blastema affects joint formation in the lateral compartment. One possibility is that high levels of Cx43 in the blastema mesenchyme acts as a joint-inhibition signal to the overlying ZNS5-positive compartment. Thus, the increased cx43 expression in alf dty86 fins leads to joint failure, and reducing Cx43 levels permits joint regularity.
Figure 6.
Targeted gene knockdown of cx43 restores joint formation in alf dty86 fins. (A) Segment length in alf dty86 fins following cx43 gene knockdown (alf KD) is similar to segment length following cx43 gene knockdown in wild-type fins (wt KD). Results of gene knockdown in both wild-type and alf dty86 are statistically different from wild-type fins treated with a non targeting morpholino (p ≪ 0.05). Wild-type and alf dty86 knockdown fins are not statistically different from one another. (B) Joints (arrows) are observed in each of three alf dty86 fin rays treated with cx43-MO2.
Discussion
This is the first report to document joint morphogenesis in zebrafish fin rays (Figure 7). We suggest that Cx43 plays two independent roles in joint development. First, an early role in the fin ray mesenchyme that may determine the location of the joint, and a later role in the ZNS5-positive cells surrounding the future joints. Support for an early role of Cx43 in the mesenchymal compartment is provided by our analyses of the fin length mutants, sof b123 and alf dty86, and by direct manipulation of cx43 levels. From these experiments, there is a clear correlation between Cx43 levels and joint location. Cx43 also appears to be upregulated in the population of condensed ZNS5-positive cells surrounding the presumptive joints. Polarization of Cx43 towards the medial surface of the ZNS5-positive cells may play a role in maturation of the joint itself.
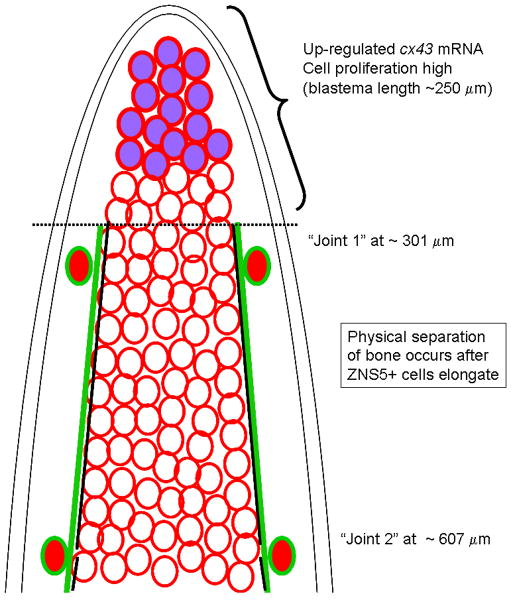
Figure 7.
Model showing relative localization of Cx43-positive cells, ZNS5 positive cells, and appearance of joints. The cx43 mRNA is upregulated in the blastema, and is coincident with proliferating cells (Hoptak-Solga et al., 2008); blastema length has been reported to be up to 250 μm (Nechiporuk and Keating, 2002). Cx43 protein is found on cell membranes throughout the mesenchyme, whereas ZNS5-positive cells are found laterally surrounding bone matrix. The increased level of Cx43 expression in the blastemal compartment may inhibit joint formation, creating a boundary for joint morphogenesis. Lateral ZNS5-positive cells may respond on the proximal side of this boundary by elongating and upregulating Cx43 in presumptive joints. Later steps of joint maturation involves separation of ZNS5-positive cells from a single row of elongated cells into two rows of rounded cells, and the formation of the physical joint in the bone matrix. Cx43 is upregulated in the elongated ZNS5-positive cells and may play a role in joint maturation.
The condensation of ZNS5-positive cells on the lateral surface of the future joint is reminiscient of the interzone in synovial joints (Pacifici et al., 2006). It has been suggested that communication among interzone cells contributes to their function as a joint signaling center. Indeed, the gap junction proteins Cx43 and Cx32 are expressed in the cells of the interzone during synovial joint formation (Archer et al., 2003), although a specific role has not been defined. Furthermore, disruption of Cx40 expression in the mouse causes defects in joint and bone morphogenesis (Pizard et al., 2005). We find Cx43 expressed in the putative interzone of zebrafish fin ray joints, and the zebrafish genome has duplicate genes representing both mammalian Cx32 and Cx40 (Eastman et al., 2006). One or more of these connexins may also contribute to joint morphogenesis. Indeed, the combination of mammalian and zebrafish studies may reveal that direct cell-cell communication is a generalized mechanism for the placement and/or maturation of skeletal articulations.
Most interesting from this work is the strong correlation between fin growth, joint formation, and Cx43 levels. For example, the sof b123 mutant exhibits short fins, premature joints, and reduced cx43. The alf dty86 mutant exhibits fin overgrowth, joint failure, and increased cx43. Previous work from our lab on sof mutants and in cx43-knockdown fins revealed that the level of Cx43 function regulates the level of cell proliferation (Hoptak-Solga et al., 2008). This can explain defects in fin length, but does not by itself explain the short segment phenotype since manipulation of growth rate and/or cell proliferation levels is not sufficient to alter segment length (Iovine and Johnson, 2000; Lee et al., 2005; Quint et al., 2002). We have suggested that Cx43 may be responsible for coordinating fin growth with segment length. This report provides deeper insight into the nature of this coordination.
The cx43 mRNA is upregulated in the rapidly dividing cells of the blastema while the protein is found throughout the fin ray mesenchyme. We suggest that high levels of Cx43, and/or increased cell proliferation, may inhibit joint formation (Figure 7). And further, that the difference of Cx43 expression levels from high to basal represents a potential boundary where joint morphogenesis begins. The lateral ZNS5-positive cells may respond to the boundary by aligning into a single row of elongated cells and by up-regulating Cx43 in this population of cells. In sof b123 mutants, the boundary between high and low Cx43 may be shifted, so joint initiation begins prematurely. In contrast, in alf dty86 mutants the Cx43 boundary may be blurred, causing joint initiation to fail most (but not all) of the time. Thus, reduced cx43 expression results in a simple shift of the joints as in sof b123 mutants, while overexpression of cx43 causes stochastic factors to play a more important role in whether or not a joint will develop in alf dty86 mutants.
It is possible that an independent requirement for joint formation exists in the lateral ZNS5-positive population. For example, perhaps only a subpopulation of ZNS5-positive cells is capable of responding to the suggested Cx43 boundary. This putative “joint field” may express a unique set of genes. One candidate for a joint field gene is evx1, which is expressed in a sub-population of bone forming cells at the level of the mature joint and also in a population of cells that may represent the future joint (Borday et al., 2001). Interestingly, the latter population of evx1-expressing cells is observed before the growing segment is completed, indicating that evx1 is expressed well-before ZNS5-positive cells begin to elongate at the future joint site (recall that condensation of elongated ZNS5-positive cells represents the location of the final joint). One possibility is that the evx1-positive/ZNS5-positive population can respond to the proposed Cx43 boundary, but evx1-negative/ZNS5-positive cells cannot. The response includes changes in cellular morphology and upregulation of Cx43, which may be required for further maturation of the joint.
An analysis of gene expression in the lateral ZNS5-positive cells, in accord with physical landmarks such as the physical separation of bone matrix, will be an important next step. A recent report on the joints of the zebrafish fin radials (which articulate the cartilagenous elements between the axial skeleton and the fin rays) revealed that the expression of genes found in mammalian joints is largely conserved in zebrafish (Crotwell and Mabee, 2007). These and other candidate genes will be examined for temporal and spatial expression patterns in the joints of wild-type, sof b123, and alf dty86 fins, providing detailed insights into joint initiation and morphogenesis.
Conclusions
This report is the first to describe joint morphogenesis in the easily accessible zebrafish caudal fin. The combination of fin structure, imaging possibilities, existence of joint mutants, and knockdown strategies establish the zebrafish caudal fin as an excellent model system for further analyses of this problem. Here, we show that direct cell-cell communication is involved in decisions regarding joint location and may play a later role in joint maturation. This work therefore provides novel insights into the role of cell-cell communication during joint morphogenesis.
Acknowledgments
The authors wish to thank Lynne Cassimeris for help with confocal microscopy and Jacob Fugazzotto for care of the zebrafish colony. Jutta Marzillier and Isha Jain facilitated the qRT-PCR experiments and analyses. Maria Brace helped with figure preparation. This work was supported by the NICHD (HD047737 to MKI). ZNS5 and alf dty86 are available from the Zebrafish International Resource Center, which is supported by P40 RR12546.
References
- Archer CW, Dowthwaite GP, Francis-West P. Development of synovial joints. Birth Defects Res C Embryo Today. 2003;69:144–55. doi: 10.1002/bdrc.10015. [DOI] [PubMed] [Google Scholar]
- Borday V, Thaeron C, Avaron F, Brulfert A, Casane D, Laurenti P, Geraudie J. evx1 transcription in bony fin rays segment boundaries leads to a reiterated pattern during zebrafish fin development and regeneration. Dev Dyn. 2001;220:91–8. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1091>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Crotwell PL, Mabee PM. Gene expression patterns underlying proximal-distal skeletal segmentation in late-stage zebrafish, Danio rerio. Dev Dyn. 2007;236:3111–28. doi: 10.1002/dvdy.21352. [DOI] [PubMed] [Google Scholar]
- Du SJ, Frenkel V, Kindschi G, Zohar Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev Biol. 2001;238:239–46. doi: 10.1006/dbio.2001.0390. [DOI] [PubMed] [Google Scholar]
- Eastman SD, Chen TH, Falk MM, Mendelson TC, Iovine MK. Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics. 2006;87:265–74. doi: 10.1016/j.ygeno.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Klein KA, Derosa AM, White TW, Iovine MK. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 2007;581:3297–302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, Iovine MK. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol. 2008;317:541–8. doi: 10.1016/j.ydbio.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–19. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal. Fin Genetics. 2000;155:1321–9. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Africa D, Horne S, Postlethwait JH. Half-tetrad analysis in zebrafish: mapping the ros mutation and the centromere of linkage group I. Genetics. 1995;139:1727–35. doi: 10.1093/genetics/139.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Ochiai T, Rountree RB, Kingsley DM, Enomoto-Iwamoto M, Iwamoto M, Pacifici M. Synovial joint formation during mouse limb skeletogenesis: roles of Indian hedgehog signaling. Ann N Y Acad Sci. 2007;1116:100–12. doi: 10.1196/annals.1402.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–83. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–17. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Shibukawa Y, Wu C, Tamamura Y, Enomoto-Iwamoto M, Iwamoto M. Cellular and molecular mechanisms of synovial joint and articular cartilage formation. Ann N Y Acad Sci. 2006;1068:74–86. doi: 10.1196/annals.1346.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–18. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl M, Cassimeris L. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol Biol Cell. 2003;14:916–25. doi: 10.1091/mbc.E02-09-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizard A, Burgon PG, Paul DL, Bruneau BG, Seidman CE, Seidman JG. Connexin 40, a target of transcription factor Tbx5, patterns wrist, digits, and sternum. Mol Cell Biol. 2005;25:5073–83. doi: 10.1128/MCB.25.12.5073-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–9. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, Keating MT. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–58. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, Akimenko MA. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99:8713–8. doi: 10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz R, Stricker S, Gazzerro E, Clor JL, Witte F, Nistala H, Zabski S, Pereira RC, Stadmeyer L, Wang X, Gowen L, Sleeman MW, Yancopoulos GD, Canalis E, Mundlos S, Valenzuela DM, Economides AN. The mutation ROR2W749X, linked to human BDB, is a recessive mutation in the mouse, causing brachydactyly, mediating patterning of joints and modeling recessive Robinow syndrome. Development. 2008;135:1713–23. doi: 10.1242/dev.015149. [DOI] [PubMed] [Google Scholar]
- Santamaria JA, Mari-Beffa M, Becerra J. Interactions of the lepidotrichial matrix components during tail fin regeneration in teleosts. Differentiation. 1992;49:143–50. doi: 10.1111/j.1432-0436.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Dev Biol. 2006;299:438–54. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Smith A, Zhang J, Guay D, Quint E, Johnson A, Akimenko MA. Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole-mount in situ hybridization method. Dev Dyn. 2008;237:417–25. doi: 10.1002/dvdy.21417. [DOI] [PubMed] [Google Scholar]
- Thummel R, Bai S, Sarras MP, Jr, Song P, McDermott J, Brewer J, Perry M, Zhang X, Hyde DR, Godwin AR. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–46. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Nusslein-Volhard C. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development. 1996;123:255–62. doi: 10.1242/dev.123.1.255. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene, OR: 1993. [Google Scholar]