Abstract
The identification of Smads as protein transcription factors in 1995 led to elucidation of the canonical transforming growth factor–β (TGF-β) signaling pathway. In the years that have followed, nuances of the pathway have been realized, and the once-simple scheme of ligand to receptor to activated transcription factor is now understood to be highly regulated at each step and riddled with crosstalk from other pathways. The Smads are also recognized as important players outside of canonical TGF-β–dependent signaling and are responsible for regulating diverse cellular processes. New evidence suggests that Smad7 plays an integral role in maintaining cell-cell adhesion through direct regulation of β-catenin. Receptor-activated Smads regulate the processing of a subset of microRNAs, particularly miR-21. The number of reports demonstrating the interactions of Smads with proteins outside of canonical TGF-β signaling is increasing, although the functional relevance of these interactions is not known. Investigating these interactions will likely yield more evidence that Smads serve important and diverse purposes beyond their original reported function as signal transducers in the TGF-β pathway.
A Canonical View
Transforming growth factor–β (TGF-β) was first isolated from serum growth factor in the early 1980s (1) and was subsequently characterized as a potent regulator of cell migration, differentiation, and proliferation (2, 3). It was not until over a decade later that receptor-activated Smads (R-Smads) were identified as the transcription factors that carry the TGF-β signal downstream (4). Three classes of Smads have since been identified: R-Smads, common Smads (co-Smads), and inhibitory Smads (I-Smads). In the presence of TGF-β or the related ligands bone morphogenetic proteins (BMPs), ligand-specific TGF-β receptors, which are composed of a type II and type I subunit), become activated, and the type I TGF-β receptor subunit (TβRI) phosphorylates R-Smads: Smad 1, 5, and 8 (BMP) or Smad 2 and 3 (TGF-β) (5). At the level of the TβRI, the I-Smads, Smad6 and 7, act as competitive inhibitors of R-Smad phosphorylation (6). Once phosphorylated, R-Smads complex with the co-Smad Smad4 and accumulate in the nucleus to mediate transcriptional events (7) (summarized in Fig. 1). The canonical transcriptional outcome of TGF-β stimulation is dependent on cell type (8, 9), secondary to which coactivators or repressors are recruited to the Smad transcriptional complex (10). TGF-β has been implicated in many aspects of tissue development and maintenance (2), and aberrant signaling causes a cadre of disease states (2); thus, understanding this pathway is a priority. Elucidation of this signaling cascade temporarily subjugated Smads to the role of mere transducers of TGF-β signaling. New studies, however, reveal that Smads control processes that are independent of TGF-β and that Smads have a complicated existence of their own.
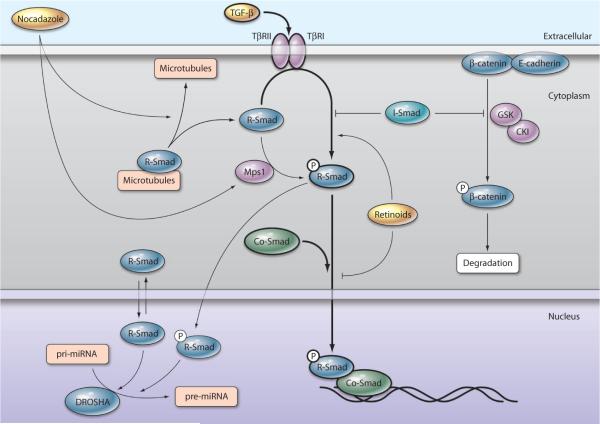
Fig. 1.
Canonical and noncanonical Smad signaling. A simplified representation of canonical TGF-β signaling is depicted centrally with thick arrows and involves ligand-mediated receptor activation, R-Smad phosphorylation, and Smad translocation to the nucleus to regulate transcription. Noncanonical pathways have thinner arrows. A noncanonical function of I-Smad is the stabilization of β-catenin, which allows it to interact with E-cadherin and increases cell-cell adhesion. Retinoids promote noncanonical activities of R-Smads by simultaneously potentiating R-Smad phosphorylation by TβRI and blocking R-Smad nuclear accumulation, thus decreasing nuclear functions of Smads. Noncanonical activation of R-Smads occurs after nocodazole treatment, which leads to disassociation of R-Smads from microtubules and activation of Mps1 kinase, which phosphorylates R-Smads. Nuclear-localized R-Smads also join the DROSHA miRNA-processing complex to increase the conversion of pri- to pre-miRNA of specific miRNAs.
Noncanonical Functions of Smads
The I-Smad Smad7, in the context of canonical TGF-β signaling, has been implicated in both inhibition (11–13) and promotion (14, 15) of tumorigenesis. In early tumorigenesis, it has been suggested that loss of Smad7 is protective, yielding more R-Smad phosphorylation and subsequently more apoptosis and growth arrest of cancerous cells (16). In contrast, increased abundance of Smad7 late during tumorigenesis can yield less R-Smad phosphorylation and consequently fewer downstream events, such as epithelialmesenchymal transformation (17) and cell migration (12), thus providing a protective effect. Tang et al. have shed new light on the role of Smad7 in cancer and how its noncanonical function can promote increased cell-cell adhesion (18), which may potentially inhibit cancer metastasis. They demonstrated that Smad7 directly interacted with β-catenin to inhibit its phosphorylation by the target kinases glycogen synthase kinase 3β (GSK-3β) and casein kinase Iα (CKIα), resulting in decreased β-catenin degradation (18). Unphosphorylated, stabilized β-catenin complexes with E-cadherin, yielding strengthened cell-cell adhesion (18). Whether Smad7 regulates the phosphorylation of other intracellular proteins remains unknown, but Smad7 does interact with other proteins that can be phosphorylated, including PIASy, protein inhibitor of activated signal transducer and activator of transcription (STAT) (19). Likewise, Smad7 binds both the R-Smad transcriptional co-activator, p300, and its activating kinase, Akt (20), creating a possible scenario for Smad7 to further regulate TGF-β signaling in a noncanonical manner. The extent to which Smad7 is a general phosphorylation inhibitor is unknown, but the door is clearly open for further studies.
Noncanonical functions of R-Smads have also been reported. The R-Smads have been implicated in the processing of RNAs. Following a yeast–two hybrid screen designed to discover novel Smad binding partners, Warner et al. determined that Smad3 directly bound Splicing factor 3b subunit2 (SF3b2), a member of the major RNA splicing complex (21). Although the functional consequences of this interaction remain unknown, R-Smads have now been reported to function in another type of RNA processing, microRNA (miRNA) processing. The R-Smads—Smads 1, 2, and 3—can facilitate the posttranscriptional processing of a subset of miRNAs, specifically miR-21 (22). These R-Smads interacted with p68, an RNA helicase that operates in the DROSHA miRNA processing complex (22). By an uncharacterized mechanism, Smad involvement in the DROSHA complex promotes cleavage of the primary precursor mRNA (pri-miRNA) into precursor miRNA (pre-miRNA) (22). Because miR-21 down-regulates the tumor suppressor PDCD4 (programmed cell death 4) (23–25), regulation of miR-21 production by Smads further implicates Smads as promoters of tumorigenesis. TGF-β and BMP ligands have indirect control over this mechanism because phosphorylated receptor Smads accumulate in the nucleus after ligand stimulation and only nuclear-localized Smads complex with DROSHA (22). However, Smad phosphorylation status has no bearing on the ability of Smads to participate in this miRNA processing (22); both phosphorylated and unphosphorylated Smads can participate. Although Smads are in a perpetual state of flux between the nucleus and the cytoplasm (26), it is unknown to what extent they are involved in basal miRNA processing in the absence of TGF-β or BMP stimulation. Reevaluation of R-Smad knockout models in light of this function in miRNA processing may reveal the importance of miR-21 in a number of biological processes. Smad2 knockout mice are embryonic lethal because of a gastrulation defect (27), and it would be of interest to determine whether misregulation of miR-21 contributes to this phenotype and whether miR-21 plays a role in early developmental processes. Likewise, Smad3 knockout mice show accelerated wound healing and increased incidence of colon carcinoma (27), and it remains to be determined whether amounts of miR-21 are altered and contribute to these phenotypes.
Noncanonical Regulation of Smad Activity
Phosphorylation of R-Smads on the canonical Ser-Ser-X-Ser (where X is any amino acid) motif can be accomplished by stimulation with agents other than TGF-β and by alternate kinases other than TβRI. Epidermal and hepatocyte growth factors stimulate the acute phosphorylation of R-Smads (that is, the effect is not secondary to transcriptional events); however, the kinase responsible for this phosphorylation remains unknown (28). Treating cells with nocodazole, a microtubule-depolymerizing agent, also increases Smad phosphorylation above basal levels in the absence of TGF-β (29). Because the R-Smads bound microtubules, Dong et al. hypothesized that microtubules sequester Smads and restrict their access to the TGF-β receptor (29). However, an alternate kinase, Mps1, which is involved in cell cycle and mitotic control, is activated by nocodazole (30). Mps1 phosphorylates R-Smads in a TGF-β–independent manner, leading Zhu et al. to propose that “cell cycle-specific activation of Smad signaling” by Mps1 might control transcription of genes that are temporally important for mitosis (30).
The activities of R-Smads can be directly regulated by retinoids (that is, nontranscriptional). One example of this interaction is the observation that retinoids can potentiate TGF-β–mediated phosphorylation of Smad2 (31). However, Smads phosphorylated in the presence of retinoids do not behave in a canonical manner. When cells are stimulated with both TGF-β and retinoids, phosphorylated R-Smads do not enter the nucleus or participate in downstream transcriptional events as effectively compared with cells stimulated with TGF-β alone (31). Thus, retinoids can influence both canonical TGF-β signaling and noncanonical functions of Smads by controlling the phosphorylation of R-Smads by TβRI, while simultaneously regulating their nuclear accumulation (31). Because the retinoid and TGF-β pathways are critical in many of the same normal and disease processes, including development (32, 33), skin disorders (34), and tumorigenesis (35), it is important to remember that these two pathways are intimately linked on many levels, including transcriptional regulation of TGF-β pathway components by retinoids (32, 33, 35) and regulation of Smad phosphorylation and nuclear localization by retinoids (31).
Many questions remain regarding Smads' activities, inside and outside of canonical TGF-β signaling. Reevaluation of previously unexplainable findings in light of these newly recognized roles for Smads might reveal that noncanonical Smad signaling contributes to the observed phenotypes and cellular responses. Likewise, several functionally uncharacterized Smad protein-protein interactions have been reported (21), and these could serve as a launching pad for further elucidating the diverse roles of Smads, particularly those that are autonomous of TGF-β.
References
- 1.Roberts AB, Anzano MA, Lamb LC, Smith JM, Sporn MB. New class of transforming growth factors potentiated by epidermal growth factor: Isolation from non-neoplastic tissues. Proc. Natl. Acad. Sci. U.S.A. 1981;78:5339–5343. doi: 10.1073/pnas.78.9.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massague J. TGF-β signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 3.Kingsley DM. The TGF-β superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 4.Heldin CH, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 5.Lutz M, Knaus P. Integration of the TGF-β pathway into the cellular signalling network. Cell. Signal. 2002;14:977–988. doi: 10.1016/s0898-6568(02)00058-x. [DOI] [PubMed] [Google Scholar]
- 6.Park SH. Fine tuning and cross-talking of TGF-β signal by inhibitory Smads. J. Biochem. Mol. Biol. 2005;38:9–16. doi: 10.5483/bmbrep.2005.38.1.009. [DOI] [PubMed] [Google Scholar]
- 7.Feng XH, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 8.Kimura A, Katoh O, Hyodo H, Kuramoto A. Transforming growth factor-β regulates growth as well as collagen and fibronectin synthesis of human marrow fibroblasts. Br. J. Haematol. 1989;72:486–491. doi: 10.1111/j.1365-2141.1989.tb04310.x. [DOI] [PubMed] [Google Scholar]
- 9.Chatani Y, Tanimura S, Miyoshi N, Hattori A, Sato M, Kohno M. Cell type-specific modulation of cell growth by transforming growth factor β 1 does not correlate with mitogen-activated protein kinase activation. J. Biol. Chem. 1995;270:30686–30692. doi: 10.1074/jbc.270.51.30686. [DOI] [PubMed] [Google Scholar]
- 10.Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 11.Azuma H, Ehata S, Miyazaki H, Watabe T, Maruyama O, Imamura T, Sakamoto T, Kiyama S, Kiyama Y, Ubai T, Inamoto T, Takahara S, Itoh Y, Otsuki Y, Katsuoka Y, Miyazono K, Horie S. Effect of Smad7 expression on metastasis of mouse mammary carcinoma JygMC(A) cells. J. Natl. Cancer Inst. 2005;97:1734–1746. doi: 10.1093/jnci/dji399. [DOI] [PubMed] [Google Scholar]
- 12.Javelaud D, Delmas V, Moller M, Sextius P, Andre J, Menashi S, Larue L, Mauviel A. Stable overexpression of Smad7 in human melanoma cells inhibits their tumorigenicity in vitro and in vivo. Oncogene. 2005;24:7624–7629. doi: 10.1038/sj.onc.1208900. [DOI] [PubMed] [Google Scholar]
- 13.Javelaud D, Mohammad KS, McKenna CR, Fournier P, Luciani F, Niewolna M, Andre J, Delmas V, Larue L, Guise TA, Mauviel A. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Lee J, Cooley M, Bhogte E, Hartley S, Glick A. Smad7 but not Smad6 cooperates with oncogenic ras to cause malignant conversion in a mouse model for squamous cell carcinoma. Cancer Res. 2003;63:7760–7768. [PubMed] [Google Scholar]
- 15.Halder SK, Rachakonda G, Deane NG, Datta PK. Smad7 induces hepatic metastasis in colorectal cancer. Br. J. Cancer. 2008;99:957–965. doi: 10.1038/sj.bjc.6604562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halder SK, Beauchamp RD, Datta PK. Smad7 induces tumorigenicity by blocking TGF-β-induced growth inhibition and apoptosis. Exp. Cell Res. 2005;307:231–246. doi: 10.1016/j.yexcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y, Liu Z, Zhao L, Clemens TL, Cao X. Smad7 stabilizes β-catenin binding to E-cadherin complex and promotes cell-cell adhesion. J. Biol. Chem. 2008;283:23956–23963. doi: 10.1074/jbc.M800351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imoto S, Sugiyama K, Muromoto R, Sato N, Yamamoto T, Matsuda T. Regulation of transforming growth factor-β signaling by protein inhibitor of activated STAT, PIASy through Smad3. J. Biol. Chem. 2003;278:34253–34258. doi: 10.1074/jbc.M304961200. [DOI] [PubMed] [Google Scholar]
- 20. http://www.thebiogrid.org and http://phospho.elm.eu.org.
- 21.Warner DR, Roberts EA, Greene RM, Pisano MM. Identification of novel Smad binding proteins. Biochem. Biophys. Res. Commun. 2003;312:1185–1190. doi: 10.1016/j.bbrc.2003.11.049. [DOI] [PubMed] [Google Scholar]
- 22.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 24.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 26.Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 28.de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev. 1998;12:1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong C, Li Z, Alvarez R, Jr., Feng XH, Goldschmidt-Clermont PJ. Microtubule binding to Smads may regulate TGF β activity. Mol. Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhu S, Wang W, Clarke DC, Liu X. Activation of Mps1 promotes transforming growth factor-β-independent Smad signaling. J. Biol. Chem. 2007;282:18327–18338. doi: 10.1074/jbc.M700636200. [DOI] [PubMed] [Google Scholar]
- 31.Hoover LL, Burton EG, O'Neill ML, Brooks BA, Sreedharan S, Dawson NA, Kubalak SW. Retinoids regulate TGFβ signaling at the level of Smad2 phosphorylation and nuclear accumulation. Biochim. Biophys. Acta. 2008;1783:2279–2286. doi: 10.1016/j.bbamcr.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmood R, Flanders KC, Morriss-Kay GM. Interactions between retinoids and TGF β s in mouse morphogenesis. Development. 1992;115:67–74. doi: 10.1242/dev.115.1.67. [DOI] [PubMed] [Google Scholar]
- 33.Yu Z, Xing Y. All-trans retinoic acid inhibited chondrogenesis of mouse embryonic palate mesenchymal cells by down-regulation of TGF-β/Smad signaling. Biochem. Biophys. Res. Commun. 2006;340:929–934. doi: 10.1016/j.bbrc.2005.12.100. [DOI] [PubMed] [Google Scholar]
- 34.Choi Y, Fuchs E. TGF-β and retinoic acid: Regulators of growth and modifiers of differentiation in human epidermal cells. Cell Regul. 1990;1:791–809. doi: 10.1091/mbc.1.11.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh B, Murphy RF, Ding XZ, Roginsky AB, Bell RH, Jr., Adrian TE. On the role of transforming growth factor-β in the growth inhibitory effects of retinoic acid in human pancreatic cancer cells. Mol. Cancer. 2007;6:82. doi: 10.1186/1476-4598-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]