Abstract
Assembly of a transcriptional and post-translational molecular interaction network in B cells, the human B-cell interactome (HBCI), reveals a hierarchical, transcriptional control module, where MYB and FOXM1 act as synergistic master regulators of proliferation in the germinal center (GC). Eighty percent of genes jointly regulated by these transcription factors are activated in the GC, including those encoding proteins in a complex regulating DNA pre-replication, replication, and mitosis. These results indicate that the HBCI analysis can be used for the identification of determinants of major human cell phenotypes and provides a paradigm of general applicability to normal and pathologic tissues.
Keywords: B cell, germinal centers, interactome, master regulator
Introduction
Comprehensive repertoires of molecular interactions (interactomes) are becoming increasingly useful in the dissection of biological processes because they provide an integrated view of regulatory programs in the cell (Rhodes et al, 2005). However, their ability to elucidate novel mechanisms of regulation of physiologic and pathologic human phenotypes is still largely undeveloped. For instance, sophisticated analyses of protein–protein (Goh et al, 2007; Lage et al, 2007; Karni et al, 2009; Wu et al, 2009), transcriptional (Ergun et al, 2007), and literature-based (Rzhetsky et al, 2004) interactomes were recently successful in recapitulating previously known disease-related genes. However, novel predictions lacked specificity and were not validated. A notable exception is constituted by recent models of obesity (Yang et al, 2009), whose predictions were confirmed in vivo. These methods, however, require transgenic and knockout mouse models and cannot be readily extended to human phenotypes. Regulatory networks in multicellular organisms are also exquisitely cell-context specific. Yet, most interactomes lack context-specificity because their interactions are supported by ex vivo assays (Rual et al, 2005) or literature data assembled from a diverse mix of cellular phenotypes. Finally, except for a few examples in yeast (Yeger-Lotem et al, 2004; Yu et al, 2006) and for literature-based attempts (Bader et al, 2003; Rzhetsky et al, 2004), available interactomes represent individual layers, such as transcriptional regulation (Rhodes et al, 2005; Palomero et al, 2006; Ergun et al, 2007) or protein complexes (Goh et al, 2007; Lage et al, 2007) rather than an integrated view of regulatory processes.
Here, we show how cell-context-specific interactomes can be efficiently and accurately assembled from high-throughput data (e.g. gene expression profiles (GEPs), yeast two-hybrid assays, etc) using an evidence integration approach by assembling a human B-cell interactome (HBCI). Furthermore, we show that its analysis elucidates both master regulator (MR) genes individually or synergistically controlling specific cellular processes and transcriptional regulation of proteins in large complexes, whose availability must be regulated in context-dependent manner. The latter is a poorly understood process, as transcriptional networks and protein–protein interaction (PPI) networks are usually studied in isolation. It specifically highlights the advantage of an integrated regulatory model, where transcriptional and post-translational interactions may be interrogated at once to discover novel complexes. Specifically, the HBCI was interrogated to discover MRs of key genetic programs in the germinal center (GC) reaction of antigen-mediated immune response, that is, genes that are required for normal progression through the GC, as well as novel physical interactions between the pre-replication complex and mitotic-control proteins. GCs are structures where antigen-stimulated B cells highly proliferate, undergo somatic hypermutation of immunoglobulin genes, and are selected based on the production of high-affinity antibodies. GC B cells (centroblasts) derive from naive B cells, from which they differ for the activation of genetic programs controlling cell proliferation, DNA metabolism, and pro-apoptotic programs and for the repression of anti-apoptotic, cell-cycle arrest, DNA repair, and signal transduction programs from cytokines and chemokines (Klein et al, 2003). A few transcriptional regulators (BACH2, BCL6, IRF8, POU2AF1, and SPIB) necessary for GC formation (Klein and Dalla-Favera, 2008) were identified by genetic and biochemical analyses. However, an unbiased and comprehensive repertoire of GC MRs is not available, and methods for the identification of MRs of human phenotypes are still lacking.
Results
The human B-cell interactome
To construct an integrated, cell-context specific, human interactome, we reverse-engineered transcriptional and post-translational interactions in mature human B cells from a large and phenotypically diverse collection of 254 B-cell GEPs representing 24 distinct phenotypes derived from normal and malignant mature B cells (Lefebvre et al, 2007). Reverse engineering was performed using validated algorithms, such as ARACNe (transcriptional) (Basso et al, 2005; Margolin et al, 2006; Palomero et al, 2006) and MINDy (post-translational) (Wang et al, 2006, 2009a, 2009b; Mani et al, 2008). An established Bayesian evidence integration algorithm (Jansen et al, 2003) further integrated evidence from experimental assays, databases, and literature data mining, filtered by context-specific criteria (full details of the method, performance analysis, and comparison with other methods can be found in Supplementary Figures S1–S3). The HBCI comprises ∼66 000 B-cell-specific molecular interactions (Supplementary Table I), including both PPIs, representing direct physical interactions and indirect ones within the same complex, and direct protein–DNA interactions (Lefebvre et al, 2007).
MAster regulator INference algorithm
To discover MRs of the GC reaction, we interrogated the HBCI using a new algorithm, MAster Regulator INference algorithm (MARINa), designed to infer transcription factors (TFs) controlling the transition between the two phenotypes, A and B, and the maintenance of the latter phenotype. Expression at the mRNA level is often a poor predictor of a TF's regulatory activity and an even worst predictor of its biological relevance in regulating phenotype-specific programs. To obviate this problem, MARINa infers TF activity from the global transcriptional activation of its regulon (i.e. its activated and repressed targets) and its biological relevance by TF-regulon overlap with phenotype-specific programs. Indeed, we reasoned that if the naive-B-cell → centroblast transition is supported by the activation (or repression) of specific TFs, then their targets should be among the most-differentially expressed genes between the two cellular phenotypes, with activated and repressed targets at the opposite end of the expression range. MARINa requires a regulatory model (the HBCI in this case) and a GEP signature of the phenotype transition (i.e. all genes ranked by their differential expression in two phenotypes). Here, the signature, SGC, was obtained by t-test analysis of GC centroblasts versus naive B-cells GEPs. MRs of GC development were then inferred in four steps. First, each TF was associated with a positive and a negative regulon, using its activated and repressed targets in the HBCI, respectively. Second, we computed for each TF its activity by gene set enrichment analysis (GSEA) (Subramanian et al, 2005) of the SGC signature in TF-regulon genes (SGC enrichment) and its biological relevance by the overall contribution of TF-regulon genes to GC-specific programs measured with the differentially expressed target odds ratio (DETOR) score (see Materials and methods). Third, MARINa corrected for TF-regulon overlap, a frequent event in mammalian networks (Margolin et al, 2009; Carro et al, 2010), leading to false-positive MR inference. Indeed, if the regulons of an MR and of a non-MR gene overlapped significantly, then the latter would also be SGC enriched, yielding a false positive. These TFs (shadow regulators or SRs) can be identified and removed by determining whether their enrichment is significantly reduced when shared targets are disregarded (see Supplementary information). We also identified synergistic TF pairs, whose common targets are more SGC enriched than their individual regulons (see Supplementary information). Finally, the GC-MRs are defined as the non-SRs, that is enriched TFs that are not a shadow of any other, and SRs that are synergistic with a non-SR (see Supplementary information).
When applied to 194 TFs displaying ⩾20 targets in the HBCI, MARINa identified 41 candidate MRs, of which 26 were GC activated and 15 were GC repressed (P<10−3, Supplementary Figure S4). Results of the different steps of MARINa can be found in Supplementary Tables II, III, IV. Note that inference of the TF regulons from a subset of the data that did not include samples used to define the SGC signature identified virtually the same MR set, as shown in the Supplementary Figures S5 and S6. This confirms that use of the SGC-related samples in the network inference analysis does not lead to data overfitting in the inference of MRs.
To preliminarily validate the results, we examined whether MARINa-inferred MRs included established GC MRs (Table I; see review Klein and Dalla-Favera, 2008). Of five such genes, three (BCL6, POU2AF1, and SPIB) were directly identified by MARINa, one (IRF8) was borderline significant (P⩽0.01), and one (BACH2) could not be analyzed because it was not represented in the GEP data. It should be noted here that POU2AF1 is not a TF, as it does not directly bind DNA, but rather a transcriptional regulator. In this case, it was included only to illustrate that it could be recovered accurately by the analysis. Ultimately, all transcriptional regulators may be analyzed by the same approach if desired. MARINa also identified other TFs reported to have a function in GCs, such as LMO2 (Natkunam et al, 2007) (GC activated), IRF4 (Klein and Dalla-Favera, 2008), BHLHE40 (Seimiya et al, 2004), and JUNB (Ci et al, 2009) (GC repressed), suggesting that the analysis recapitulates known regulators of GC development. MARINa-inferred activity was generally qualitatively (i.e. activated versus repressed) but not quantitatively consistent with TF differential expression (Supplementary Figure S4). However, not all the MRs identified by the method could have been identified by GEP analysis alone (Supplementary Figure S7).
Table 1. Known MRs of germinal center formation and maintenance and MARINa inference.
| Master regulator | MARINa-inferred TF activity | MARINa P-value |
|---|---|---|
| A master regulator is either ‘activated’ or ‘repressed’, that is showing, respectively, increased or decreased activity in centroblasts compared with naive B cells. If activated, an MR shows more activation and/or repression of its targets, whereas if repressed, an MR will show less activation and/or repression. | ||
| BACH2 | Not tested | NA |
| BCL6 | Activated | <10−4 |
| IRF8 | Activated | 0.01 |
| POU2AF1 | Activated | <10−4 |
| SPIB | Repressed | 4 × 10−4 |
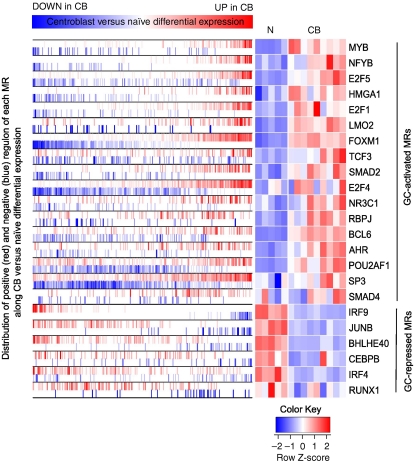
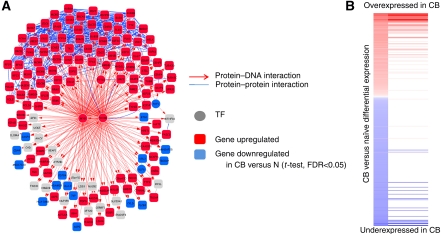
MARINa identified 23 candidate GC-MRs controlling large GC-specific programs (n⩾100 targets) including several new ones (Figure 1). MYB was the most significant GC-activated MR, with >64% of its targets differentially expressed in GCs (FDR⩽0.05 by t-test). Interestingly, regulon analysis (steps 1–2) suggested strong GC activation of the MYC-regulon genes (P⩽10−4). However, MYC was identified and removed as an SR (Supplementary Table III), consistent with the observation that its expression is low in the majority of GC B cells (Klein et al, 2003). FOXM1 provided the largest contribution to this result, by regulating ⩾35% of the MYC-regulon genes that were GC activated. This suggests that FOXM1 may have a critical function in GC formation by regulating specific MYC targets necessary for rapid growth and proliferation. Further analysis of candidate synergistic TF pairs by MARINa revealed that FOXM1 appeared in 7 of the 10 pairs that regulate large GC programs (Supplementary Table IV), specifically co-regulating 150 MYB targets, 121 of which (81%) were differentially expressed in GCs (FDR⩽0.05 by t-test, Figure 2). Taken together, these results suggest that MYB and FOXM1 are synergistic MRs of proliferation in the GC, as also supported by the GC-specific downregulation of mir150 (Basso et al, 2009), a tight repressor of MYB (Xiao et al, 2007), and the fact that MYB protein is expressed in centroblasts despite low MYB mRNA levels (Supplementary Figure S8), suggesting specific stabilization.
Figure 1.
GC-MRs regulating >100 targets. Left side of the plot shows the distribution of the positively (red) and negatively (blue) correlated targets of the MR on the gene list ranked by differential mRNA expression in centroblast compared with naive samples. The right side of the plot shows a heatmap of the mRNA expression level of the MRs in naive (N) and centroblast (CB) samples. Source data is available for this figure at www.nature.com/msb.
Figure 2.
FOXM1/MYB network. (A) FOXM1/MYB subnetwork from the HBCI. (B) Distribution of the MYB/FOXM1 regulon against a list of genes ranked by their differentially expression in centroblast compared with naive B cells. Source data is available for this figure at www.nature.com/msb.
MYB and FOXM1 form a feed-forward loop
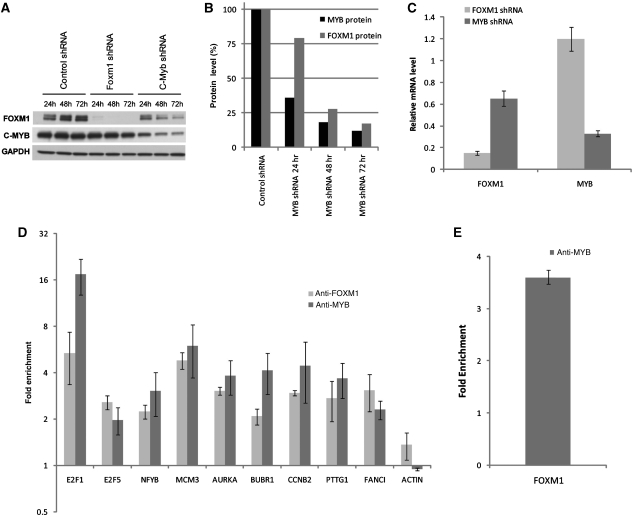
We first tested whether MYB and FOXM1 may regulate each other as predicted in the HBCI. Synergistic TFs often participate in feed-forward loops (Palomero et al, 2006; Carro et al, 2010), increasing the resilience of regulatory networks to transient perturbations by the maintenance of the cell in a phenotype that could be affected by the fluctuation in the levels of either TF alone. MYB and FOXM1 were silenced in the ST486 Burkitt's lymphoma line, representative of transformed GC B cells, by lentiviral-mediated transduction of MYB/FOXM1-specific and control shRNAs (Figure 3). Decrease of endogenous MYB and FOXM1 protein levels in specific versus control shRNA-transduced samples was confirmed by western blot (Figure 3A and B). qPCR assays confirmed mRNA-level reduction of both TFs at 24 h (Figure 3C), persisting at 48 and 72 h (data not shown). MYB mRNA and protein levels were not affected by FOXM1 silencing, whereas FOXM1 mRNA and protein levels were both reduced in a time-dependent manner after MYB silencing (Figure 3A–C). These results suggest that MYB is a transcriptional activator of FOXM1. Quantitative chromatin IP (qChIP) assays showed that MYB binds in vivo to the FOXM1 promoter (Figure 3D). Together, these findings suggest that MYB and FOXM1 form a feed-forward loop, involved in the synergistic activation of a large subset of GC-specific genes.
Figure 3.
MYB directly regulates the transcription of FOXM1. (A) Western blot analysis of ST486 total cell lysates obtained at 24, 48, and 72 h after lentiviral-mediated transduction of control, FOXM1, or MYB shRNA. Viral load was set for complete silencing of FOXM1 at 24 h but only partial (>50%) silencing of MYB, as complete silencing induced >80% apoptosis at 48 h (data from previous experiment). GAPDH was used as a loading control. (B) Percentage of FOXM1 protein level on MYB silencing were calculated by densitometric analysis (in ImageJ) and normalized to GAPDH protein level. (C) qPCR analysis showing the mRNA expression level of FOXM1 and MYB at 24 h. Similar results were observed at 48 and 72 h (data not shown). (D) qChIP with control IgG or anti-MYB antibody at FOXM1-binding site. Data shown in (A, C) are representative of one of three independent experiments. Error bars were calculated (C) from samples run in duplicate or (D) from two sets of three independent real-time PCR reactions. (E) qChIP with control IgG, anti-FOXM1, and anti-MYB antibodies performed in the nuclear extract of ST486 cells. Samples from two independent experiments were pooled together and real-time PCR reactions were run in duplicate. Error bars represent the standard error.
MYB and FOXM1 silencing affects the expression of their common predicted targets and other MRs
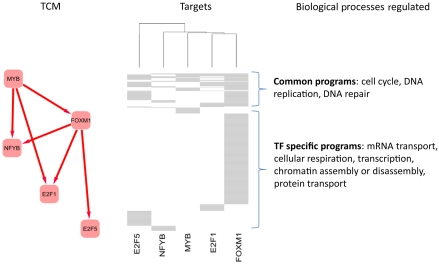
To test whether common MYB/FOXM1 targets and other MRs were affected by silencing of either TF, Affymetrix HG-U95A GeneChip GEP were obtained in triplicate from ST486 cells after transduction with FOXM1, MYB, and control shRNA. To eliminate indirect regulation effects, a 24 h time point was selected with a 25% lower viral load for MYB shRNA than for the time-course experiment (Figure 3A), ensuring that FOXM1 protein levels were not yet affected by MYB silencing (Supplementary Figure S9). Silencing was confirmed by western blot and qPCR (Supplementary Figure S9). GSEA analysis of predicted MYB/FOXM1 targets against differentially expressed genes after silencing confirmed that positive MYB/FOXM1-regulon targets were downregulated by silencing either TF (P<10−4 in both cases, Supplementary Figure S10). Although, the negative regulon was too small for GSEA analysis, having only 19 genes, five of these were significantly upregulated after FOXM1 or MYB silencing. Accordingly, qChIP assays for six inferred MYB/FOXM1 targets (AURKA, BUBR1, CCNB2, FANCI, MCM3, and PTTG1), whose mRNA levels decreased significantly after silencing of either TF (Supplementary Table V) confirmed that both TFs bind to their promoters (Figure 3E). GEP analysis, confirmed by qPCR, identified TFs downregulated after MYB/FOXM1 silencing, including NFYB, E2F5, and E2F1, three of the most significant MRs after MYB (Supplementary Figure S11), whereas qChIP assays confirmed binding of both FOXM1 and MYB to their promoters (Figure 3E). Taken together, these data suggest a highly connected, hierarchical transcriptional control module (TCM), with MYB and FOXM1 as synergistic top-level MRs (Figure 4).
Figure 4.
TCM showing GC-MRs controlled by MYB and FOXM1 and their targets. The figure describes the relationship between MYB and FOXM1 and three other MRs downregulated after silencing the two TFs independently (see qPCR results in Supplementary Figure S11). We used hierarchical clustering to cluster the five MRs according to their common targets. The distance between each MR was computed using their percentage of common targets and we used the ward agglomeration method. Gene Ontology enrichment was computed for common MRs targets as well as specific targets of each MR.
MYB and FOXM1 silencing induce apoptosis and reduce cell proliferation
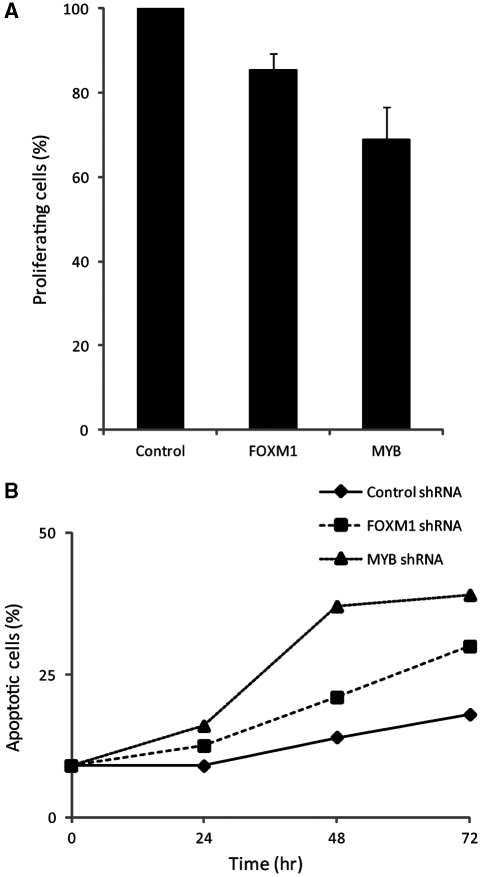
To gain insight into the biological significance of the programs controlled by the MYB/FOXM1 pair, we performed Gene Ontology (GO) (Ashburner et al, 2000) term enrichment analysis of their common target genes, which showed that they regulate cell-cycle dependent programs, including replication, replication initiation, cell proliferation, and mitosis (Supplementary Table VI). We thus investigated the physiological changes induced by FOXM1 and MYB silencing. BrdU assays at 24 h in ST486 cells showed a 15 and 30% decrease in proliferating cells, after FOXM1 and MYB silencing, respectively (Figure 5A), and a similar decrease at 48 and 72 h (data not shown). Annexin V and propidium iodide staining at the same time points showed gradual increase of apoptotic cells reaching 30 and 40% (Figure 5B) and dead cells reaching 67 and 77% (data not shown) at 72 h after FOXM1 and MYB silencing, respectively. At that time point, MYB protein was 90% silenced, suggesting that loss of MYB protein induces apoptosis both in a FOXM1-dependent and a FOXM1-independent way. These results indicate that MYB and FOXM1 are necessary for viability and rapid proliferation of GC-related B cells.
Figure 5.
FOXM1 and MYB are required for cell proliferation and viability of ST486 cells. (A) Proliferating ST486 cells were quantified by BrdU labeling of cells transduced with control, FOXM1, or MYB shRNA, and collected at 24 h. Error bars represent standard error calculated from the samples run in triplicate. (B) The percentage of apoptotic ST486 cells was assessed by Annexin V staining of cells transduced with control, FOXM1, or MYB shRNA and collected at 24, 48, and 72 h post-infection.
Pre-replication and mitosis-related targets of MYB and FOXM1 are involved in a protein complex
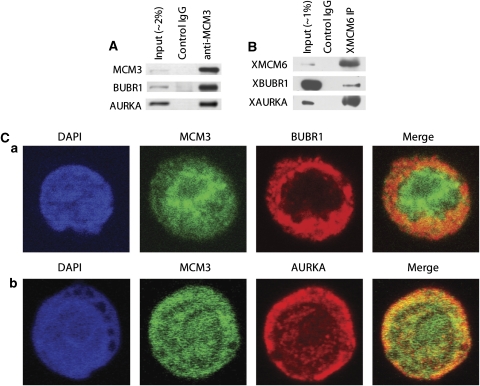
To gain more insight into the control of GC-proliferation phenotype by MYB and FOXM1, we further examined specific targets involved in the coordinated formation of a large and previously uncharacterized protein complex (Figure 2A). This type of analysis was not afforded by earlier approaches that considered the transcriptional and post-translational interaction layers in isolation and highlights a key novelty of such an integrated regulatory model. Approximately half of MYB/FOXM1 targets cluster within an HBCI-inferred protein complex, which is poorly represented in existing protein–protein interactomes (Rual et al, 2005). Surprisingly, these proteins are virtually all members of only three biological process categories in GO: replication initiation (P⩽10−6), replication (P⩽10−19), and mitosis (P⩽10−32) (Supplementary Figure S12). For instance, the HBCI-inferred complex included four of the six members of the replicative helicase complex MCM2–7 (Ying and Gautier, 2005), suggesting that missing proteins (MCM2 and MCM5) may be false negatives in the HBCI and that assembly of the replicative helicase in GCs may be regulated by the MYB/FOXM1 module. This was experimentally confirmed since five (MCM3–7) out of the six MCM genes (80%), and 58% of all pre-replication complex (pre-RC) proteins were downregulated after MYB or FOXM1 silencing (Supplementary Figure S13). As pre-RC and mitotic proteins have not been previously reported to physically interact, we tested whether two mitotic kinases (BUBR1 and AURKA) in the inferred complex physically interact with MCM3, a core protein of the MCM2–7 replicative helicase, chosen based on reagent availability. GEP, qPCR, and qChIP assays showed that their mRNA level was affected by silencing of either FOXM1 or MYB (Supplementary Figure S11A) and that both TFs bind to their promoters (Figure 3E), confirming that they are direct targets of FOXM1 and MYB. BUBR1/MCM3, AURKA/MCM3 interactions were confirmed by endogenous co-immunoprecipitation (co-IP) assays in Ramos (Figure 6A) and ST486 Burkitt's lymphoma lines (data not shown). Ramos cells were included as their large size is optimal for confocal microscopy analysis. The novel AURKA/BUBR1 interaction was also confirmed (data not shown). Co-IP assays of homologues of these proteins in Xenopus extracts, which are devoid of genomic DNA, confirmed that these interactions are not chromatin mediated (Figure 6B). Similar results were obtained when the MCM complex was immunoprecipitated with XMCM3 and XMCM6 antibodies (data not shown). These experiments establish that MCM3/6-BUBR1 and MCM3/6-AURKA interactions are either direct or protein mediated, as predicted by the HBCI, but not DNA mediated. High-resolution confocal imaging showed that these proteins are co-localized either in the nucleus or perinuclearly (Figure 6C). Finally, analysis of E-Map data (Collins et al, 2007) (see Supplementary information) showed a strong association between pre-RC and mitotic control in yeast, suggesting that the interaction between these proteins may be evolutionarily conserved (Supplementary Tables VII, VIII; Supplementary Figure S14).
Figure 6.
MCM3 and MCM6 interact with BUBR1 and AURKA. (A) MCM3 co-immunoprecipitates with BUBR1 and AURKA. Cell lysates of Ramos cell line were immunoprecipitated with control IgG or anti-MCM3 antibodies and probed with anti-BUBR1 and anti-AURKA antibodies. (B) MCM6 binds to BUBR1 and AURKA in Xenopus cell-free extracts. Interphase extracts from Xenopus cytosolic fraction were immunoprecipitated with anti-XMCM6 or control IgG antibodies and probed with anti-XMCM6, anti-XBUBR1, and anti-XAURKA antibodies. (C) Co-localization of MCM3 with BUBR1 and AURKA in Ramos cells shown by confocal dual-color indirect immunofluorescence with (a) anti-MCM3 and anti-BUBR1 antibodies or (b) anti-MCM3 and anti-AURKA antibodies. DNA was stained with DAPI. Data shown are representative of one of three independent experiments.
Discussion
In summary, these results document that coordinated analysis of both transcriptional and post-translational interactions in the HBCI can identify synergistic MRs of human phenotypes, as well as provide insight on the functional regulatory role of these proteins. Specifically, analysis of the MRs’ regulatory targets led to the discovery of a large complex that includes both the pre-RC and mitotic-control proteins and whose functional role is narrowly defined. This would not have been possible if the transcriptional and post-translational regulation layer had been independently analyzed. Indeed, biochemical and functional validation suggests that the HBCI constitutes an informative and accurate model of regulation, providing a useful framework to investigate normal B-cell biology and its dysregulation in B-cell lymphomas, which derive from GC B cells (Mani et al, 2008).
Regulon-based clustering of MRs (Supplementary Figure S15) shows that the regulatory programs controlled by the MYB/FOXM1 TCM and by other MRs, including experimentally validated ones are virtually non-overlapping. This suggests that GC regulation is implemented by the coordinated activation and repression of several individual MRs and MR modules, each one controlling specific GC-related programs, and thus not individually sufficient for GC formation. Moreover, the network reconstruction and MR identification are not dependent on the inclusion of a specific subset of B-cell samples (Supplementary information). Indeed, excluding 14 naive and centroblast samples or any 14 samples did not significantly affect the TCM inference (Supplementary Figures S5 and S6).
The HBCI constitutes only an initial draft of human B-cell regulatory networks. For instance, some interactions may be missed (false negatives, FN) and some incorrect interactions may be included in the model (false positives, FP). In addition, analysis of multivariate regulation is only in its infancy and better models will be developed over the next few years. The HBCI must thus be used with the full understanding of its limitations but with the expectation that, as such models improve over time, they will provide increasingly more accurate model of cell-specific behavior. In this, evolutionary constraints to achieve functional canalization (Waddington, 1959) help dramatically. For instance, as shown here and in earlier work (Palomero et al, 2006; Margolin et al, 2009; Carro et al, 2010), the dominant use of feed-forward loops to stabilize and specialize TF regulation in human cells creates critical redundancy that obviates FNs. Similarly, regulon size of TFs associated with regulation of key function makes these models relatively insensitive to FPs as, statistically, activity can be better estimated from large number of targets due to their combinatorial regulation. It is also difficult to evaluate the accuracy of the network reconstruction with traditional metrics, mainly because of the highly incomplete nature of gold standard sets, both positive and negative. These metrics can be effectively used to compare relative performance between different approaches but not to estimate the absolute FP and FN rates of the methods. Therefore, experimental validation is the ultimate criterion to measure the method's precision. As shown, experimental validation confirmed the accuracy of the reverse-engineered network and suggested that the use of such a model can capture important regulatory processes such as cell proliferation.
Taken together, our data suggest that the assembly of context-specific interactomes, such as the HBCI, coupled with algorithms for their interrogation, such as MARINa, constitute valuable resources in the elucidation of physiologic phenotypes. In particular, the approach can be applied to any phenotypic transition thus establishing a novel paradigm for the study of context-specific cellular regulation.
Materials and methods
Differentially expressed target odds ratio
Results were classified using the DETOR score for differentially expressed target odds ratio and computed as
where GSiLE and RSiLE are the number of genes before the leading edge in GSEA for the gene set (or regulon) and for the reference set, whereas GSi and RSi are the sizes of the gene set and the reference set.
Cell lines and cell culture
ST486 and Ramos cells were maintained in Iscove's modified Dulbecco's medium supplemented with 10% FBS and antibiotics. 293FT cells were maintained in Dulbecco's minimum essential medium supplemented with 10% FBS and antibiotics.
Lentiviral infection of B-cell lymphoma cell lines
Control shRNA (SHC002), FOXM1 shRNA (TRCN0000015546), or MYB shRNA (TRCN0000040062) cloned into pLKO.1-puro lentiviral vector was purchased from Sigma. To generate lentiviral particles, the individual shRNA clones (2.8 μg) were co-transfected with VSVG envelope plasmid (280 ng) and Δ8.9 packaging vector (2.5 μg) into subconfluent 100 mm plate of 293FT cells using Fugene 6 (Roche). The viral particles were collected at 48 and 72 h post-transfection, filtered and concentrated by ultracentrifugation in Beckman SW28 rotor at 25 000 rpm for 1.5 h 5 × 106 ST486 cells were transduced with the viral particles in the presence of 8 μg/ml polybrene and centrifuging at 450 g for 1.5 h.
Immunoblot analysis
For immunoblotting, whole-cell lysates were prepared from ST486 cells by using RIPA buffer (Teknova) with protease inhibitor cocktail (Roche). Proteins were fractionated by SDS–PAGE and analyzed by standard immunoblotting procedures using the following antibodies: anti-FOXM1 (sc-502; Santa Cruz), anti-MYB (sc-517, Santa Cruz), anti-MCM3 (559543, BD Pharmingen), anti-BUBR1 (ab4637, Abcam), anti-AURKA (gift from Dr PT Stukenberg), anti-XBubR1 (gift from Dr Y Mao), and anti-GAPDH (sc-32233; Santa Cruz).
Co-immunoprecipitation
Whole-cell lysates of 40 × 106 Ramos cells were prepared by using cell lysis buffer (50 mM Tris–HCl pH 8.1, 150 mM NaCl and 0.5% NP-40) with protease inhibitor cocktail. The lysate was incubated overnight at 4°C with mouse IgG or anti-MCM3 antibody (M038-3, MBL Intl), followed by incubation in protein-G agarose beads (17-0618, Amersham) for 2 hours. The beads were washed in buffer (20 mM Tris–HCl pH 8.1, 150 mM NaCl, 10% glycerol, 0.2% NP-40 and 1 mM EDTA) for four times and incubated in SDS-loading buffer at 100°C for 10 min. Similarly, cell-free extract (LSS) was prepared from unfertilized Xenopus eggs as described earlier(Smythe and Newport, 1991). Co-IPs were performed by incubating extracts with anti-XMCM6 antibodies as described earlier (Ying and Gautier, 2005)) or control antibodies (Rabbit IgG, Sigma) for 30 min at 4°C, followed by addition of Protein A Sepharose beads (GE Healthcare) for 1 h at 4°C. Beads were washed with ELB-0.2%NP-40 and ELB-0.5MNaCl, then boiled to release bound proteins. Proteins were fractionated by SDS–PAGE and analyzed by standard immunoblotting procedures.
Indirect immunofluorescence
Ramos cells were fixed in 10% formalin for 20 min and post-fixed in ice-cold 100% methanol for 20 min. Cells were then blocked in 5% BSA for 1 h and incubated with anti-MCM3 (559543, BD Pharmingen) and anti-BUBR1 (ab4637, Abcam) or anti-MCM3, and anti-AURKA (ab13824, Abcam) antibody for 2 h at room temperature. The cells were washed three times in PBS with 0.01% Tween-20 followed by incubation with biotinylated goat anti-mouse IgG for 30 min. It was then incubated with FITC-conjugated goat anti-rabbit antibody and Cy3-conjugated Streptavidin antibody (Jackson ImmunoResearch) for 30 min. Stained cells were acquired at × 100 magnification in LSM510 Multiphoton confocal microscope (Zeiss).
RNA isolation, microarray analysis, and RT–PCR
Total RNA extracts was extracted with Trizol (Invitrogen) and purified using RNeasy kit (Qiagen). For qPCR, cDNA was prepared from the total RNA extract by using Reverse Transcriptase kit (Qiagen), and the amount of each gene was determined by using SYBR Green (Qiagen) on an Applied Biosystems 7300 thermal cycler. The reactions were run in duplicate, and the number of copies of each gene was normalized to the housekeeping gene GAPDH. For microarray analysis, total RNA was amplified and converted into labeled double-stranded cDNA by reverse transcription using Superscript II reverse transcriptase (Invitrogen) and poly-dT oligonucleotide containing T7 RNA polymerase initiation site. Biotinylated cRNA was generated from the double-stranded cDNA by using the one-cycle cDNA synthesis protocol (Affymetrix, 701025 Rev.6). A measure of 15 μg of fragmented cRNA was hybridized to HG-U95Av2 microarrays (Affymetrix). For oligonucleotide sequences, see Supplementary Table IX. GEPs were normalized using a modified GC robust multiarray average (Lim et al, 2007) and differential expression was assessed by t-test (Supplementary Table V). GEO accession number is GSE17172.
Quantitative chromatin IP
Promoter sequences of the target genes 2 kb upstream and 2 kb downstream of the transcription start site were retrieved by using UCSC Golden Path database (Build35, May 2004) (Karolchik et al, 2008). We identified sites, as given by TRANSFAC, in 4 kb promoters using a scoring threshold corresponding to 0.9 specificity across all known promoters (Smith et al, 2007). ChIP analysis was done in ST486 cell line by following the protocol described in the literature (Frank et al, 2001). In brief, the protein–DNA complexes were cross-linked using formaldehyde, followed by IP with anti-MYB (mixture of sc-7874 and sc-517 (Santa Cruz)), FOXM1 (sc-502, Santa Cruz), or Rabbit IgG. The samples were reverse cross-linked and DNA was purified by phenol-chloroform extraction. DNA pellet resuspended in 30 μl of water, and the total input chromatin taken before IP was diluted in 150 μl of water. A measure of 3 μl of the DNA samples were taken and binding of the TFs to the targets was determined by qPCR using the primers designed to contain the putative-binding sites. For oligonucleotide sequences, see Supplementary Table X.
Apoptotic analysis
The number of apoptotic and dead cells were determined by using the Annexin V-FITC Apoptosis Kit (BD Pharmingen) according to the manufacturer's instructions. Samples were analyzed by using FACS Calibur (Becton Dickinson).
Proliferation assay
The number of proliferating cells was determined by using the BrdU Cell Proliferation Assay (Calbiochem) according to the manufacturer's protocol.
Supplementary Material
Supplementary text, Supplementary figures S1–15, Supplementary tables SI–X
Acknowledgments
This work was supported by the National Cancer Institute (R01CA109755), the National Institute of Allergy and Infectious Diseases (R01AI066116), the In Silico Research Centre of Excellence (NCI-caBIG 29XS192), the National Centers for Biomedical Computing NIH Roadmap Initiative (U54CA121852), the National Institute of General Medical Sciences (5R01GM085659-02), and National Institute on Drug Abuse (5R01DA013821-06). We thank Klaus Rajewsky for his advice and critical suggestions. We also thank Andrey Rzhetsky and Raul Rodriguez-Esteban for providing us GeneWays interactions and for their helpful comments, Anshul Kundaje for helping us for the implementation of Adaboost and Alberto Ambesi-Impiombato for useful discussions on qPCR results.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res 31: 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A (2005) Reverse engineering of regulatory networks in human B cells. Nat Genet 37: 382–390 [DOI] [PubMed] [Google Scholar]
- Basso K, Sumazin P, Morozov P, Schneider C, Maute RL, Kitagawa Y, Mandelbaum J, Haddad J Jr, Chen CZ, Califano A, Dalla-Favera R (2009) Identification of the human mature B cell miRNome. Immunity 30: 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A (2010) The transcriptional network for mesenchymal transformation of brain tumours. Nature 463: 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W, Polo JM, Cerchietti L, Shaknovich R, Wang L, Yang SN, Ye K, Farinha P, Horsman DE, Gascoyne RD, Elemento O, Melnick A (2009) The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood 113: 5536–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Ergun A, Lawrence CA, Kohanski MA, Brennan TA, Collins JJ (2007) A network biology approach to prostate cancer. Mol Syst Biol 3: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL (2007) The human disease network. Proc Natl Acad Sci USA 104: 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M (2003) A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science 302: 449–453 [DOI] [PubMed] [Google Scholar]
- Karni S, Soreq H, Sharan R (2009) A network-based method for predicting disease-causing genes. J Comput Biol 16: 181–189 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Kober KM, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A et al. (2008) The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res 36: D773–D779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R (2008) Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol 8: 22–33 [DOI] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R (2003) Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci USA 100: 2639–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, Moreau Y, Brunak S (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol 25: 309–316 [DOI] [PubMed] [Google Scholar]
- Lefebvre C, Lim WK, Basso K, Dalla-Favera R, Califano A (2007) A context-specific network of protein-DNA and protein-protein interactions reveals new regulatory motifs in human B cells. Lect Notes Bioinform 4532: 42–56 [Google Scholar]
- Lim WK, Wang K, Lefebvre C, Califano A (2007) Comparative analysis of microarray normalization procedures: effects on reverse engineering gene networks. Bioinformatics 23: i282–i288 [DOI] [PubMed] [Google Scholar]
- Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, Califano A (2008) A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol 4: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Favera D, Califano A (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform 7 (Suppl 1): S1–S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G (2009) ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci USA 106: 244–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, Hammer AS, Hamilton Dutoit S, Lossos IS, Levy R (2007) The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood 109: 1636–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA (2006) NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA 103: 18261–18266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM (2005) Mining for regulatory programs in the cancer transcriptome. Nat Genet 37: 579–583 [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Rzhetsky A, Iossifov I, Koike T, Krauthammer M, Kra P, Morris M, Yu H, Duboue PA, Weng W, Wilbur WJ, Hatzivassiloglou V, Friedman C (2004) GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform 37: 43–53 [DOI] [PubMed] [Google Scholar]
- Seimiya M, Wada A, Kawamura K, Sakamoto A, Ohkubo Y, Okada S, Hatano M, Tokuhisa T, Watanabe T, Saisho H, Tagawa M, O-Wang J (2004) Impaired lymphocyte development and function in Clast5/Stra13/DEC1-transgenic mice. Eur J Immunol 34: 1322–1332 [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ (2007) Tissue-specific regulatory elements in mammalian promoters. Mol Syst Biol 3: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C, Newport JW (1991) Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol 35: 449–468 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1959) Canalization of development and genetic assimilation of acquired characters. Nature 183: 1654–1655 [DOI] [PubMed] [Google Scholar]
- Wang K, Alvarez MJ, Bisikirska BC, Linding R, Basso K, Dalla Favera R, Califano A (2009a) Dissecting the interface between signaling and transcriptional regulation in human B cells. Pac Symp Biocomput 264–275 [PMC free article] [PubMed] [Google Scholar]
- Wang K, Banerjee N, Margolin AA, Nemenman I, Califano A (2006) Genome-wide discovery of modulators of transcriptional interactions in human B lymphocytes. Lect Notes Comput Sci 3909: 348–362 [Google Scholar]
- Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, Rajbhandari P, Shen Q, Nemenman I, Basso K, Margolin AA, Klein U, Dalla-Favera R, Califano A (2009b) Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol 27: 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu Q, Jiang R (2009) Align human interactome with phenome to identify causative genes and networks underlying disease families. Bioinformatics 25: 98–104 [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131: 146–159 [DOI] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, Torosyan G, Majid S, Falkard B, Kleinhanz RR, Karlsson J, Castellani LW, Mumick S, Wang K, Xie T, Coon M, Zhang C, Estrada-Smith D, Farber CR, Wang SS et al. (2009) Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet 41: 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem E, Sattath S, Kashtan N, Itzkovitz S, Milo R, Pinter RY, Alon U, Margalit H (2004) Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc Natl Acad Sci USA 101: 5934–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying CY, Gautier J (2005) The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J 24: 4334–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xia Y, Trifonov V, Gerstein M (2006) Design principles of molecular networks revealed by global comparisons and composite motifs. Genome Biol 7: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary text, Supplementary figures S1–15, Supplementary tables SI–X