Abstract
Tissue inhibitors of metalloproteinases (TIMPs) are a family of closely related proteins that inhibit matrix metalloproteinases (MMPs). In the central nervous system (CNS), TIMPs 2, 3, and 4 are constitutively expressed at high levels, whereas TIMP1 can be induced by various stimuli. Here, we studied the effects of constitutive expression of TIMP1 in the CNS in transgenic mice. Transgene expression started prenatally and persisted throughout lifetime at high levels. Since MMP activity has been implicated in CNS development, in proper function of the adult CNS, and in inflammatory disorders, we investigated Timp1-induced CNS alterations. Despite sufficient MMP inhibition, high expressor transgenic mice had a normal phenotype. The absence of compensatory up-regulation of MMP genes in the CNS of Timp1 transgenic mice indicates that development, learning, and memory functions do not require the entire MMP arsenal. To elucidate the effects of strong Timp1 expression in CNS inflammation, we induced experimental allergic encephalomyelitis. We observed a Timp1 dose-dependent mitigation of both experimental allergic encephalomyelitis symptoms and histological lesions in the CNS of transgenic mice. All in all, our data demonstrate that (1) long-term CNS expression of TIMP1 with complete suppression of gelatinolytic activity does not interfere with physiological brain function and (2) TIMP1 might constitute a promising candidate for long-term therapeutic treatment of inflammatory CNS diseases such as multiple sclerosis.
Tissue-inhibitor of matrix metalloproteinase-1 (TIMP1), a tightly regulated 28.5-kDa glycoprotein,1,2 belongs to a family of multifunctional secreted proteins (TIMPs 1 to 4) that regulate the proteolytic activity of matrix metalloproteinases (MMPs). Together, MMPs and TIMPs control the pericellular environment, including the turnover of extracellular matrix proteins, bioavailability of growth factors and cytokines, and shedding of membrane receptors.3 In the central nervous system (CNS), expression of MMP and TIMP genes is highly orchestrated during development and normal brain function.4,5,6 The spatio-temporal expression pattern of TIMP1 indicates a developmental role in the hippocampus and the cerebellum.4,7,8,9 TIMP1 was also proposed as a candidate plasticity protein in learning and memory.10,11 Several MMPs on the other hand are expressed in the developing cerebellum,4 and in migrating neural precursors,12,13 and MMP activity was localized to the growth cone of cultured sympathetic neurons.14,15,16 In the normal adult CNS, however, MMP expression is low.17,18 Alterations of the MMP/TIMP ratio are tightly associated with pathological processes and clearly influence the course of various diseases.12,17,18,19 In autoimmune diseases such as multiple sclerosis and the animal model experimental allergic encephalomyelitis (EAE), elevated activity of several MMPs was detected and suspected to influence the outcome of the disease.17,20,21,22,23 Accordingly, synthetic MMP inhibitors blocked the blood-brain-barrier disruption and improved the clinical condition of the animals,23,24,25 and Timp1 deficient mice revealed increased leukocyte infiltration as well as enhanced spinal cord demyelination.26 Due to serious side-effects, however, only few synthetic MMP-inhibitors have been introduced to human therapy.27
Since high levels of Timp1 were expressed by astrocytes around the inflammatory lesions in EAE17 and to further investigate the role of Timp1 in the CNS, we targeted its expression to astrocytes in a transgenic mouse model.
Materials and Methods
Generation of Timp1 Transgenic Mice
The coding sequences for the murine Timp1 gene (nucleotide, GenBank accession number NM_011593.2, CDS 165-782) was amplified by RT-PCR from mouse spleen RNA. The PCR primers (5′-ATGCGGCCGCATGATGGCCCCCTTTGCATC-3′ and 3′-ATCCCGGGTCATCGGGCCCCAAGGGATC-5′) amplified 635 nucleotides specific for the Timp1 gene. The amplified fragments were cloned into a vector consisting of the mouse GFAP promoter, an SV40 splice donor/acceptor site upstream, and a polyadenylation signal of the human growth hormone followed by the genomic sequence for GFAP downstream as described before.28 To confirm the identity of the amplified fragments, overlapping sequence analysis of both DNA strands was performed. The resulting fusion gene constructs were excised from the vector and purified, and the TIMP1 construct was microinjected into fertilized eggs of (C57BL/6J) mice. Transgenic offspring were identified by PCR analysis of DNA from tail biopsies. The PCR primers were designed to generate fragments of 700 bases for the Timp1 transgene. By using a downstream primer homologous to the hGH polyadenylation signal, amplification was restricted to transgenic Timp1 sequences. Three transgenic mouse lines termed T1F10, T1F18, and T1F60 were established. All experimental procedures were approved by the local authorities (Regierungspräsidien [regional boards], Freiburg, and Giessen).
RNA Preparation
Female mice, 7 to 8 weeks of age, were mated with males to generate pregnant animals for collection of staged embryonic and postnatal tissues. Twenty-four hours after mating, a plug check was performed and in the case of a positive result defined as E 0.5. Female animals were sacrificed at days E 17.5 to recover embryos, whereas newborn mice were sacrificed at various days post partum. Animals were euthanized with isofluran-baxter (Baxter Deutschland GmbH, Unterschleißheim, Germany), and the brain was removed and the two hemispheres were separated following the rostro-caudal middle line. One hemisphere was dissected into the forebrain (cerebrum and olfactory bulb) and hindbrain (cerebellum and brain stem). The spinal cord was removed from the spine. All tissues were then immediately snap-frozen in liquid nitrogen and stored at −80°C pending RNA extraction. Total RNA was extracted with Trizol reagent (Invitrogen GmbH, Darmstadt, Germany) according to the manufacturer’s protocol.
Histology and Immunohistochemistry
The remaining brain hemisphere was either embedded in Tissue Tek (Sakura Finetek Europe B.V., Zoeterwoude, The Netherlands) and shock-frozen in liquid nitrogen chilled isopentane or fixed in 4% paraformaldehyde in PBS and embedded in paraffin. Paraffin sections (4-μm thick) were processed according to standard procedures and used for routine staining and immunohistochemistry. Cryostat sections (10-μm thick) were air-dried and fixed in cold (−20°C) acetone-methanol (1:1) for 45 seconds. Endogenous peroxidase was blocked for 18 minutes with 0.3% H2O2 in PBS. Subsequently, sections were incubated with normal serum (avidin-biotin complex kit; Vector Laboratories, Burlingame, CA) for 30 minutes. Primary antibodies against GFAP (Dako, Hamburg, Germany), Neurofilament (Sigma-Aldrich Chemie GmbH, Steinheim, Germany), NeuN (Chemicon International, Temecula, CA), and Calbindin (Swant, Bellinzona, Switzerland) on paraffin embedded slices were incubated overnight at 4°C. Primary rat monoclonal antibodies against infiltrating lymphocytes (CD4, CD8, and CD45R; BD Biosciences, Heidelberg, Germany), activated microglia, and macrophages (MAC-1, membrane attack complex 1, generous gift from M. Simon, Max Planck Institute for Immunbiology, Freiburg, Germany) were incubated overnight at 4°C on fixed cryostat sections. Bound primary antibody was detected by using either a mouse-adsorbed, biotinylated anti-rat antibody for the cell surface molecules (Southern Biotechnology Associates, Birmingham, AL), or the biotinylated secondary antibody from the avidin-biotin complex kit followed by horseradish peroxidase-coupled avidin-biotin complex kit and 3–3-diaminobenzidine (Merck, Darmstadt, Germany) as a substrate. Sections were counterstained with Mayer’s hematoxylin for 15 seconds, dehydrated in graded ethanol and xylene, and mounted. Perivascular and diffuse infiltration of the CNS by lymphocytes was evaluated semiquantitatively (0 to +++) by two independent observers blinded for the genotype and the disease score of the mice.
Enzyme-Linked Immunosorbent Assay
Tissue was snap-frozen in liquid nitrogen and stored at −70°C until protein extraction. Samples were homogenized in 10 times volume to weight of extraction buffer (50 mmol/L Tris, pH 7.4; 1% Nonidet; 5 mmol/L EDTA; 5 μg/ml Pepstatin A and 4% complete [Roche Diagnostics, Mannheim, Germany]). Protein concentration was determined with a Bradford assay. Mouse Timp1 enzyme-linked immunosorbent assay was purchased from R&D systems and used following the manufacturer’s protocol.
RNase Protection Assays
RNase protection assays (RPAs) for the detection of TIMP and MMP RNAs were performed as described previously.17 To distinguish transgenic Timp1 mRNA from endogenous Timp1 mRNA, we established a probe that contained the 3′ portion of the Timp1 cDNA and the adjacent 60 bases of the hGH polyadenylation signal that does not occur in wild-type mice. Thus, this probe revealed a protected band of 264 bases from endogenous TIMP-1 RNA (T1) and a protected fragment of 324 bases from transgenic TIMP-1 RNA (T1TG). For the detection of the MT-MMP RNA species, a novel probe set was established according to the method described before.17 In brief, specific sections of MT1-, MT2-, MT3-, MT4-, and MT5-MMP genes were determined by alignment (MegAlign, DNAStar, Madison, WI). These were amplified by RT-PCR, and the resulting RT-PCR products were cloned into pGEM4Z. Plasmids were linearized with EcoRI and each probe was initially tested separately to verify the protected RNA. Subsequently, the multi probe set was established. The RNA samples were hybridized with labeled probe sets as described previously.29 For quantification, densitometric analysis of each band was performed on scanned autoradiographs by using the ImageJ software as described previously.29 Band density of the individual mRNAs was normalized to that of L32.
In Situ Hybridization
Mice were dissected as described above and the brain was fixed in 4% buffered paraformaldehyde. Subsequently, the brains were dehydrated through graded alcohol solutions and embedded in paraffin. In situ hybridization was performed on tissue sections with a labeled antisense TIMP-1 RNA probe as described before.30 For immunohistochemical detection of GFAP, sections were blocked for 1 hour at room temperature in PBS containing 3% goat serum. Immunohistochemistry was performed as described above. Finally, the sections were counterstained with Mayer’s hematoxylin, coverslipped, and examined by bright-field microscopy. Controls for reaction specificity included incubation of the sections with a nonimmune rabbit serum, as well as omission of the primary antibody.
In Situ Zymography
In situ zymography was performed as described before.18 Briefly, cryostat sections (8 to 10 μm) of the CNS of control and transgenic EAE mice were overlaid with assay solution (50 mmol/L Tris-HCl, 5 mmol/L CaCl, pH 7.5) containing 40 mg/ml fluorescein isothiocyanate-labeled DQ gelatin (Molecular Probes, Eugene, OR). Sections were incubated at 37°C for 48 hours and examined by fluorescence microscopy. Gelatinolytic proteases cleave quenched DQ gelatin resulting in fluorescent breakdown products that thereby allow for the localization of net gelatinolytic activity in the tissue. Addition of 10 mmol/L EDTA, a chelating agent that inhibits metal ion dependent proteases such as the MMPs, to the assay solution or omission of DQ gelatin served as negative controls and resulted in lack of fluorescence.
Behavioral Tests
Behavioral procedures were approved by the Cantonal Veterinary Office of Zurich. Mice (16 TG of line T1F60, 16 littermates wild-type, balanced numbers of males and females) were housed singly and allowed to adapt for 1 week to a reverse light cycle (lights off at 8:00 AM; on at 8:00 PM). They were then analyzed in a blinded manner in the following order of tests under diffuse dim light (approximately 12 lux): water-maze place navigation, open field, O-maze, emergence test, object exploration, and rotarod. Their age was 3.6 months (3.0 to 3.9) at the start of the experiments. Except for the rotarod test, experiments were recorded by using a Noldus EthoVision video-tracking system (version 3.0, Leesburg, VA), and recorded data were exported to the analysis software Wintrack (version 2.4, http://www.dpwolfer.ch/wintrack, last accessed May 28, 2010) for further analysis.31
Water-maze place navigation was tested as described in detail previously32 by using a white circular pool (150 cm diameter) filled with milky water (24°C to 26°C). Acquisition training consisted of 18 trials (six per day; intertrial interval, 30 to 60 minutes) during which the submerged platform (14 × 14 cm) was left in the same position. Trials lasted a maximum of 120 seconds. To monitor reversal learning, the platform was moved to the opposite position for two additional days of training (six trials per day). The first 30 seconds of the first reversal trial served as probe trial to test for spatial retention.
Anxiety-related and exploratory behavior was analyzed as described previously.33 The round open-field arena had a diameter of 150 cm, a white plastic floor, and 35-cm high sidewalls. The animals were observed during 10 minutes each on two subsequent days. For analysis, the arena was divided into a wall zone (7 cm wide), a transition zone, and a center field (50% of the arena surface). The O-maze had a 5.5 cm wide annular runway with an outer diameter of 46 cm and was elevated 40 cm above the floor. Two opposing 90° sectors were protected by 16-cm high inner and outer walls (height, 16 cm). The animals were observed for 10 minutes. For analysis, the runway was divided into protected (50°) and unprotected (70°) zones, separated by transition zones (30°) from which the mice could perform exploratory head dips (recorded manually by using the key board) while keeping their body protected between the side walls. In the emergence test, four mice were tested concurrently in adjacent quadratic arenas (50 × 50 cm) with 35-cm high side walls. A small home box was introduced to the home cage 24 hours before the test. The mice were transferred to the test arena with the home box and observed for 30 minutes. For analysis, the arena was divided in a wall zone (5 cm wide), a center field, and a home zone (5 cm around the box). The object exploration was performed in the same arena on the following day. After 30 minutes of further habituation to the empty arena, a Falcon tube was placed in the center of the arena and behavior was recorded for another 30 minutes. For analysis, the arena was divided into an object zone (14 cm diameter), a transition zone, and four corner zones (5 × 5 cm). Object exploration was estimated as total duration of small nonlocomotor movements in the object zone, corrected by subtraction of the values obtained in absence of the object during the habituation period.
The rotarod test was done on an apparatus from Ugo Basile (Comeria VA, Italy). Each mouse did five trials (intertrial interval, 30 minutes) during which the speed increased linearly from 4 to 40 rpm during 5 minutes. A trial was ended when the mouse fell down or clung to the rod and rode a full revolution.
Statistical Analysis
A two-way factorial analysis of variance design with genotype and sex as between subject factors was used (Statview 5.0, Cary, NC, no longer sold). Where appropriate, the model was complemented by within subject factors to explore the dependence of genotype effects on place or time. The sex factor was only introduced to reduce unexplained variance and to check genotype effects for sex-dependence. Because there were no significant sex effects or interactions, results of this factor are not shown.
Induction of EAE
Active immunization was performed with myelin oligodendrocyte glycoprotein (MOG35–55) peptide (Sigma). On day 0, 8- to 12-week-old wild-type mice or transgenic T1F60 littermates of both sexes were immunized s.c. into the rear flanks with a 1:1 emulsion of 200 μg MOG peptide in 100 μl incomplete Freund’s adjuvant (Difco, Detroit, MI) supplemented with 500 μg Mycobacterium tuberculosis H37RA (Difco). In addition, each mouse received an i.p. injection of 250 ng pertussis toxin in saline (Alexis Biochemicals, Lausen, Switzerland) on days 0 and 2. Animals were observed for up to 6 weeks on a daily basis, and disease severity was graded from 0 to 5 as follows: 0, no disease; grade 0.5, partial loss of tail tonus; grade 1, complete tail atony; grade 2, hind limb paresis, waddling gait; grade 3, hind limb paralysis; grade 4, moribund; and grade 5, death. Statistical analyses were performed for four experiments with a total of n1 = 31 wild-type; n2 = 31 TG animals. Since variances were heterogeneous over time, an analysis of variance is problematic for such data. Hence, we used summary measures to compare disease courses between the two groups as proposed for example by Altman.34 For each mouse, we computed three measures: the mean, the peak of the disease course, and the area under the curve. All measures were compared by a stratified Mann-Whitney U test, and statistical significance was calculated with the Approximative Wilcoxon Mann-Whitney Rank Sum test.
Results
Pattern of Timp1 Expression
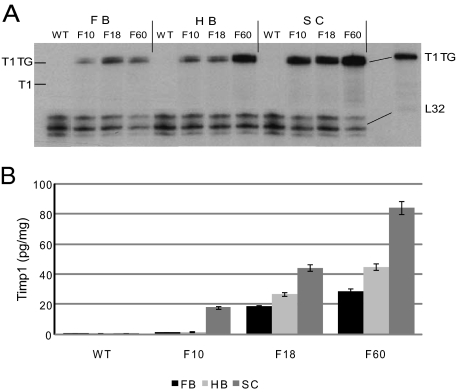
Adult wild-type mice did not show constitutive expression of Timp1 in the CNS (Figure 1A). Adult animals of all three transgenic mouse lines expressed transgenic Timp1 in all regions of the CNS (Figure 1A). Compared with T1F10 mice, T1F18 and T1F60 mice showed higher and similar levels of Timp1 RNA expression in the forebrain, respectively. In the hindbrain and the spinal cord, mice of the T1F10 and T1F18 showed similar levels of transgene expression, whereas mice of the T1F60 line revealed maximal expression in these regions (Figure 1A). The absence of signals below the T1TG bands demonstrated that constitutive transgenic expression of Timp1 did not induce the expression of endogenous Timp1 (Figure 1A). RT-PCR analysis of DNase treated RNA revealed restriction of transgene expression to CNS organs (brain, spinal cord, and eyes), whereas none of the peripheral organs tested (liver, spleen, thymus, lymph node, kidney, skeletal muscle, heart, lung, and ovary) showed a PCR product (data not shown). In good correlation with the RNA expression, there was no detectable Timp1 protein in the CNS of wild-type mice (Figure 1B), whereas mice of the T1F10, T1F18, and T1F60 lines showed minimum expression in the forebrain, intermediate expression in the hindbrain, and a maximum of 18 pg/mg, 45 pg/mg, and 85 pg/mg Timp1 protein, respectively, in the spinal cord. These results demonstrated that the RNA was translated into protein at levels corresponding to RNA expression levels.
Figure 1.
A: Timp1 mRNA expression in the CNS. An RPA was used to detect Timp1 mRNA in the forebrain (FB), hindbrain (HB), and the spinal cord (SC) of adult animals of the three mouse lines, T1F10, T1F18, and T1F60. Healthy wild-type (WT) animals had no constitutive Timp1 expression. All transgenic mice of the three mouse lines expressed the transgene in every region of the CNS (FB, HB, and SC). The probe consisted of the 3′ portion of Timp1 and an adjacent portion of the hGH polyadenylation signal. This probe allowed for the differentiation of wild-type (T1 264 b) and transgenic Timp1 mRNA (T1 TG 324 b). The RPA was performed as described in Materials and Methods. B: Timp1 concentrations (pg/mg wet tissue) in various CNS regions of transgenic mice of the T1F10, T1F18, and T1F60 lines and WT C57/BL6 mice were determined by enzyme-linked immunosorbent assay. Error bars = SEM.
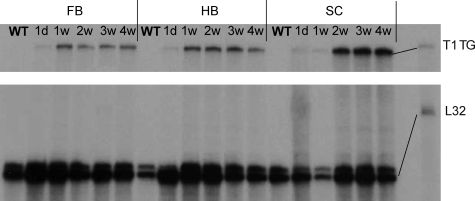
Since the highest level of transgenic Timp1 expression was observed in the T1F60 mouse line, we performed further detailed expression-pattern analysis on this mouse line. In mice with the highest levels of transgene expression (T1F60), Timp1 RNA was detectable from P1 and reached a maximum level after 1 week (Figure 2). Wild-type Timp1 RNA was detectable neither in transgenic nor in wild-type mice at any time point from 1 day to 4 weeks postnatally (Figure 2).
Figure 2.
Postnatal time course of transgenic Timp1 expression in the T1F60 mouse line. Transgene expression (T1 TG) was detectable in every region of the CNS (forebrain [FB], hindbrain [HB], and spinal cord [SC]) on the first postnatal day and reached maximum levels within the first postnatal week. There was no gene expression of wild-type Timp1. The L32 band served as a loading control. RPA was performed as described in Materials and Methods.
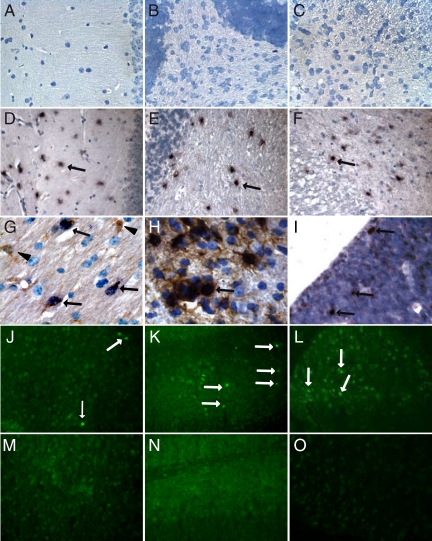
In situ hybridization confirmed the presence and distribution pattern of transgenic Timp1 mRNA in the forebrain, the hindbrain, and the spinal cord of transgenic T1F60 mice (Figure 3). In wild-type animals, no Timp1 mRNA was detectable in any CNS region (Figure 3, A–C), whereas in T1F60 animals all regions showed Timp1 mRNA-positive cells (Figure 3, D–F, arrows). Combined in situ hybridization and immunohistochemistry demonstrated in all transgenic T1F60 mice the colocalization of Timp1 RNA exclusively with GFAP-immunoreactivity (Figure 3, G and H, arrows). Thus, transgenic Timp1 was expressed only by astrocytes. A large percentage of GFAP-immunoreactive astrocytes was double stained throughout the CNS with virtually all astrocytes Timp1-positive in the hypothalamus, the hippocampus, corpus callosum, the cerebellar white matter, the brain stem, and the spinal cord. Few cells revealed Timp1 mRNA expression as early as embryonic stage 17.5 days (Figure 3I, arrows) in the CNS of T1F60 animals, whereas there was no staining in the CNS of wild-type animals (data not shown). To test whether transgenic Timp1 had an MMP-inhibitory activity in the CNS, we performed in situ zymography on brain sections of 10-day-old wild-type and TG T1F60 mice. This analysis demonstrated gelatinolytic activity in numerous cells in the forebrain, the hindbrain, and the spinal cord of wild-type mice (Figure 3, J–L, arrows), whereas in the respective brain areas of TG T1F60 mice no gelatinolytic activity was observed (Figure 3, M–O). Thus, transgenic Timp1 completely suppressed gelatinolytic activity in the CNS tissue.
Figure 3.
In situ hybridization for Timp1. No Timp1 RNA was detected in the brains of adult wild-type animals (A–C), whereas strong Timp1 signals were observed in all CNS regions of adult TG T1F60 mice (D–F, arrows). Hippocampus (A, D), cerebellum (B, E), and spinal cord (C, F). The Timp1 RNA (arrows) co-localized with GFAP immunoreactive cells (brown staining) demonstrating that transgenic Timp1 was synthesized by astrocytes (G, H). A minority of astrocytes in the T1F60 mouse line were Timp1 RNA negative (G, arrowheads). At embryonic stage E 17.5, few cells showed transgene expression in transgenic mice of the T1F60 mouse line (I, arrows). Wild-type mice of this stage revealed no positive staining in the CNS (data not shown). Original magnification: ×40 (A–F); ×100 (G–I). In situ zymography on brain sections of P10 mice showed gelatinolytic activity in the forebrain (J), the hindbrain (K), and the spinal cord (L) of wild-type animals (J–L, arrows). No activity was detected in the same regions of transgenic littermates of the T1F60 mouse line (M–O). Control slices showed that the enzymatic activity was inhibited by 10 mmol/L EDTA and that substrate was necessary to obtain a fluorescent signal (data not shown). Original magnification: ×20.
Constitutive Timp1 Expression Has No Direct Impact on the CNS Morphology
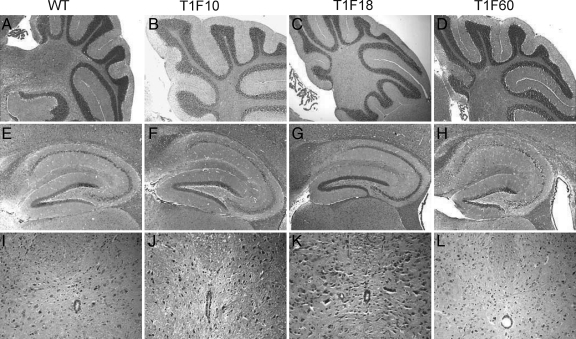
The potential impact of constitutive transgenic Timp1 expression on the morphology of the brain was analyzed by histology. H&E staining revealed an overall normal CNS morphology of the three mouse lines (T1F10, T1F60, and T1F18; Figure 4). Cerebellum (wild-type: Figure 4A, TG: Figure 4, B–D), hippocampus (wild-type: Figure 4E, TG: Figure 4, F–H), as well as spinal cord (wild-type: Figure 4I, TG: Figure 4, J–L) sections of the transgenic mice revealed a morphology that was indistinguishable from wild-type control sections. To further characterize the cellular subpopulations in the CNS of the T1F60 mouse line, we performed immunohistochemistry for NeuN, Calbindin, and GFAP (Supplemental Figure S1, see http://ajp.amjpathol.org). This analysis revealed a normal morphology and distribution of neurons and glial cells in the hippocampus, hindbrain, and spinal cord. On a histological basis, transgenic animals were undistinguishable from their wild-type littermates.
Figure 4.
Histology of the CNS. The morphology of the cerebellum (A–D), hippocampus (E–H), and spinal cord (I–L) of adult animals of all three mouse lines revealed no alterations as compared with WT mice. H&E magnification: ×5 (A–H); ×20 (I–L).
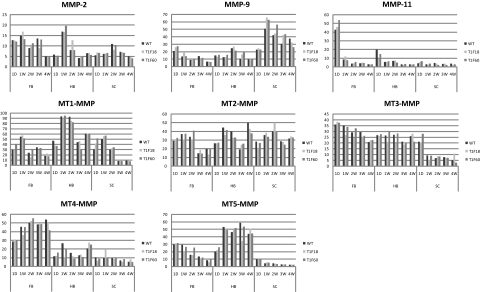
To investigate whether the inhibition of MMPs by transgenic Timp1 led to increased MMP gene expression, we performed RPA with probes for MMPs and MT-MMPs (Figure 5). Neither control mice nor transgenic animals showed mRNAs for MMP-1, -3, -7, -8, -10, -12, -13, and -27 in the CNS. In contrast, RPA showed distinct expression patterns of several MMP species: although MMP-2 and MT3-MMP were predominantly expressed in the forebrain and the hindbrain, MMP-9 showed maximum expression in the spinal cord. MMP-11 was predominantly expressed during the first day postnatal in the forebrain and the hindbrain. High levels of MT1-MMP and MT5-MMP RNA were observed in the postnatal cerebellum, whereas MT4-MMP revealed maximum expression the forebrain. MT2-MMP was homogenously expressed in all three CNS areas. Taken together, no up-regulation of MMP-genes occurred despite constitutive high expression of Timp1 and complete suppression of gelatinolytic activity in the CNS of transgenic mice of the T1F60 line.
Figure 5.
Postnatal MMP RNA expression in the CNS. RPA was performed on CNS tissue from forebrain (FB), hindbrain (HB), and spinal cord (SC) of 1 day (1D), 1-, 2-, 3-, and 4-week-old (1W, 2W, 3W, and 4W, respectively) animals. The amount of specific RNA is depicted as arbitrary units (y axis). Patterns of regulation are represented for MMP-2, MMP-9, MMP-11, and MT1-MMPs to MT5-MMPs. RPA was performed as described in Materials and Methods.
Behavior
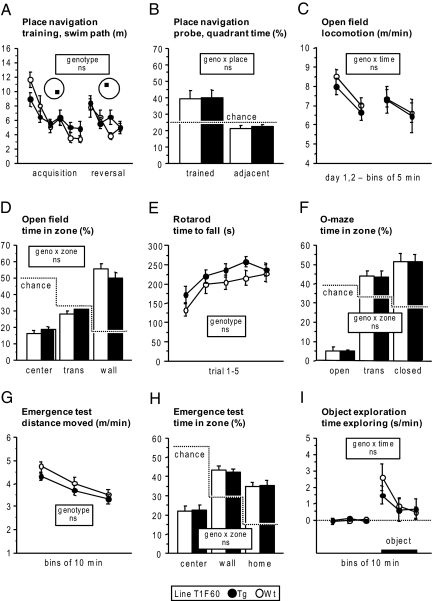
To further analyze potential alterations of the CNS in TIMP-1 expressing mice that might have been missed by histological examination, we performed behavioral tests with the most strongly expressing line T1F60. Spatial learning and memory was assessed by using a water-maze place navigation task. Wild-type and TG mice acquired the task at similar rates and were also equally efficient in adapting to a new platform position during reversal training (Figure 6A). There was no mutation effect on swim speed or thigmotaxis (wall hugging), and TG mice showed normal spatial selectivity during training according to several measures, including Wishaw’s error (Table 1).35,36 During the probe trial, selectivity for the trained quadrant was unaffected by genotype with scores overall typical of the C57BL/6 strain in this protocol (Figure 6B). This could be confirmed by more stringent measures of spatial selectivity such as zone time and annulus crossings (Table 1).35,36
Figure 6.
Behavioral tests. A and B: Water-maze place navigation: A: Overall performance and learning rates were similar in wild-type (WT) and TG mice of line T1F60. Each point represents three subsequent trials: genotype F(1,25) = 0.025 ns; time F(9225) = 8.775, P < 0.0001; genotype × time F(9225) = 1.598 ns. B: Irrespective of genotype, mice of line T1F60 showed a robust preference for the target quadrant in comparison with the adjacent quadrants: place F(1,25) = 16.108, P < 0.0005; genotype × place F(1,25) = 0.013 ns. C–I: Anxiety-related behaviors, exploratory activity, and locomotion: C: Wild-type and TG of line T1F60 did not differ with respect to level and time course of activity: genotype F(1,25) = 0.271 ns; time F(3,75) = 9.293, P < 0.0001; genotype × time F(3,75) = 0.150 ns. D: Time spent in the center, transition, and wall zone of the open field. Independently of genotype, mice avoided the center zone in favor of the wall zone: zone F(2,52) = 68.500, P < 0.0001; genotype × zone F(2,52) = 1.198 ns. E: TG and wild-type were indistinguishable with respect to both performance level and learning rate in the rotarod test: genotype F(1,26) = 2.162 ns; trial F(4104) = 11.875, P < 0.0001; genotype × trial F(4104) = 0.492 ns. F: Time spent in the open, transition, and closed zone of the elevated O-maze. Independently of genotype, mice strongly avoided the open zone: zone F(2,52) = 89.770, P < 0.0001; genotype × zone F(2,52) = 0.007 ns. G: Although TG moved on average slightly shorter distances than wild-type, there was no genotype effect on level or habituation rate of activity: genotype F(1,26) = 1.692 ns; time F(2,52) = 47.435, P < 0.0001; genotype × time F(2,52) = 0.570 ns. H: Irrespective of genotype, mice preferred the home and wall zones: zone F(2,52) = 27.386, P < 0.0001; genotype × zone F(2,52) = 0.089 ns. I: Exploratory activity toward a novel object was also significant and unaffected by genotype: genotype F(1,26) = 0.233 ns; time F(5130) = 8.079, P < 0.0001; genotype × time F(5130) = 0.668 ns.
Table 1.
Water-Maze Place Navigation, Factorial Analysis of Variance
| Water-maze place navigation | Genotype effect T1F60 | P |
|---|---|---|
| Training | ||
| Escape latency/swim time (s) | F(1,25) = 0.1 | <0.7621 |
| Swim path (m) | F(1,25) = 0.1 | <0.8772 |
| Time in current goal quadrant (%) | F(1,25) = 0.1 | <0.8822 |
| Wishaw’s error (%) | F(1,25) = 0.1 | <0.9626 |
| Path efficiency (%) | F(1,25) = 0.1 | <0.7606 |
| Cumulative search error (m × s) | F(1,25) = 0.2 | <0.6316 |
| Time near wall (%) | F(1,25) = 0.2 | <0.6794 |
| Swim speed (m/s) | F(1,25) = 0.3 | <0.5673 |
| Time floating (log s) | F(1,25) = 0.2 | <0.6828 |
| Probe | ||
| Time in trained quadrant (% of first 30 s) | F(1,25) = 0.1 | <0.9927 |
| Time in trained zone (% of first 30 s) | F(1,25) = 0.2 | <0.6601 |
| Annulus crossing index (x/minutes in first 30 s) | F(1,25) = 0.1 | <0.8196 |
Training variables were averaged across all trials. Whishaw’s error = % path outside a 0.1856-m wide corridor connecting release point and goal, adapted from Wishaw.35 Path efficiency = % path during which speed vector component toward goal is 75% or more. Cumulative search error = sum of distances to target measured at 1 second intervals minus value that would be obtained for an ideal direct swim.36 Width of wall zone = 7 cm. Swim speed calculated after exclusion of time spent floating = episodes of immobility or decelerations with speed minimum <0.06 m/s. Trained zone: circle, 1/8 of pool surface. Annulus: 16 × 16 cm.
Anxiety-related behaviors and exploratory activity of TG mice were assessed in several tests and found to be normal. In the open field test, they showed unaltered activity level and rate of habituation (Figure 6C; Table 2).37 Their avoidance of the center field was indistinguishable from wild-type mice (Figure 6D; Table 2).37 The number of fecal boli deposited during the test, often taken as a measure of stress, was not affected by the transgene (Table 2). In addition, the open field test revealed normal speed and acceleration during locomotion. There was no evidence for unstable gait, abnormal path tortuosity, or circling, indicating normal motor coordination in TG mice (Table 2).37 This was confirmed by the rotarod test (Figure 6E). On the O-maze, TG mice showed normal avoidance of the open sectors and head dipping behavior, with overall activity indistinguishable from controls (Figure 6F; Table 2).37 The emergence test (open field with home box) confirmed the results previously obtained in the larger open field arena: TG mice could not be distinguished from wild-type with respect to activity level, habituation rate (Figure 6G; Table 2),37 and avoidance of the center field in favor of the wall and home zones (Figure 6H; Table 2).37 Finally, when a novel object was introduced into the empty arena after further familiarization, TG and wild-type mice explored it with similar intensity and time course (Figure 6I; Table 2).37
Table 2.
Anxiety-Related Behaviors, Exploratory Activity, and Locomotion, Factorial Analysis of Variance
| Exploration tests | Genotype effect T1F60 | Tendancy | P |
|---|---|---|---|
| Open field | |||
| Time immobile (%) | F(1,26) = 1.6 | + | <0.2142 |
| Velocity while progressing (m/s) | F(1,26) = 0.3 | <0.6078 | |
| Median absolute acceleration (m/s2) | F(1,26) = 1.6 | + | <0.2150 |
| Circling index (°/m) | F(1,26) = 2.7 | + | <0.1101 |
| Tortuosity index (°/m) | F(1,26) = 2.3 | − | <0.1432 |
| Wobbling index (°/m) | F(1,26) = 0.1 | <0.9264 | |
| Bolus count (x) | F(1,26) = 1.3 | − | <0.2603 |
| Distance moved, wall—center zone (m/min) | F(1,26) = 0.8 | − | <0.3772 |
| Average distance to center (m) | F(1,26) = 1.2 | − | <0.2826 |
| O-maze | |||
| Closed sector entries (x/min) | F(1,26) = 1.5 | − | <0.2329 |
| Distance moved (m/min) | F(1,26) = 0.1 | <0.9382 | |
| Bolus count (x) | F(1,26) = 0.2 | <0.6498 | |
| Open sector entries (%) | F(1,26) = 0.7 | + | <0.4194 |
| Head dips (x/min) | F(1,26) = 1.1 | + | <0.3066 |
| Protected head dips (%) | F(1,26) = 0.4 | <0.5456 | |
| Emergence test | |||
| Time immobile (% visible time) | F(1,26) = 1.1 | + | <0.3138 |
| Velocity while progressing (m/s) | F(1,26) = 0.1 | <0.9621 | |
| Average distance to center (m) | F(1,26) = 0.3 | <0.6126 | |
| Time in box (%) | F(1,26) = 0.3 | <0.5893 | |
| Object exploration | |||
| Time immobile (%) | F(1,26) = 0.2 | <0.6984 | |
| Velocity while progressing (m/s) | F(1,26) = 0.6 | + | <0.4344 |
| Corner time (%) | F(1,26) = 0.4 | <0.5299 | |
| Distance to object while exploring (m) | F(1,24) = 0.1 | <0.8658 | |
| Visits to object zone (x/min) | F(1,26) = 1.0 | − | <0.3182 |
Variables were averaged across the entire observation period of each test. Immobility = 2 seconds or longer with smoothed speed values (averaging frame 0.5 s) <2.5 cm/s (noise level). Progressive movement = 8.5 cm/s for >5 cm. Median absolute acceleration = median across progression episodes of maximal velocity divided by episode duration.37 Circling index = sum of signed direction changes during curves, divided by total path length. Path tortuosity = sum of unsigned direction changes during curves with consistent direction change during >1 second, divided by total path length. Wobbling = sum of unsigned direction changes between successive segments of the unsmoothed path, divided by the distance moved. Protected head dips: body center in transition zone, unprotected head dips: body center on open sector. Tendancy; + indicates increased value; −indicates reduced value in TG vs. WT.
Timp1-Dependent Alteration of the EAE Disease Course
Although Timp1 expression had no impact on the anatomical and behavioral phenotype of transgenic animals, we wondered whether the presence of Timp1 would influence the outcome in an inflammation paradigm. Since Timp1 is up-regulated in astrocytes around inflammatory lesions in the spinal cord during EAE, we investigated whether high levels of transgenic Timp1 in the CNS of transgenic mice of the T1F60 line could affect the clinical course of this disease. Analysis by RPA showed a similar up-regulation of the GFAP gene in wild-type and TG mice (Supplemental Figure S2, see http://ajp.amjpathol.org).
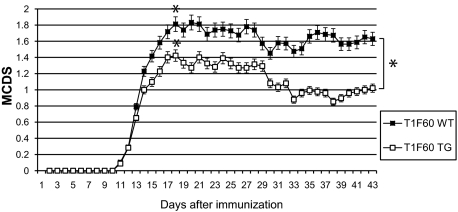
In both animal groups, disease onset was on day 11 after immunization (Figure 7). The mean clinical disease score (MCDS) of the wild-type group (n = 31) showed a first peak of 1.8 at day 18, whereas the transgenic T1F60 mice (n = 31) had a maximal MCDS of 1.4 at day 18. Both transgenic and wild-type T1F60 groups revealed a slight remission during the observation period with a mean clinical disease score (MCDS) of approximately 1.6 for the wild-type compared with 1 for the transgenic mice after 43 days of monitoring. The mean clinical disease course analyzed in four independent experiments as well as the disease peak were significantly different from the respective wild-type values (area under the curve: Approximative Wilcoxon Mann-Whitney Rank Sum Test: Z = −2.3239, P = 0.01919; Mean: Approximative Wilcoxon Mann-Whitney Rank Sum Test: Z = −2.4209, P = 0.01478; Peak: Approximative Wilcoxon Mann-Whitney Rank Sum Test: Z = −2.5538, P = 0.009502). In both wild-type and TG groups, the minimal and maximal disease score observed was 0 and 3, respectively. There was a difference of disease incidence between the groups (wild-type: 84%; TG: 71%) and a higher percentage (77%) of the wild-type animals developed a clinical score of 2 or higher as compared with 55% of the TG mice. Hence, transgenic animals of the T1F60 mouse line are more resistant to induction of clinical disease in the EAE paradigm as compared with wild-type littermates. Table 3 summarizes the clinical findings in EAE of the T1F60 mouse line.
Figure 7.
Disease course in the EAE model of wild-type (WT) and transgenic mice of the T1F60 (nWT = 31; nTG = 31) line. 0: no symptoms; 1: limp tail; 2: ataxia; 3: hind limb paresis; 4: hind limb and fore limb paresis; 5: moribund; *P ≤ 0.05.
Table 3.
Characteristics of the EAE Disease Course Observed for the T1F60 Mouse Line. The Minimal and Maximal Disease Scores for Both Wild-Type and TG Were 0 and 3, Respectively
| T1F60 | Minimal clinical score | Maximal clinical score | Mice with a clinical score ≥2 | Incidence |
|---|---|---|---|---|
| Wild type | 0 | 3 | 24/31 (77%) | 26/31 (84%) |
| TG | 0 | 3 | 17/31 (55%) | 22/31 (71%) |
High Doses of Timp1 Inhibit Migration of Lymphocytes into the CNS Parenchyma
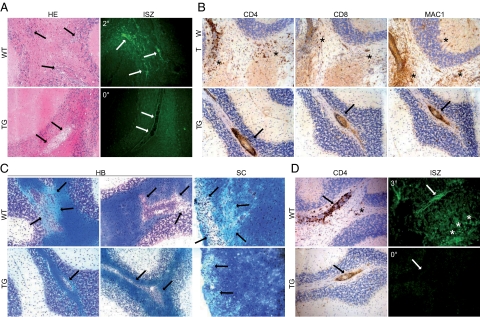
To further elucidate the mechanism of altered EAE-severity, we analyzed the distribution of lymphocytes in the spinal cord and the hindbrain of wild-type and transgenic mice at 13, 15, and 40 days after immunization. Immunization led to the extravazation of CD4+ and CD8+ lymphocytes as well as Mac-1+ macrophages into the brain and the spinal cord of all mice with disease symptoms and immunized healthy transgenic T1F60 animals that were histologically evaluated (Figure 8). Healthy wild-type mice did not show lymphocytic infiltration. In wild-type animals there was a close correlation of clinical symptoms and the degree of lymphocytic infiltration in the CNS, that is, sick mice revealed diffuse infiltration of lymphocytes in the CNS, whereas healthy mice had no lymphocytes in this organ. Immunized transgenic T1F60 animals, in contrast, regularly revealed perivascular cuffs in the spinal cord and the cerebellum and were, however, free of clinical symptoms or showed only low level disease scores. To further dissect the infiltration pattern in the CNS, the localization of lymphocytes was evaluated semiquantitatively by two observers blinded to the genotype and disease score of the animals at day 40 after immunization. In wild-type mice we observed perivascular cuffing and diffuse infiltration of the brain and spinal cord parenchyma by lymphocytes. Transgenic mice of the T1F60 line showed predominantly perivascular accumulation of lymphocytes and only mice with the highest disease scores (2.5 to 3) revealed diffuse infiltration of lymphocytes in the CNS parenchyma. Together, diffuse infiltration by lymphocytes correlated to severe disease while perivascular cuffing of lymphocytes around vessels alone related to lower disease scores or no sickness at all (Table 4).
Figure 8.
Pattern of leukocyte infiltration, gelatinolytic activity, and demyelination in the hindbrain of wild-type (WT) and transgenic (TG) mice of the T1F60 mouse line after induction of EAE. At day 15 after immunization wild-type animals with a clinical score of 2 (2°) showed diffuse lymphocytic infiltration (A, HE arrows) correlating with strong gelatinolytic activity detected by in situ zymography (ISZ) in serial sections (A, ISZ arrows). Despite a clinical score of 0 (0°), TG animals revealed perivascular accumulation of lymphocytes (A, HE arrows) and no gelatinolytic activity (A, ISZ arrows). At 40 days there was diffuse infiltration of CD4+ and CD8+ lymphocytes as well as MAC1+ macrophages and reactive microglia in symptomatic control animals (B, wild-type, asterisks). In transgenic mice these cells were predominantly confined to perivascular cuffs (B, TG, arrows). Klüver-Barrera (KB) staining revealed demyelinated areas in the white matter of hindbrain (HB) and spinal cord (SC) of wild-type mice (C, wild-type, arrows). In transgenic T1F60 mice there was an overall normal myelinization around inflammatory lesions in the cerebellum and spinal cord (C, TG, arrow). At day 40 wild-type animals with a high clinical disease score of 3 (3°) showed strong perivascular and diffuse infiltration of CD4+ lymphocytes (D, wild-type CD4, arrow and asterisk, respectively) correlating to perivascular and diffuse gelatinolytic activity (D, wild-type ISZ arrow and asterisks, respectively). In sharp contrast, CD4+ lymphocytes accumulated around the vessels in TG hindbrain and no gelatinolysis was observed (D, TG CD4 and ISZ arrows). Addition of 10 mmol/L EDTA or omission of the quenched gelatin resulted in no fluorescent signal (not shown). Original magnification: ×40 (A and B); ×20 (C and D).
Table 4.
Spatial Distribution of Lymphocytes in the Spinal Cord and the Cerebellum of Wild-Type and TG Mice of the T1F60 Line 40 After Injection (Seven Wild-Type and Nine TG T1F60 Animals)
| EAE score | 0° | 0.5° | 1° | 2° | 2.5° | 3° |
|---|---|---|---|---|---|---|
| T1F60 WT | No infiltrates | NA | PV + | PV + | PV + | PV + |
| D +++ | D +++ | D +++ | D ++++ | |||
| T1F60 TG | PV +++ | PV +++ | PV + | PV ++ | PV ++ | PV + |
| D ++++ | D ++++ |
Levels of lymphocytic infiltration were semiquantitatively evaluated from + to +++ by two observers blinded to the genotype and clinical score of the mice. In WT mice we observed perivascular cuffing (PV) and diffuse infiltration (D) of the cerebellum and spinal cord parenchyma by lymphocytes. Transgenic mice of the T1F60 line showed predominantly perivascular accumulation of lymphocytes and only the highest disease scores (2.5 to 3) concurred with diffuse infiltration of lymphocytes in the CNS parenchyma. NA, no wild-type mouse with a clinical disease score of 0.5° was observed at that time point.
To determine the cause for the reduced migration of lymphocytes into the CNS parenchyma of T1F60 mice, we performed hematoxylin and eosin staining (Figure 8A, HE) as well as in situ zymography on serial brain slices to detect gelatinolytic activity at 15 days after immunization (Figure 8A, ISZ). At this time point we observed inflammatory lesions correlating to strong gelatinolytic activity in the hindbrain of sick wild-type mice (Figure 8A, wild-type HE and ISZ, arrows). Transgenic mice of the T1F60 line revealed perivascular cuffs along with complete absence of gelatinolytic activity in the CNS (Figure 8A; TG, HE, and ISZ arrows, respectively). At day 40 after immunization, diffuse infiltration of the CNS parenchyma by lymphocytes occurred regularly in wild-type mice (Figure 8B, wild-type asterisks), whereas TG mice showed predominantly perivascular cuffs (Figure 8B, TG arrows). Only occasionally diffuse infiltration occurred in severely sick transgenic animals. Myelin damage was evaluated by Klüver-Barrera staining (Figure 8C). In wild-type animals, extensive myelin loss was observed around inflammatory lesions in the hindbrain and the spinal cord (Figure 8C, wild-type arrows). In sharp contrast, transgenic T1F60 littermates showed no signs of demyelination around perivascular cuffs (Figure 8C, TG arrows), a finding that correlated well with the significantly alleviated disease course in T1F60 mice. Immunostaining for CD4 and in situ zymography on serial sections at day 40 after EAE induction revealed perivascular and diffuse gelatinolytic activity in the tissue of wild-type animals (Figure 8D, wild-type CD4 and ISZ, arrows and asterisks, respectively), whereas several healthy T1F60 mice (disease score of 0) revealed massive perivascular infiltrates in the hindbrain (Figure 8D; TG and CD4, arrow) and no gelatinolytic activity (Figure 8D; TG and ISZ, arrow). Similar findings were observed in the CNS of transgenic T1F18 mice and less pronounced observations were made in the lower expressing transgenic mice of the T1F10 line (data not shown). Thus, high levels of Timp1 in the CNS of T1F18 and T1F60 TG mice were sufficient to completely inhibit gelatinolytic activity in the CNS tissue. Nevertheless, a few transgenic T1F60 mice had high clinical disease scores and showed diffuse lymphocytic infiltration in the spinal cord and hindbrain despite complete loss of gelatinolytic activity (data not shown). This indicates that inhibition of the gelatinolytic activity alone is not sufficient for complete protection from EAE.
Together, our results demonstrate that high levels of Timp1 alleviate EAE symptoms by reducing diffuse infiltration of lymphocytes and myelin damage in the CNS parenchyma while otherwise maintaining a normal phenotype of transgenic mice.
Discussion
The activity of MMPs is pivotal for a myriad of physiological and pathological conditions including development, tissue remodeling, inflammation, and wound healing.38,39 Tight regulation of MMP-activity is achieved on several levels, one of which comprises the inhibition of mature enzymes by inhibitors such as the TIMPs. The activity of MMPs has been implied in the development and physiological functioning of the CNS.12,21 A number of in vitro studies demonstrated that several MMPs enabled neuronal processes to penetrate three dimensional collagen matrices.14,15,40,41 Similarly, process outgrowth of cultured oligodendrocytes was associated with MMP-9 activity.42 Ex vivo experiments on cerebellar slice cultures showed that MMP-9 was expressed in P12 developing mouse cerebellum and facilitated process outgrowth and migration of granular precursor cells.40 MMP null mutant mice, however, had to be thoroughly scrutinized to reveal only minor constitutive alterations of CNS-morphology.43,44 Several injury models showed impaired remyelination after lesioning and decreased scar formation along with macrophage influx.39,44,45,46 The apparent discrepancy between the in vitro findings and the observations in MMP null mice suggests that the outgrowth of neuronal processes into collagen matrices is different from the behavior of neurites in the CNS. Although process outgrowth in the CNS concurs with MMP-expression, proteolytic activity by certain MMPs seems not to be a prerequisite for proper CNS development most likely because brain tissue contains very low amounts of extracellular matrix. Alternatively, the lack of a single MMP might have been counter-regulated by the action of other MMPs in vivo. To shed further light on this conundrum, we examined the effect of long-term constitutive expression of Timp1 in the CNS in a transgenic mouse model. The nanomolar concentrations of Timp1 in the CNS of our transgenic mice were several magnitudes above those needed for the inhibition of most MMPs and several members of the A disintegrin and metalloprotease family.47 Accordingly, the complete inhibition of gelatinolysis in areas of transgene expression demonstrated the inhibitory activity of Timp1 in the CNS of transgenic mice. Therefore, we expected that transgenic expression of Timp1 starting from embryonic stage (E 17.5) would have dramatic consequences for brain morphology and function. In the transgenic mice described here, however, we observed a physiological phenotype. The normal formation of the cerebrum in Timp1 transgenic mice is in line with only moderate prenatal MMP-inhibition since the fetal transgenic Timp1 expression was relatively low and TIMP1 is physiologically present in early stages of CNS-development.5,6,9,43,48 The murine cerebellum and parts of the dentate gyrus, in contrast, develop postnatally in a period of time when transgenic Timp1 expression reached its maximum. Alongside, in the T1F60 mouse line, in situ zymography of P10 cerebellum revealed complete lack of gelatinolytic activity in transgenic mice demonstrating sufficient inhibitory activity of Timp1. Hence, the normal morphology and function of the brain and in particular the cerebellum were unexpected findings, and we therefore investigated whether there was a counter-regulatory up-regulation of MMP-gene expression in the CNS of transgenic animals. We observed specific spatiotemporal mRNA expression patterns of MMP2, MMP9, and MMP11, as well as of all five MT-MMPs during postnatal CNS development. These findings corroborate and extend previous reports on MMP expression in the prenatal and postnatal brain.6,48 Interestingly, we observed particularly high RNA levels of the membrane type MMPs MT1-, MT2-, MT3-, and MT5-MMP in the postnatal cerebellum. Overall, there was a highly orchestrated expression of various MMP species in the course of postnatal brain development. Comparison of transgenic and wild-type mice, however, revealed no counter-regulatory up-regulation of MMP genes in postnatal CNS development. The observations in animals with a disruption of MMP genes and our present findings demonstrate that although MMPs are tightly regulated in the CNS, absence of individual MMPs, or inhibition of many MMPs by Timp1 do not lead to a constitutively pathological phenotype of the respective mutants.10,11,43,45 Together, this could be an indication that the effects of individual MMPs or—in our case—many MMPs and several members of the A disintegrin and metalloprotease family might be substituted by other proteases that are not sensitive to Timp1. The lack of up-regulated MMP expression in the Timp1 producing mice, however, renders the question in how far MMP activity is necessary for proper brain function and cerebellar development. Our present study and the findings in MMP null mice indicate that many MMP activities although helpful are not crucial for proper CNS development. Since morphological examination cannot reveal all possible alterations of the CNS, we tested the behavior and memory formation of the transgenic mice. This analysis revealed no differences between wild-type and transgenic mice of the T1F60 line that expressed the highest dose of TIMP1. Although mice with a disruption of the Timp1 gene showed impaired learning, Chaillan and co-workers10,11 demonstrated even improved learning of Timp1 transgenic mice in the olfactory tubing maze. Thus, these earlier and our own findings demonstrate that Timp1 expression in the CNS is a prerequisite for learning and increased expression at excessive levels does not impair memory formation.
Induction of TIMP1 expression in the adult brain has been found in diverse pathological conditions such as ischemia,12,19,49 cancer,49,50,51 neurodegenerative diseases, and inflammatory processes such as multiple sclerosis and its animal model EAE.17,52,53,54,55,56 However, the physiological role of TIMP1 in autoimmunity and demyelination is not yet fully understood. The expression of Timp1 in astrocytes around inflammatory lesions together with amelioration of the clinical symptoms by synthetic inhibitors in EAE suggested a protective role for this inhibitor in neuroinflammation.17,23,57 Here, we observed a partial protection from EAE in the T1F60 line. Although in wild-type mice lymphocytes were observed in the forebrain, the hindbrain, and the spinal cord as perivascular cuffs and diffuse infiltrates in the white matter, in transgenic mice the infiltrates were predominantly found around vessels and only in higher disease scores diffuse infiltration occurred. In particular the immunized healthy transgenic mice with perivascular infiltrates in the white matter of the cerebellum and the spinal cord demonstrated that expression of Timp1 in astrocytes might suppress the clinical appearance of EAE symptoms. This finding suggests that Timp1 secreted by astrocytes acts as a firewall that inhibits the migration of lymphocytes into the CNS and thereby exerts its protective effect through the course of the disease (acute inflammation, days 13 to 15 after immunization; chronic inflammation, day 40 after immunization). Despite complete inhibition of gelatinolytic activity in high expressors, few sick transgenic T1F60 mice revealed diffuse infiltration of T-cells suggesting that the inhibitory effect of Timp1 was circumvented by proteases that are not inhibited by Timp1. Moreover, these results indicate that the diffuse infiltration of CNS tissue by lymphocytes is crucial in inducing clinical symptoms while the perivascular cuffs are not necessarily accompanied by disease symptoms. Our results are consistent with observations in animals with a disruption of the Timp1 gene where both the clinical symptoms as well as the CD4+ T-cell response were not altered as compared with the control group. Remarkably, there was more myelin damage in Timp1−/− mice than in wild-type animals.26 Moreover, the increased disruption of myelin correlated with a higher lymphocyte infiltration rate and persistent microglia/macrophage activation. This model shows that the lack of Timp1, in the context of neuroinflammation, enhances lymphocyte infiltration and hence the impact on the tissue that surrounds the inflammatory lesion.
Taken together, the GFAP-TIMP1 transgenic mouse lines present an eminent example for a potential therapeutic functionality of TIMP1. Continuous high dose expression of Timp1 during development and adulthood disturbed neither tissue remodeling nor the behavior of the animals. In EAE, however, Timp1 acted as a neuroprotectant and lead to an alleviated disease course. Together, our results suggest TIMP1 as a promising endogenous therapeutic candidate for inflammatory CNS disorders.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Ginette Bortolussi and Tina Hagena.
Footnotes
Address reprint requests to Axel Pagenstecher, M.D., Department of Neuropathology, University of Marburg, Baldingerstrasse, 35043 Marburg, Germany. E-mail: pagenste@med.uni-marburg.de.
Supported by a grant of the Deutsche Forschungsgemeinschaft (to A.P.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Edwards D, Waterhouse P, Holman M, Denhardt D. A growth-responsive gene (16C8) in normal mouse fibroblasts homologous to a human collagenase inhibitor with erythroid-potentiating activity: evidence for inducible and constitutive transcripts. Nucleic Acids Res. 1986;14:8863–8878. doi: 10.1093/nar/14.22.8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leco K, Hayden L, Sharma R, Rocheleau H, Greenberg A, Edwards D. Differential regulation of TIMP-1 and TIMP-2 mRNA expression in normal and Ha-ras-transformed murine fibroblasts. Gene. 1992;117:209–217. doi: 10.1016/0378-1119(92)90731-4. [DOI] [PubMed] [Google Scholar]
- Sternlicht M, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant C, Didier-Bazès M, Hutter A, Belin M, Thomasset N. Spatiotemporal expression patterns of metalloproteinases and their inhibitors in the postnatal developing rat cerebellum. J Neurosci. 1999;19:4994–5004. doi: 10.1523/JNEUROSCI.19-12-04994.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich R, Gerhauser I, Seeliger F, Baumgärtner W, Alldinger S. Matrix metalloproteinases and their inhibitors in the developing mouse brain and spinal cord: a reverse transcription quantitative polymerase chain reaction study. Dev Neurosci. 2005;27:408–418. doi: 10.1159/000088455. [DOI] [PubMed] [Google Scholar]
- Fager N, Jaworski D. Differential spatial distribution and temporal regulation of tissue inhibitor of metalloproteinase mRNA expression during rat central nervous system development. Mech Dev. 2000;98:105–109. doi: 10.1016/s0925-4773(00)00437-8. [DOI] [PubMed] [Google Scholar]
- Rivera S, Tremblay E, Timsit S, Canals O, Ben-Ari Y, Khrestchatisky M. Tissue inhibitor of metalloproteinases-1 (TIMP-1) is differentially induced in neurons and astrocytes after seizures: evidence for developmental, immediate early gene, and lesion response. J Neurosci. 1997;17:4223–4235. doi: 10.1523/JNEUROSCI.17-11-04223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine-Aizawa Y, Hama E, Watanabe K, Tsubuki S, Kanai-Azuma M, Kanai Y, Arai H, Aizawa H, Iwata N, Saido T. Matrix metalloproteinase (MMP) system in brain: identification and characterization of brain-specific MMP highly expressed in cerebellum. Eur J Neurosci. 2001;13:935–948. doi: 10.1046/j.0953-816x.2001.01462.x. [DOI] [PubMed] [Google Scholar]
- Ayoub A, Cai T, Kaplan R, Luo J. Developmental expression of matrix metalloproteinases 2 and 9 and their potential role in the histogenesis of the cerebellar cortex. J Comp Neurol. 2005;481:403–415. doi: 10.1002/cne.20375. [DOI] [PubMed] [Google Scholar]
- Jourquin J, Tremblay E, Bernard A, Charton G, Chaillan F, Marchetti E, Roman F, Soloway P, Dive V, Yiotakis A, Khrestchatisky M, Rivera S. Tissue inhibitor of metalloproteinases-1 (TIMP-1) modulates neuronal death, axonal plasticity, and learning and memory. Eur J Neurosci. 2005;22:2569–2578. doi: 10.1111/j.1460-9568.2005.04426.x. [DOI] [PubMed] [Google Scholar]
- Chaillan F, Rivera S, Marchetti E, Jourquin J, Werb Z, Soloway P, Khrestchatisky M, Roman F. Involvement of tissue inhibition of metalloproteinases-1 in learning and memory in mice. Behav Brain Res. 2006;173:191–198. doi: 10.1016/j.bbr.2006.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong V, Power C, Forsyth P, Edwards D. Metalloproteinases in biology and pathology of the nervous system. Nat Rev Neurosci. 2001;2:502–511. doi: 10.1038/35081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber C, Hocking J, Yong V, Stange C, McFarlane S. Metalloproteases and guidance of retinal axons in the developing visual system. J Neurosci. 2002;22:8091–8100. doi: 10.1523/JNEUROSCI.22-18-08091.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman R, Williams A. Neurite penetration into collagen gels requires Ca2+-dependent metalloproteinase activity. Dev Neurosci. 1989;11:41–51. doi: 10.1159/000111884. [DOI] [PubMed] [Google Scholar]
- Sheffield J, Krasnopolsky V, Dehlinger E. Inhibition of retinal growth cone activity by specific metalloproteinase inhibitors in vitro. Dev Dyn. 1994;200:79–88. doi: 10.1002/aja.1002000108. [DOI] [PubMed] [Google Scholar]
- Nordstrom L, Lochner J, Yeung W, Ciment G. The metalloproteinase stromelysin-1 (transin) mediates PC12 cell growth cone invasiveness through basal laminae. Mol Cell Neurosci. 1995;6:56–68. doi: 10.1006/mcne.1995.1006. [DOI] [PubMed] [Google Scholar]
- Pagenstecher A, Stalder A, Kincaid C, Shapiro S, Campbell I. Differential expression of matrix metalloproteinase and tissue inhibitor of matrix metalloproteinase genes in the mouse central nervous system in normal and inflammatory states. Am J Pathol. 1998;152:729–741. [PMC free article] [PubMed] [Google Scholar]
- Pagenstecher A, Wussler E, Opdenakker G, Volk B, Campbell I. Distinct expression patterns and levels of enzymatic activity of matrix metalloproteinases and their inhibitors in primary brain tumors. J Neuropathol Exp Neurol. 2001;60:598–612. doi: 10.1093/jnen/60.6.598. [DOI] [PubMed] [Google Scholar]
- Khrestchatisky M, Jourquin J, Ogier C, Charton G, Bernard A, Tremblay E, Rivera S. Matrix metalloproteinases and their inhibitors, modulators of neuro-immune interactions and of pathophysiological processes in the nervous system. J Soc Biol. 2003;197:133–144. [PubMed] [Google Scholar]
- Gijbels K, Masure S, Carton H, Opdenakker G. Gelatinase in the cerebrospinal fluid of patients with multiple sclerosis and other inflammatory neurological disorders. J Neuroimmunol. 1992;41:29–34. doi: 10.1016/0165-5728(92)90192-n. [DOI] [PubMed] [Google Scholar]
- Yong V, Krekoski C, Forsyth P, Bell R, Edwards D. Matrix metalloproteinases and diseases of the CNS. Trends Neurosci. 1998;21:75–80. doi: 10.1016/s0166-2236(97)01169-7. [DOI] [PubMed] [Google Scholar]
- Leppert D, Ford J, Stabler G, Grygar C, Lienert C, Huber S, Miller K, Hauser S, Kappos L. Matrix metalloproteinase-9 (gelatinase B) is selectively elevated in CSF during relapses and stable phases of multiple sclerosis. Brain. 1998;121 (Pt 12):2327–2334. doi: 10.1093/brain/121.12.2327. [DOI] [PubMed] [Google Scholar]
- Gijbels K, Galardy R, Steinman L. Reversal of experimental autoimmune encephalomyelitis with a hydroxamate inhibitor of matrix metalloproteases. J Clin Invest. 1994;94:2177–2182. doi: 10.1172/JCI117578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson A, Smith T, Leonard J, Cuzner M. Suppression of experimental allergic encephalomyelitis in the Lewis rat by the matrix metalloproteinase inhibitor Ro31–9790. Inflamm Res. 1995;44:345–349. doi: 10.1007/BF01796266. [DOI] [PubMed] [Google Scholar]
- Liedtke W, Cannella B, Mazzaccaro R, Clements J, Miller K, Wucherpfennig K, Gearing A, Raine C. Effective treatment of models of multiple sclerosis by matrix metalloproteinase inhibitors. Ann Neurol. 1998;44:35–46. doi: 10.1002/ana.410440110. [DOI] [PubMed] [Google Scholar]
- Crocker S, Whitmire J, Frausto R, Chertboonmuang P, Soloway P, Whitton J, Campbell I. Persistent macrophage/microglial activation and myelin disruption after experimental autoimmune encephalomyelitis in tissue inhibitor of metalloproteinase-1-deficient mice. Am J Pathol. 2006;169:2104–2116. doi: 10.2353/ajpath.2006.060626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormán G, Kocsis-Szommer K, Spadoni C, Ferdinandy P. MMP inhibitors in cardiac diseases: an update. Recent Pat Cardiovasc Drug Discov. 2007;2:186–194. doi: 10.2174/157489007782418964. [DOI] [PubMed] [Google Scholar]
- Pagenstecher A, Lassmann S, Carson M, Kincaid C, Stalder A, Campbell I. Astrocyte-targeted expression of IL-12 induces active cellular immune responses in the central nervous system and modulates experimental allergic encephalomyelitis. J Immunol. 2000;164:4481–4492. doi: 10.4049/jimmunol.164.9.4481. [DOI] [PubMed] [Google Scholar]
- Van den Steen PE, Van Aelst I, Starckx S, Maskos K, Opdenakker G, Pagenstecher A. Matrix metalloproteinases, tissue inhibitors of MMPs and TACE in experimental cerebral malaria. Laboratory Investigation. 2006;86:873–888. doi: 10.1038/labinvest.3700454. [DOI] [PubMed] [Google Scholar]
- Rauer M, Pagenstecher A, Schulte-Mönting J, Sauder C. Upregulation of chemokine receptor gene expression in brains of Borna disease virus (BDV)-infected rats in the absence and presence of inflammation. J Neurovirol. 2002;8:168–179. doi: 10.1080/13550280290049741. [DOI] [PubMed] [Google Scholar]
- Wolfer D, Madani R, Valenti P, Lipp H. Extended analysis of path data from mutant mice using the public domain software Wintrack. Physiol Behav. 2001;73:745–753. doi: 10.1016/s0031-9384(01)00531-5. [DOI] [PubMed] [Google Scholar]
- Mohajeri M, Madani R, Saini K, Lipp H, Nitsch R, Wolfer D. The impact of genetic background on neurodegeneration and behavior in seizured mice. Genes Brain Behav. 2004;3:228–239. doi: 10.1111/j.1601-1848.2004.00073.x. [DOI] [PubMed] [Google Scholar]
- Madani R, Kozlov S, Akhmedov A, Cinelli P, Kinter J, Lipp H, Sonderegger P, Wolfer D. Impaired explorative behavior and neophobia in genetically modified mice lacking or overexpressing the extracellular serine protease inhibitor neuroserpin. Mol Cell Neurosci. 2003;23:473–494. doi: 10.1016/s1044-7431(03)00077-0. [DOI] [PubMed] [Google Scholar]
- Altman DG. Practical statistics for medical research. London: Chapman and Hall,; 1991 [Google Scholar]
- Whishaw I. Cholinergic receptor blockade in the rat impairs locale but not taxon strategies for place navigation in a swimming pool. Behav Neurosci. 1985;99:979–1005. doi: 10.1037//0735-7044.99.5.979. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Burwell R, Burchinal M. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav Neurosci. 1993;107:618–626. doi: 10.1037//0735-7044.107.4.618. [DOI] [PubMed] [Google Scholar]
- Kafkafi N, Pagis M, Lipkind D, Mayo C, Bemjamini Y, Golani I, Elmer G. Darting behavior: a quantitative movement pattern designed for discrimination and replicability in mouse locomotor behavior. Behav Brain Res. 2003;142:193–205. doi: 10.1016/s0166-4328(03)00003-2. [DOI] [PubMed] [Google Scholar]
- Vu T, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A, Ewald A, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C, Rodland K, Matrisian L, Magun B, Ciment G. NGF induction of the gene encoding the protease transin accompanies neuronal differentiation in PC12 cells. Neuron. 1989;2:1587–1596. doi: 10.1016/0896-6273(89)90047-0. [DOI] [PubMed] [Google Scholar]
- Zuo J, Neubauer D, Dyess K, Ferguson T, Muir D. Degradation of chondroitin sulfate proteoglycan enhances the neurite-promoting potential of spinal cord tissue. Exp Neurol. 1998;154:654–662. doi: 10.1006/exnr.1998.6951. [DOI] [PubMed] [Google Scholar]
- Oh L, Larsen P, Krekoski C, Edwards D, Donovan F, Werb Z, Yong V. Matrix metalloproteinase-9/gelatinase B is required for process outgrowth by oligodendrocytes. J Neurosci. 1999;19:8464–8475. doi: 10.1523/JNEUROSCI.19-19-08464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant C, Meissirel C, Mutin M, Belin M, Lund L, Thomasset N. MMP-9 deficiency affects axonal outgrowth, migration, and apoptosis in the developing cerebellum. Mol Cell Neurosci. 2003;24:395–408. doi: 10.1016/s1044-7431(03)00196-9. [DOI] [PubMed] [Google Scholar]
- Larsen P, DaSilva A, Conant K, Yong V. Myelin formation during development of the CNS is delayed in matrix metalloproteinase-9 and -12 null mice. J Neurosci. 2006;26:2207–2214. doi: 10.1523/JNEUROSCI.1880-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P, Wells J, Stallcup W, Opdenakker G, Yong V. Matrix metalloproteinase-9 facilitates remyelination in part by processing the inhibitory NG2 proteoglycan. J Neurosci. 2003;23:11127–11135. doi: 10.1523/JNEUROSCI.23-35-11127.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J, Bourguignon L, Adams C, Peyrollier K, Zhang H, Fandel T, Cun C, Werb Z, Noble-Haeusslein L. Matrix metalloproteinase-9 facilitates glial scar formation in the injured spinal cord. J Neurosci. 2008;28:13467–13477. doi: 10.1523/JNEUROSCI.2287-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troeberg L, Tanaka M, Wait R, Shi Y, Brew K, Nagase H. E. coli expression of TIMP-4 and comparative kinetic studies with TIMP-1 and TIMP-2: insights into the interactions of TIMPs and matrix metalloproteinase 2 (gelatinase A). Biochemistry. 2002;41:15025–15035. doi: 10.1021/bi026454l. [DOI] [PubMed] [Google Scholar]
- Nuttall R, Sampieri C, Pennington C, Gill S, Schultz G, Edwards D. Expression analysis of the entire MMP and TIMP gene families during mouse tissue development. FEBS Lett. 2004;563:129–134. doi: 10.1016/S0014-5793(04)00281-9. [DOI] [PubMed] [Google Scholar]
- Elezkurtaj S, Kopitz C, Baker A, Perez-Cantó A, Arlt M, Khokha R, Gansbacher B, Anton M, Brand K, Krüger A. Adenovirus-mediated overexpression of tissue inhibitor of metalloproteinases-1 in the liver: efficient protection against T-cell lymphoma and colon carcinoma metastasis. J Gene Med. 2004;6:1228–1237. doi: 10.1002/jgm.637. [DOI] [PubMed] [Google Scholar]
- Michael M, Babic B, Khokha R, Tsao M, Ho J, Pintilie M, Leco K, Chamberlain D, Shepherd F. Expression and prognostic significance of metalloproteinases and their tissue inhibitors in patients with small-cell lung cancer. J Clin Oncol. 1999;17:1802–1808. doi: 10.1200/JCO.1999.17.6.1802. [DOI] [PubMed] [Google Scholar]
- Thomas P, Khokha R, Shepherd F, Feld R, Tsao M. Differential expression of matrix metalloproteinases and their inhibitors in non-small cell lung cancer. J Pathol. 2000;190:150–156. doi: 10.1002/(SICI)1096-9896(200002)190:2<150::AID-PATH510>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Toft-Hansen H, Nuttall R, Edwards D, Owens T. Key metalloproteinases are expressed by specific cell types in experimental autoimmune encephalomyelitis. J Immunol. 2004;173:5209–5218. doi: 10.4049/jimmunol.173.8.5209. [DOI] [PubMed] [Google Scholar]
- Teesalu T, Hinkkanen A, Vaheri A. Coordinated induction of extracellular proteolysis systems during experimental autoimmune encephalomyelitis in mice. Am J Pathol. 2001;159:2227–2237. doi: 10.1016/S0002-9440(10)63073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg R, De Groot C, Montagne L, Freitag P, van der Valk P, Kappos L, Leppert D. The expression profile of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in lesions and normal appearing white matter of multiple sclerosis. Brain. 2001;124:1743–1753. doi: 10.1093/brain/124.9.1743. [DOI] [PubMed] [Google Scholar]
- Boz C, Ozmenoglu M, Velioglu S, Kilinc K, Orem A, Alioglu Z, Altunayoglu V. Matrix metalloproteinase-9 (MMP-9) and tissue inhibitor of matrix metalloproteinase (TIMP-1) in patients with relapsing-remitting multiple sclerosis treated with interferon beta. Clin Neurol Neurosurg. 2006;108:124–128. doi: 10.1016/j.clineuro.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Ichiyama T, Kajimoto M, Suenaga N, Maeba S, Matsubara T, Furukawa S. Serum levels of matrix metalloproteinase-9 and its tissue inhibitor (TIMP-1) in acute disseminated encephalomyelitis. J Neuroimmunol. 2006;172:182–186. doi: 10.1016/j.jneuroim.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Gran B, Tabibzadeh N, Martin A, Ventura E, Ware J, Zhang G, Parr J, Kennedy A, Rostami A. The protease inhibitor, Bowman-Birk Inhibitor, suppresses experimental autoimmune encephalomyelitis: a potential oral therapy for multiple sclerosis. Mult Scler. 2006;12:688–697. doi: 10.1177/1352458506070769. [DOI] [PubMed] [Google Scholar]