Abstract
XPC, the main damage-recognition protein responsible for nucleotide excision repair of UVB damage to DNA, is lost or mutated in xeroderma pigmentosum group C (XP-C), a rare inherited disease characterized by high incidence and early onset of non-melanoma and melanoma skin cancers. The high incidence of skin cancers in XP-C patients suggests that loss of expression of XPC protein might also provide a selective advantage for initiation and progression of similar cancers in non XP-C patients in the general population. To test whether XPC is selectively lost in squamous cell carcinomas from non XP-C patients, we examined XPC expression by immunohistochemistry on a tissue microarray with 244 tissue cores, including in situ and invasive squamous-cell carcinomas (SCCs), keratoacanthoma (KA), and normal skin samples from both immunocompetent and immunosuppressed patients. We found that XPC expression was lost in 49% of invasive squamous cell carcinomas from immunocompetent patients and 59% from immunosuppressed patients. Loss of expression was correlated with deletions of chromosomal 3p and mutations in the XPC gene. The XPC gene is consequently inactivated or lost in almost half of squamous cell carcinomas from non XP-C patients. Loss or mutation of XPC may be an early event during skin carcinogenesis that provides a selective advantage for initiation and progression of squamous cell carcinomas in non XP-C patients.
Nonmelanoma skin cancer (NMSC) is the most common type of human malignancy in the United States, with over one million new cases annually.1 The escalating incidence of NMSC due to increased sun exposure over the last decades has made this disease a major public health issue and warrants a better understanding of the molecular pathogenesis of NMSC. NMSC includes basal-cell carcinoma, kerathoacanthoma (KA), and squamous cell carcinoma (SCC), with SCC constituting approximately 20% of all NMSCs.1 Basal-cell carcinoma is a locally invasive tumor, it does not metastasize, and its molecular pathogenesis has been extensively characterized.2 KA is also listed as NMSC but this classification is debatable; although some authorities consider it to be a benign neoplasm that regresses spontaneously, most regard it as a low-grade SCC. In contrast, SCC characteristically exhibits a propensity for invasion and may be lethal.1 Metastasis of SCCs is dependent on the tumor thickness; tumors less than 2 mm rarely metastasize, but tumors greater than 6 mm show a metastatic frequency of 16%.3 Surprisingly, despite the interesting ability of KA to regress spontaneously and despite the potential for lethality of SCC, the molecular pathogenesis of these NMSCs remains poorly understood. Invasive SCC is associated with UV-induced mutations in the p53 tumor suppressor gene,4 Ras activation, NF-κB blockade5 and activation of Src family kinases.6 However, these factors do not constitute a clear-cut cause or diagnostic or prognostic indicators of tumor aggressiveness. Consequently, there is a continuing need for the identification of target genes or pathways that are relevant to the molecular pathogenesis of SCC.
Over the last 40 years, important new insights into the molecular pathogenesis of skin cancers has come from the study of xeroderma pigmentosum (XP), a rare human, autosomally inherited, skin disease characterized by early onset in childhood of a high incidence of UV-induced NMSCs.7,8,9,10,11 The mechanism involved in XP is a failure of nucleotide excision repair, the cellular DNA repair mechanisms that act on UV-induced DNA photoproducts. UV light from the sun generates DNA photoproducts that distort the DNA structure and interrupt normal replication or transcription. Nucleotide excision repair consists of a sequential series of reactions through which UV-induced DNA photoproducts are excised and the DNA strand repaired.12 The main damage-recognition protein involved in nucleotide excision repair of UVB damage to DNA is specifically lost or mutated in XP group C (XP-C) patients, who display an extremely high and early incidence of skin cancers. In the absence of functional XPC, unrepaired photoproducts accumulate and generate replication errors and C to T mutations, including especially CC to TT mutations, which are found at very high frequencies in p53 in sun-induced skin cancers.13,14,15,16
The high incidence of SCCs in XP-C patients suggests that loss of expression of the XPC gene might provide a selective advantage for initiation and progression of SCCs in non XP-C patients (general population) and, consequently, that loss of XPC protein could be a novel biomarker of SCCs. If so, XPC protein should be lost in SCCs from non XP-C patients. To test this hypothesis, we examined XPC protein expression by immunohistochemistry on a tissue microarray of 244 tissue cores including in situ and invasive SCCs, KA, and normal skin samples from both immunocompetent and immunosuppressed patients. Our tissue microarray was stained with an antibody to XPC protein, which allowed detection of the nuclear protein by IHC from paraffin sections of normal and tumor tissue.
Materials and Methods
Tissue Microarray
We obtained tissue specimens from paraffin-embedded tumors from the UCSF Department of Dermatology archives after their primary use. Biopsies had been removed surgically for diagnosis of skin tumors in patients with varied etiologies undergoing dermatological treatment, fixed in paraformaldehyde, and embedded in paraffin. The patients were classified as having received an organ transplant (OTR) or were not a transplant recipient (non-OTR). The OTR patients had a varied history of different immunosuppressive agents, and we did not stratify according to treatment because the results did not appear to be influenced by the OTR status. All other identifiers were removed. Plugs (0.6 mm) were removed for re-embedding to create an array of 244 plugs of alternating normal and tumor tissues. Where possible adjacent normal tissue was included, but this was not always possible as in some cases there was insufficient normal tissue.
Immunohistochemistry
Sections were cut and processed for immunohistochemistry using a mouse monoclonal antibody to the XPC protein (clone 3.26; GeneTex, San Antonio, TX). Cultures of XPC-deficient cells and skin biopsies from XP-C families were previously used for validation of the method, after informed consent.17 In this previous study we showed that the antibody consistently detected XPC loss in all of the known XP-C patients analyzed and identified several previously undiagnosed patients. XPC immunostaining was performed on 5-μm sections of paraffin-embedded tissue using the XPC mouse monoclonal antibody. Deparaffinized sections underwent heat-mediated antigen retrieval by microwaving (900 watts for 10 minutes in 10 mmol/L citrate buffer, pH 6.0). Slides were incubated for 1 hour with a 1:100 to 1:1000 dilution of the antibody in PBS and 1% BSA. Antibody staining was visualized using biotinylated horse anti-mouse antibody (Vector Laboratories, Burlingame, CA) and ABC-HRP Elite (Vector Laboratories, Burlingame, CA), followed by diaminobenzidine reaction. The XPC antibody stained the nucleus exclusively. Additional sections were stained for mutant p53 (DO-1, Santa Cruz Biotechnology, Santa Cruz, CA), which is a frequent marker of UV-induced keratoses and SCCs in sun-damaged skin. Tissue culture pellets were stained with XPC antibody or with an antibody against normal p53 (DO-8, Thermo Scientific). Sections were counterstained with light hematoxylin and then dehydrated and mounted with coverslips. All figures are shown as native images scanned from gel photographs or photographed at ×10 to ×40 from slides with no further image processing.
Comparative Genome Hybridization (CGH)
Genomic DNA (1000 ng) from the paraffin-embedded tumors was labeled using random primers as previously described.18,19 Hum 3.2 Bacterial Artificial Chromosome (BAC) arrays that contain 2464 BAC clones were used. All statistical analyses were performed using the freely available R/Bioconductor software. After correction for BAC clone-specific GC content and geometrical dependence of the ratios on the array,20 the data were analyzed using circular binary segmentation to translate noisy intensity measurements into regions of equal copy number.21 The median absolute deviation was calculated for each aCGH profile, and the gain and loss status for each probe was defined using the merged level procedure.22 Clones on the arrays with absolute base-2 logarithm (log 2) ratios greater than 2.5 times the median absolute deviation were deemed aberrant (having gains or losses in numbers of copies). The status of nine clones spanning 3p24 to 3p25 was examined for tissue plugs for which the XPC expression status was established by immunohistochemistry. These clones included the clone containing XPC (3p24.3-3p25) and the four clones distal and proximal to XPC. The χ2 test was used to compare the tumors showing XPC loss and 3p chromosomal loss.
DNA Sequencing
DNA samples were PCR amplified to produce amplicons containing exons of the XPC gene for sequence analysis.23 PCR conditions were as follows: 8 ng of genomic DNA was incubated in a 10-μl reaction composed of 1 μl of Buffer, 2 μl of Q-mix, 0.4 μl of dNTP (2.5 mmol/L), 0.06 μl of Qiagen Taq polymerase, 2 μl of Forward primer (1 μmol/L), 2 μl of Reverse primer (1 μmol/L; Buffer, Q-Mix, and enzyme were from Qiagen hot start kit, QIAGEN, Inc., Valencia, CA), with cycling conditions of denaturation at 95°C for 5 minutes followed by 35 cycles of 94°C for 1 minute, annealing at 58°C for 1 minute, and extension at 72°C either for 1 minute (Amplicon 1) or for 2 minutes (Amplicon 2). At the end of the 35 cycles, the reaction mixture was held at 72°C for 10 minutes before being cooled to 4°C until the next step. The 10-μl PCR product was purified by Exo-SAP treatment, which consisted of incubation with 0.4 μl of PCR Clean-up Reagent, 10× and 3.6 μl of PCR Clean-Up Dilution Buffer (PerkinElmer Life Sciences, Inc., Boston, MA) at 37°C for 1 hour to degrade the excess primers and dNTP followed by heating the reaction mixture at 90°C for 15 minutes to inactivate the enzymes. The purified PCR product was sequenced in both directions using ABI PRISM BigDye terminator sequencing Version 3.1 on an ABI Prism 3730xl DNA analyzer (Applied Biosystems, Inc., Foster City, CA). The 12-μl sequencing reaction was made up of 2.5 μl of purified PCR product, 4.5 μl of sequencing primer (1 μmol/L), 1 μl BigDyeV3.1, 2 μl of 5× buffer, and 2 μl water. Cycling conditions were 96°C for 2 minutes followed by 25 cycles of 96°C for 15 seconds, 50°C for 1 second, 60°C for 4 minutes. After sequencing, the DNA sequence files were imported into and aligned with SEQUENCHER 4.8 (Gene Codes Corporation, Ann Arbor, MI) for variant identification.
Cell Culture
Normal (GM637) and XP-C (GM15983, GM16093) cells were obtained from Coriell Cell Repository, Camden NJ, and grown in minimal essential medium with 10% fetal calf serum. Survival was determined by irradiating cells in 96-well plates with increasing doses of UVB (1.2 J · m−2 · sec) and measuring colorimetric activation of MTT (3-(4,5-Dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide) 7 days later. Pellets of cells were fixed in paraformaldehyde, embedded in paraffin, and analyzed by immunohistochemistry as described for tissues.
Western Blot
Cells were washed with PBS and lysed using radioimmunoprecipitation (RIPA) buffer [150 mmol/L NaCl, 1% (v/v) NP40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, in 50 mmol/L Tris–HCl, pH 8.0] supplemented with protease (1836153, Roche) and phosphatase inhibitors (78420, Pierce) according to the manufacturer’s protocol. Protein concentrations were determined using the BCA Protein Assay Kit (23225, Pierce). Total protein in 1× Laemmli buffer with 10% 2-mercaptoethanol was separated by SDS–PAGE, transferred overnight to a polyvinylidene difluoride membrane (IPVH00010, Millipore) by electro-blotting, and blocked for 1 hour in 5% (w/v) dry milk/TBS/0.1%(v/v) Tween-20. Membranes were incubated overnight at 4°C with primary antiserum followed by incubation with a horseradish-peroxidase-conjugated secondary antiserum for 1 hour and developed using enhanced chemiluminescence (32106, Pierce or 64–201BP, Millipore). Membranes were probed with antibodies to XPC (clone 3.26; GeneTex, San Antonio, TX), β-actin (A5441; Sigma); and wild-type p53 (DO-7 Thermo Scientific).
Results
Validation of XPC Antibody by Western Blot and Immunohistochemistry
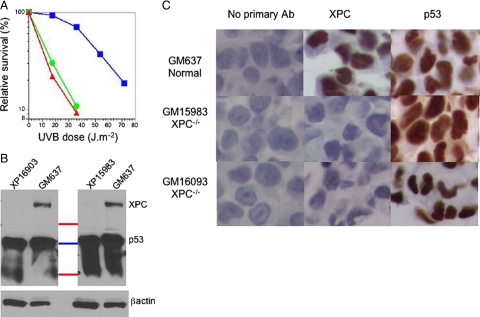
To evaluate possible cross-reaction of the XPC antibody with other proteins we used human SV40-transformed cells from a normal donor and two XP-C patients. These cells have high levels of p53, due to stabilization by the SV40 large T antigen.24 The manufacturer’s information states that this XPC antibody cross-reacts with p53. The XP-C cell lines showed characteristic increased sensitivity to irradiation with UVB light (Figure 1A). In Western blots the XP-C cell lines lacked a protein band at the size expected for the XPC protein that was present in the normal cells (Figure 1B). There was, however, an additional large signal at a size corresponding to p53 and lower molecular weights in all of the cell lines. After stripping and reprobing with antibody to p53 we confirmed that this size region corresponded to p53 present in all cell lines. The XPC antibody therefore cross-reacts to proteins in the size range of p53, as well as reacting with the XPC protein. Immunohistochemical analysis of pellets of these cells processed in an identical fashion as for tissue sections showed strong expression of XPC only in the normal cells but strong expression of p53 in all of the cells (Figure 1C).
Figure 1.
Validation of XPC antibody. A: Survival of SV40 transformed normal (GM637, blue squares) and two XP-C cell lines (GM15983 green circles, GM16093 red triangles) irradiated with UVB and grown for seven days before assay with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide). B: Western blot stained with XPC antibody for GM637, GM16093, and GM15983 cells and marker lane (top to bottom: red 75 Kd, blue 50 Kd, red 25 Kd). Positions shown for XPC (105 kDa) and lower bands in region of p53 (53 kDa) and below. Blot was stripped and reprobed with antibody to β actin. C: Immunohistochemical staining of sectioned formalin-fixed normal (top row) and XPC-deficient (center and bottom rows) cell pellets stained with either no primary antibody (left column) or with antibody to XPC (center column) or p53 (right column).
These experiments indicate that although the XPC antibody strongly reacts with proteins of p53 size range under the denaturing conditions of a Western blot, it does not react with these proteins under the milder antigen retrieval conditions of immunohistochemistry. Our standard antigen retrieval procedure involves mild microwaving that exposes epitopes but does not denature the proteins to the extent that occurs during Western analysis. The XPC epitopes were therefore exposed using our standard IHC procedures, but p53 was not denatured to become antigenic to the XPC antibody. In additional preliminary experiments we demonstrated that increased microwaving exposed the antigenic activity of p53 in the XP-C cells, which then showed positive signals with this antibody. We subsequently analyzed human SCCs under standardized antigen retrieval to minimize cross-reactions.
Loss of XPC Expression in UV-Induced SCCs
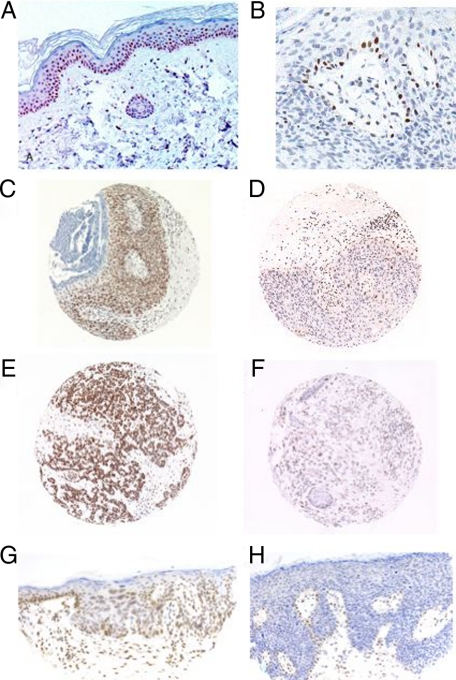
Sections of normal epidermis from non-XPC patients showed expression of XPC in the basal layer, which diminished as keratinization proceeded in the upper layers (Figure 2A).17 When we examined SCCs from non XP-C patients, we found that some cases showed continued expression of XPC in normal basal keratinocytes adjacent to the tumor but loss of XPC expression in the tumor tissue itself, except focally in rare keratinocytes (Figure 2, B, D, F and H). We examined XPC expression by immunohistochemistry in a tissue microarray of 244 tissue cores, of which a total of 221 could be analyzed (the remainder were lost or degraded and gave no information). We analyzed 35 in situ SCCs, 69 invasive SCCs, 29 KA, and 88 normal skin samples (Table 1). We found that the tissue cores displayed either diffuse XPC expression (example shown on Figure 2, C and E) or loss of XPC expression (Figure 2, D and F). Specifically, 26 to 49% of the SCCs from immunocompetent patients (Table 1) and 50 to 59% of SCCs from immunosuppressed patients showed loss of XPC expression (Table 1). When XPC expression was lost in SCC cells, XPC was still expressed in the lymphocytic infiltrates and focally in rare keratinocytes (Figure 2, B, D, F, and H). In these tissue cores, the loss of XPC in keratinocytes was therefore specific for the tumor. The difference between the OTR and non-OTR patients was small and not statistically significant (Table 1).
Figure 2.
XPC expression by immunohistochemistry in non-tumoral skin and squamous cell carcinomas from non XP-C patients. A: XPC expression in a nontumoral skin (normal control skin) (×20). B: Tangential section of SCC in situ showing loss of XPC with focal retained expression in rare keratinocytes and in lymphocytes. C: Histospot of SCC in situ showing retained XPC expression (×10). D: Histospot of SCC in situ showing loss of XPC expression except focally, in rare keratinocytes and in lymphocytes (×10). E: Histospot of invasive SCC with retained XPC expression (×10). F: Histospot of invasive SCC with loss of XPC expression (×10). G: SCC in situ with retained XPC expression throughout the epidermis (×20). H: SCC in situ (Bowen’s disease) with loss of XPC expression throughout the epidermis except focally in rare cells (×20).
Table 1.
Frequencies of XPC Loss of Expression by Immunohistochemistry in Normal Skin and Squamous Cell Carcinomas from Non XP-C Patients
| Normal donors | Organ transplant recipients | |
|---|---|---|
| Normal tissue* | 2/48 = 4.2% | 1/40 = 2.5% |
| Keratoacanthoma† | 7/15 = 46.7% | 8/14 = 57.1% |
| Squamous cell carcinoma in situ† | 5/19 = 26.3% | 9/18 = 50% |
| Invasive squamous cell carcinoma† | 17/35 = 48.6% | 20/34 = 58.8% |
The XPC negative samples are shown for the cores scored, and subsequent analysis on the original blocks showed only one XPC negative result for all the normal tissues examined.
The differences between normal donors and OTR patients is not regarded as significant. Pooling the tumor types, the difference by paired t-test gave a P = 0.08.
Only 3/88 (3.4%) samples of normal skin were negative on the array (Table 1), which might be due to poor fixation and loss of antigenicity. We re-examined these in sections from the original paraffin block, and two were positive for XPC and one remained negative. This one remaining negative skin biopsy among over 100 examined may represent a staining artifact or an occult XP-C patient, but we have not returned to the original patient at this time for further examination.
The presence of multiple tumors in individual immunosupressed patients allowed us to compare the frequency of XPC loss in 2 distinct SCCs from the same patient. We did not find any significant difference between the frequency of XPC loss in the first SCC and the frequency of XPC loss in the second, although the numbers available were small (Table 2). There was no correlation in XPC loss in multiple tumors from the same person, implying that the host genotype, such as XPC polymorphisms, does not invariably determine whether XPC is lost or retained during SCC formation.
Table 2.
Frequency of XPC Loss in Two SCCs from the Same Organ Transplant Patients in Cases of Multiple Tumors in Individual Patients
| Frequency of XPC loss in first tumor | Frequency of XPC loss in second tumor when first was negative for XPC | |
|---|---|---|
| XPC Negative (% and ratio) | 50.7% (35/69) | 42.9% (6/14) |
Loss of XPC Expression Does Not Correlate with p53 Expression
Having found a loss of XPC expression in 26 to 59% of SCC, we sought to identify the cause of that loss. XPC is regulated by p53 at the transcriptional level,25 and UV-induced SCCs typically show increased levels of mutated p53.13,26 To determine whether the loss of XPC expression may be due to the loss of regulation of XPC by a mutant p53 at the transcriptional level, we compared expression of XPC with that of wild-type or mutant p53. In general, mutations in p53 stabilize the protein and give rise to higher levels of p53 staining. When all samples were pooled and the frequency of XPC loss compared with p53 expression by immunohistochemistry, we found no obvious correlation: in tumors with low levels of p53, 37 were XPC-positive and 36 were XPC-negative; in tumors with high levels of p53, 31 were XPC-positive and 31 were XPC-negative. When we stratified the samples according to both p53 expression levels and p53 mutations, there was a pattern in which tumors that contained mutant p53 expressed at high levels were more likely to be XPC-negative (Table 3).
Table 3.
Frequencies of Tumor Samples with or without XPC Expression in Comparison with the Expression Level and Sequence of p53
| P53 expression level | P53 sequence | Positive expression of XPC by IHC | Absence of XPC expression by IHC |
|---|---|---|---|
| Low | Normal | 9 | 10*† |
| Low | Mutant | 6 | 4* |
| High | Normal | 6 | 1†‡ |
| High | Mutant | 7 | 12‡ |
Superscript symbols refer to pair-wise comparisons for significance using Fisher exact. *P < 0.7, †P < 0.17, ‡P < 0.07.
Copy Number Changes on Chromosome 3p
We compared the loss of expression of XPC with the DNA copy number using CGH, by analyzing specifically the loss of chromosome 3p that contains the XPC gene. Chromosome instability in SCCs is characterized by the loss and gain of whole chromosome arms (Ridd K., unpublished data). Therefore it is not surprising that the 9 clones we specifically analyzed on chromosome 3p24–25 were either all present or all lost. We found that 45.5% of the XPC-negative tumors, as assessed by immunohistochemistry, had reduced copy number at 3p, although not homozygous loss of both alleles (Table 4). Only 10% of XPC-positive tumors showed 3p loss (Table 4). This small percentage suggests that a partial reason for loss of expression of XPC was the loss of chromosome 3p. The remainder of the tumors that were XPC-negative but retained chromosome 3p may represent smaller regions of loss around 3p25 or mutated XPC that may be difficult to resolve using our tissue arrays. Sequencing results for the XPC gene from a small number of tumors10 that were negative by immunohistochemistry showed a number of mutations in protein-coding exons that could affect function (Tables 5 and 6).
Table 4.
Loss of Chromosome 3p in Tumors That Were Positive or Negative for XPC
| XPC status | Number with loss of 3p | Number with 3p present |
|---|---|---|
| XPC negative* | 20 (45.5%) | 24 (54.5%) |
| XPC positive | 5 (10%) | 45 (90%) |
Difference between XPC negative and positive is significant, P < 0.001.
Table 5.
Primers Used in PCR/Sequencing Reactions
| Primer name | Amplicon size (BP) | Exon covered | Sequence* |
|---|---|---|---|
| 4906_F | 599 | 1 | 5′-TGTAAAACGACGGCCAGTCACAAATAACTGCCTTCTCCGAGTTC-3′ |
| 4906_R | 5′-CAGGAAACAGCTATGACCTGTTGTGCTCTTTCCTGCTTCCC-3′ | ||
| 2005_F | 441 | 2 | 5′-TGTAAAACGACGGCCAGTGCCATGCCCACCACCTGATA-3′ |
| 2005_R | 5′-CAGGAAACAGCTATGACCTGGGAAGTGGCCAATGCTAGTG-3′ | ||
| 3368_F | 500 | 4 | 5′-TGTAAAACGACGGCCAGTCCTTGGCACACAGGAGTTCCC-3′ |
| 3368_R | 5′-CAGGAAACAGCTATGACCCAGGGCAAGGCATGTCTAGGG-3′ | ||
| 0222_F | 548 | 5 | 5′-TGTAAAACGACGGCCAGTCCAGTGCCTGCAGCTGGGAT-3′ |
| 0222_R | 5′-CAGGAAACAGCTATGACCTGGCTTGCAGACGGACTTGA-3′ | ||
| 2024_F | 518 | 6 | 5′-TGTAAAACGACGGCCAGTGCTCTTACCGGTCTGAGTTGTACCT-3′ |
| 2024_R | 5′-CAGGAAACAGCTATGACCCGCAGGTGGGAAGAAGCTGA-3′ | ||
| 7879_F | 479 | 7 | 5′-TGTAAAACGACGGCCAGTGGGAGGAAAGCATACAGGCCC-3′ |
| 7879_R | 5′-CAGGAAACAGCTATGACCTAGGACAGAGGCAGGCGGGT-3′ | ||
| 5902_F | 530 | 8 | 5′-TGTAAAACGACGGCCAGTAGCACTCCACGGAGCAAGCA-3′ |
| 5902_R | 5′-CAGGAAACAGCTATGACCTGTCTGGAGTTTCCGTCGCC-3′ | ||
| 2805_F | 571 | 9–11 | 5′-TGTAAAACGACGGCCAGTTGGGCTTGGTGGCGTACTTG-3′ |
| 2805_R | 5′-CAGGAAACAGCTATGACCCTGCAGGCCAAGTGCCAAAG-3′ | ||
| 8256_F | 571 | 11 | 5′-TGTAAAACGACGGCCAGTGCTGTGACAATTAAATGAGACAACCCA-3′ |
| 8256_R | 5′-CAGGAAACAGCTATGACCTGGGTCCAAGAGTGCCTCCA-3′ | ||
| 1999_F | 458 | 12–13 | 5′-TGTAAAACGACGGCCAGTCAGCACACGCACACTGGCTC-3′ |
| 1999_R | 5′-CAGGAAACAGCTATGACCGGCCGGAGGTACAGATGCGA-3′ | ||
| 6341_F | 580 | 14 | 5′-TGTAAAACGACGGCCAGTCAGGGCTCTGACCTCCCAGG-3′ |
| 6341_R | 5′-CAGGAAACAGCTATGACCCCACGTTCAAGGCTGTTTGCC-3′ | ||
| 4564_F | 599 | 17 | 5′-TGTAAAACGACGGCCAGTTGCCATGATGTCAGAGAGGGC-3′ |
| 4564_R | 5′-CAGGAAACAGCTATGACCCGGAGCCAATGAAACTGGCA-3′ | ||
| 1287_F | 566 | 18 | 5′-TGTAAAACGACGGCCAGTCCAGGGACACCTTTCCCAACA-3′ |
| 1287_R | 5′-CAGGAAACAGCTATGACCCTGCCCACACCTGCCTCTGT-3′ | ||
| 7586_F | 459 | 19 | 5′-TGTAAAACGACGGCCAGTTGGGAACAGGTGGGAAGCTG-3′ |
| 7586_R | 5′-CAGGAAACAGCTATGACCCCTGCCCAGCCATCATCCTT-3′ | ||
| 6412_F | 591 | 19 | 5′-TGTAAAACGACGGCCAGTGGTCCTAGGTCCGCAACCGA-3′ |
| 6412_R | 5′-CAGGAAACAGCTATGACCTACTGGCTGCCTCCTGGCCT-3′ |
Forward primers contain the M13 forward primer sequence 5′-TGTAAAACGACGGCCAGT-3′, while reverse primers contain the M13 reverse sequence 5′-CAGGAAACAGCTATGACC-3′.
Table 6.
Sequence Changes Identified in the XPC Gene from XPC Negative Tumors*
| Sample with variant | Location† | Nucleotide sequence | Amino acid change |
|---|---|---|---|
| SP35 | Exon 2 (rs3731062) | 5′-GTGAAACTTTGGAGA[G/A]AAGGCTCTTCTTTGG-3′ | Leu48Phe |
| SP21 SP25 SP53.1 SP71 | Exon 11 (rs2227999) | 5′-AAGCTTGGGTCCTTA[C/T]GATGGCTCCCACGAT-3′ | Arg492His |
| SP21‡ SP25 SP35 SP53.1 SP37 SP71 | Exon 11 (rs2228000) | 5′-GAGCTTGAGGATGCC[G/A]CTGGCAAGCTTGGGT-3′ | Ala499Val |
| SP36§ SP35 SP71 | Exon 14 (rs2227998) | 5′-CTTCAGCCACGTGTC[C/T]CTGGAATGCAGAGTG-3′ | Arg687Arg¶ |
| SP15∥ SP25 SP37 SP54 | Exon 19 (rs2228001) | 5′-GCTCAGCTCACAGCT[T/G]CTCAAATGGGAACAG-3′ | Lys939Gln |
A total of 10 tumors were analyzed that were negative for XPC by immunohistochemistry.
Reference SNP number in parentheses; based on dbSNP Build 130.
SP21 is homozygous A/A, the rest are heterozygous G/A.
SP36 is homozygous T/T, the rest are heterozygous C/T.
Nucleotide change had no effect on amino acid sequence.
SP15 is homozygous G/G, the rest are heterozygous T/G.
Discussion
In this study, we show that XPC protein is absent from up to 59% of invasive SCCs from the general population (non XP-C patients). The loss of XPC expression must involve inactivation of both alleles of XPC because our IHC does not discriminate obligate XP-C heterozygotes from normal donors.17 The analysis of XPC expression by immunohistochemistry on a tissue microarray of 221 SCCs from immunocompetent and immunosuppressed patients showed that XPC expression was lost in 26 to 49% of SCCs from immunocompetent patients and in 50 to 59% of SCCs from immunosuppressed patients. Thus, XPC is a novel target gene for inactivation in NMSC. SCCs from immunosuppressed patients exhibited a slightly higher percentage of XPC loss than did SCCs from immunocompetent patients, which was not statistically significant in our sample size but may be worth further investigation.
Loss of XPC expression occurred in carcinomas in situ and well-differentiated invasive SCCs (Table 1). Loss of XPC expression therefore does not appear to be associated with degree of differentiation, suggesting that it may be an early event in tumor development. The loss was also independent for each tumor in cases of multiple tumors in individual patients and was not driven by unique features of the patients, such as XPC polymorphisms (Table 2).
Several molecular mechanisms could account for the loss of XPC expression in SCCs. SCCs display a high rate of genomic instability.27,28 Loss of XPC expression in UV-induced SCCs could therefore be caused by the genomic instability in this region, as we demonstrated by loss of the XPC locus, 3p25, by CGH (Table 3), and by mutations in the XPC gene (Table 4). Chromosome 3p may therefore be a vulnerable region for inactivation during certain kinds of solar-induced carcinogenesis. Another factor that may be involved in the loss of XPC expression in SCCs could be increased frequency of mutated p53 in UV-induced NMSC.13,26 Indeed, genomic instability is significantly greater in tumors with mutated p53 compared with wild-type tumors (Ridd K., unpublished data), as also occurs in cell cultures after inactivation of p53 by shRNA or SV40 transformation.29 XPC is regulated by p53 at the transcriptional level, and expression of wild-type p53 is required for nucleotide excision repair.30,31 However, we found no correlation between p53 and XPC expression by IHC (Table 3). Another molecular mechanism for the loss of XPC expression in SCCs could be the inactivation of both alleles of XPC, by either mutagenesis or promoter methylation. Promoter methylation could down-regulate both XPC promoters, as has been observed for the p16 gene during early stage carcinogenesis in breast epithelial cells.32 The focal persistence of XPC expression in basal keratinocytes in SCC in situ (Figure 2, B and H) and its subsequent loss as the transformed keratinocytes ascent in the epidermis (Figure 2, B and H), would be consistent with XPC loss in SCC being the consequence of an early regulatory event.
Our observation raises technical issues if IHC were to be used on an SCC for the diagnosis of XP-C patients. Our findings indicate, however, that XPC could still be detected in infiltrating lymphocytes and nonmalignant skin that flanked tumors. If only archival blocks of tumor biopsies were available for a particular XP patient, diagnosis would need to be based on XPC expression in flanking normal skin or lymphocyte infiltrations.17 Our previous results indicated apparent reappearance of XPC antigenicity in tumor tissue of XP-C patients.17 We now recognize that this observation may represent cross-reaction of the XPC antibody (Figure 1B) to denaturated p53 that is highly mutated in XP-C patients and may therefore denature more easily.15,33,34 In 99 of 100 samples, plugs of nonmalignant skin showed positive staining for XPC permitting discrimination between tumor-specific loss of XPC in SCCs from non XP-C donors and genetic loss in an XP-C patient.17
In conclusion, our study shows that XPC expression is lost in up to 59% of SCCs from non XP-C patients, demonstrating that XPC is a target for high frequency of inactivation. Because XPC is the main damage-recognition protein for repair of UV damage to DNA, this loss is expected to have major consequences for genomic stability, especially under continued solar exposure. Our study supports the idea that loss of XPC provides a selective advantage for initiation of UV-induced SCCs. Our results raise the possibility that XPC polymorphisms might impact SCC risks, as has been reported for melanoma.35,36 The authors found a correlation between three XPC polymorphisms35 but no correlation with three others.36 Patients with multiple SCCs did not, however, show a consistent pattern of loss or retention of XPC, implying that host genotypes such as inherited XPC polymorphisms are not major factors in XPC inactivation. The loss of XPC in SCCs represents a vulnerability of the tumors to chemotherapy that targets repair deficiency but might predispose the tumor cells to further genomic instability with solar exposure, as occurs in XP-C patients. Further investigation of chromosomal loss, mutation, and XPC expression is therefore warranted to understand the mechanism of XPC loss in SCC and the potential impact of immunosuppressive therapy on XPC expression.
Acknowledgments
We are grateful to the XP Society (Poughkeepsie, NY) and the XP Family Support group (Sacramento, CA) for their continued support and encouragement for the University of California, San Francisco (UCSF) research program (J.E.C). Immunohistochemistry was performed by the Tissue Core (Loretta Chan) of the UCSF Helen Diller Family Comprehensive Cancer Center, and the aCGH data were analyzed by Ritu Roy from the UCSF Helen Diller Family Comprehensive Cancer Center.
Footnotes
Address reprint requests to James E. Cleaver, Ph.D., Department of Dermatology, University of California, San Francisco, CA 94143-0808. E-mail: jcleaver@cc.ucsf.edu.
Supported originally by a grant from the National Institutes of Environmental Health Sciences grant 1 RO1 ES 8061 (J.E.C.) and a program project grant P01 AR050440-01 (PI: Ervin Epstein) and completed with the Marcia Robbins-Wilf Research Award from the Skin Cancer Foundation. Work by D.H.O. was supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, and National Institutes of Health Grant R01/CA105958.
References
- Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344:975–983. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- Rubin A, Chen E, Ratner D. Basal-cell carcinoma. N Engl J Med. 2005;353:2262–2269. doi: 10.1056/NEJMra044151. [DOI] [PubMed] [Google Scholar]
- Brantsch KD, Meisner C, Schönfisch B, Trilling, Wehner-Caroli J, Röcken M, Breuninger H. Analysis of risk factors determining prognosis of cutaneous squamous-cell carcinoma: a prospective study. Lancet Oncol. 2008;9:713–720. doi: 10.1016/S1470-2045(08)70178-5. [DOI] [PubMed] [Google Scholar]
- Bolshakov S, Walker C, Strom S, Selvan MS, Clayman GL, El-Naggar A, Lippman SM, Kripke ML, Ananthaswamy HN. p53 mutations in human aggressive and nonaggressive basal and squamous cell carcinomas. Clin Cancer Res. 2003;9:228–234. [PubMed] [Google Scholar]
- Ridky TW, Khavari PA. Pathways sufficient to induce epidermal carcinogenesis. Cell Cycle. 2004;3:621–624. [PubMed] [Google Scholar]
- Ayli EE, Li W, Brown TT, Witkiewicz A, Elenitsas R, Seykora JT. Activation of Src-family tyrosine kinases in hyperproliferative epidermal disorders. J Cutan Pathol. 2008;35:273–277. doi: 10.1111/j.1600-0560.2007.00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. Defective repair replication in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JHJ. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Vogelstein B, Kinzler K W, editors. McGraw-Hill,; The Genetic Basis of Human Cancer. 1998:245–274. [Google Scholar]
- Cleaver JE, Crowley E. UV damage. DNA repair and skin carcinogenesis. Front Biosci. 2002;7:d1024–d1043. doi: 10.2741/A829. [DOI] [PubMed] [Google Scholar]
- Thompson LH. Nucleotide excision repair: Its relation to human disease. Nickoloff J A, Hoekstra M, editors. Totowa, NJ: Humana Press,; 1998:pp 335–393. [Google Scholar]
- Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum: cutaneous, ocular and neurological abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin GA, McKenna A, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: uV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataraj AJ, Trent JC, Ananthaswamy HN. p53 gene mutations and photocarcinogenesis. Photochem Photobiol. 1995;62:165–177. doi: 10.1111/j.1751-1097.1995.tb05262.x. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Drougard C, Sarasin A, Daya-Grosjean L. Specific UV-induced mutation spectrum in the p53 gene of skin tumors from DNA-repair-deficient xeroderma pigmentosum patients. Proc Natl Acad Sci USA. 1993;90:10529–10533. doi: 10.1073/pnas.90.22.10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. The biology of xeroderma pigmentosum: lessons learned to explain the development of melanoma. Hearing V J, Leong S P L, editors. Humana Press,; 2005 [Google Scholar]
- de Feraudy S, Boubakour-Azzouz I, Fraitag S, Berneburg M, Chan L, Chew K, Clericuzio CL, Cunningham B, Tope WD, Cleaver JE. Diagnosing xeroderma pigmentosum group C by immunohistochemistry. Am J Dermatopath. 2010;32:109–117. doi: 10.1097/DAD.0b013e3181af0a5e. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowee ME, Fridlyand J, Piek JM, Dorsman JC, Jain AN, Pinkel D, van Diest PJ, Verheijen RH, Albertson DG. Genome-wide-array-based comparative genomic hybridization reveals genetic homogeneity and frequent copy number increases encompassing CCNE1 in fallopian tube carcinoma. Oncogene. 2003;22:4281–4286. doi: 10.1038/sj.onc.1206621. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Gajduskova P, Snijders AM, Kwek S, Roydasgupta R, Fridlyand J, Tokuyasu T, Pinkel D, Albertson DG. Genome position and gene amplification. Genome Biol. 2007;8:R120. doi: 10.1186/gb-2007-8-6-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Feeney L, Tang JY, Tuttle P. Xeroderma pigmentosum group C in an isolated region of Guatemala. J Invest Dermatol. 2007;127:493–496. doi: 10.1038/sj.jid.5700555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe JPG, Busch D, Yu Y, Nagasawa H, Little JB. Increased UV sensitivity and chromosomal instability related to p53 function in the xeroderma pigmentosum variant. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- Ford JM. Role of p53 in the mammalian cellular response to UV damage. Photochem Photobiol. 1998;67:73S–74S. [Google Scholar]
- Ananthaswamy HN, Ouhtit A, Evans RL, Gorny A, Khaskina P, Sands AT, Conti CJ. Persistence of p53 mutations and resistance of keratinocytes to apoptosis are associated with the increased susceptibility of mice lacking the XPC gene to UV carcinogenesis. Oncogene. 1999;18:7395–7398. doi: 10.1038/sj.onc.1203147. [DOI] [PubMed] [Google Scholar]
- Burnworth B, Arendt S, Muffler S, Steinkraus V, Bröcker EB, Birek C, Hartschuh W, Jauch A, Boukamp P. The multi-step process of human skin carcinogenesis: a role for p53, cyclin D1, hTERT, p16, and TSP-1. Eur J Cell Biol. 2007;86:763–780. doi: 10.1016/j.ejcb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Clausen OP, Aass HC, Beigi M, Purdie KJ, Proby CM, Brown VL, Mattingsdal M, Micci F, Kolvraa S, Bolund L, Deangelis PM. Are keratoacanthomas variants of squamous cell carcinomas? A comparison of chromosomal aberrations by comparative genomic hybridization. J Invest Dermatol. 2006;126:2308–2315. doi: 10.1038/sj.jid.5700375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposa RR, Feeney L, Crowley E, de Feraudy S, Cleaver JE. p53 suppression overwhelms DNA polymerase h deficiency in determining the cellular UV DNA damage response. DNA Repair. 2007;6:1794–1804. doi: 10.1016/j.dnarep.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- Ford JM, Hanawalt PC. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc Nat Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean GR, Bryson AD, Pilie PG, Goldenberg V, Baker JC, Jr, Ibarra C, Brander DM, Paisie C, Case NR, Gauthier M, Reynolds PA, Dietze E, Ostrander J, Scott V, Wilke LG, Yee L, Kimler BF, Fabian CJ, Zalles CM, Broadwater G, Tlsty TD, Seewaldt VL. Morphologically normal-appearing mammary epithelial cells obtained from high-risk women exhibit methylation silencing of INK4a/ARF. Clin Cancer Res. 2007;13:6834–6841. doi: 10.1158/1078-0432.CCR-07-0407. [DOI] [PubMed] [Google Scholar]
- Giglia G, Dumaz N, Drougard C, Avril MF, Daya-Grosjean L, Sarasin A. p53 mutations in skin and internal tumors of xeroderma pigmentosum patients belonging to the complementation group C. Cancer Res. 1998;58:4402–4409. [PubMed] [Google Scholar]
- Spatz A, Giglia-Mari G, Benhamou S, Sarasin A. Association between DNA repair-deficiency and high level of p53 mutations in melanoma of xeroderma pigmentosum. Cancer Res. 2001;61:2480–2486. [PubMed] [Google Scholar]
- Blankenburg S, König IR, Moessner R, Laspe P, Thoms KM, Krueger U, Khan SG, Westphal G, Berking C, Volkenandt M, Reich K, Neumann C, Ziegler A, Kraemer KH, Emmert S. Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case-control study. Carcinogen. 2005;26:1085–1090. doi: 10.1093/carcin/bgi055. [DOI] [PubMed] [Google Scholar]
- Blankenburg S, König IR, Moessner R, Laspe P, Thoms KM, Krueger U, Khan SG, Westphal G, Volkenandt M, Neumann C, Ziegler A, Kraemer KH, Reich K, Emmert S. No association between three xeroderma pigmentosum group C and one group G gene polymorphisms and risk of cutaneous melanoma. Eur J Hum Genet. 2005;13:253–255. doi: 10.1038/sj.ejhg.5201296. [DOI] [PubMed] [Google Scholar]