Abstract
In human pancreatic cancer, integral membrane proteins of tight junction claudins are abnormally regulated, making these proteins promising molecular diagnostic and therapeutic targets. However, the regulation of claudin-based tight junctions remains unknown not only in the pancreatic cancer cells but also in normal human pancreatic duct epithelial (HPDE) cells. To investigate the regulation of tight junction molecules including claudins in normal HPDE cells, we introduced the human telomerase reverse transcriptase (hTERT) gene into HPDE cells in primary culture. The hTERT-transfected HPDE (hTERT-HPDE) cells were positive for the pancreatic duct epithelial markers such as CK7, CK19, and carbonic anhydrase isozyme 2 and expressed epithelial tight junction molecules claudin-1, -4, -7 and, -18, occludin, JAM-A, ZO-1, ZO-2, and tricellulin. By treatment with fetal bovine serum or 12-O-tetradecanoylphorbol 13-acetate (TPA), the tight junction molecules were up-regulated at the transcriptional level via a protein kinase C (PKC) signal pathway. A PKC-α inhibitor, Gö6976, prevented up-regulation of claudin-4 by TPA. Furthermore, a PKC-δ inhibitor, rottlerin, prevented up-regulation of claudin-7, occludin, ZO-1, and ZO-2 by TPA. By GeneChip analysis, up-regulation of the transcription factor ELF3 was observed in both fetal bovine serum- and TPA-treated cells. Treatment with small interfering RNAs of ELF3 prevented up-regulation of claudin-7 by TPA. These data suggest that tight junctions of normal HPDE cells were at least in part regulated via a PKC signal pathway by transcriptional control.
Because pancreatic cancer is the fourth leading cause of cancer death in the United States, and its death rate has not decreased over the past few decades,1 useful new biomarkers for diagnosis and new molecular targets for therapeutic approaches must be developed to improve the poor conventional outcome of the disease. Recently, it was revealed that in several human cancers, some tight junction protein claudins were abnormally regulated and are promising molecular targets for diagnosis and therapy.2,3 Tight junctions are the most apical components of intercellular junctional complexes in epithelial and endothelial cells. They separate the apical and basolateral cell surface domains maintaining cell polarity (termed the “fence” function) and selectively control solute and water flow through the paracellular space (termed the “barrier” function).4,5,6,7 Recent evidence suggests that they also participate in signal transduction processes that regulate cell proliferation, gene expression, differentiation, and morphogenesis.3,8,9 Tight junctions are formed by not only the integral membrane proteins claudins, occludin, and JAMs but also peripheral membrane proteins, including the scaffold PDZ-expressing proteins, ZO-1, ZO-2, ZO-3, MUPP1, and MAGI-1, the cell polarity molecules ASIP/PAR-3, PAR-6, and PATJ, and the non-PDZ-expressing proteins, cingulin, symplekin, atypical protein kinase C, Rab3b, Rab13, PTEN, and 7H6 antigen.10,11 More recently, tricellulin was identified at tricellular contacts where there are three epithelial cells and was shown to have a barrier function.12
The claudin family, which consists of at least 24 members, is solely responsible for forming tight junction strands and has four transmembrane domains and two extracellular loops.10 The first extracellular loop is the coreceptor of hepatitis C virus13,14 and influences the paracellular charge selectivity,15 and the second extracellular loop is the receptor of Clostridium perfringens enterotoxin (CPE).16 In pancreatic cancer, claudin-4 and -18 are highly expressed17,18 and are diagnostic or therapeutic targets of monoclonal antibodies against their extracellular loops.19,20 In addition, because claudin-4 is also a high-affinity receptor of CPE,21 full-length CPE with a direct cytotoxic effect or the C-terminal receptor binding domain of CPE without a cytotoxic effect are used for selective treatment or drug delivery against claudin-4-expressing tumors.17,22,23 However, the regulatory mechanisms of claudin-based tight junctions remain unknown even in normal human pancreatic duct epithelial (HPDE) cells. Thus, analyses of the regulation of tight junction molecules, including claudin-4, in normal HPDE cells in vitro are essential to develop safer and more effective diagnostic and therapeutic methods targeting claudins in pancreatic cancer.
Protein kinase C (PKC) is a family of serine-threonine kinases known to regulate epithelial barrier function.24,25,26 PKC has been shown to induce both assembly and disassembly of tight junctions, depending on the cell type and conditions of activation.24,27 The activation of PKC causes an increase in permeability in the renal epithelial cell lines LLC-PK1 and MDCK,28,29 whereas it causes a decrease in permeability in the human colon carcinoma cell line HT29.30 Bryostatin enhances tight junction barrier function in T84 through a PKC signaling pathway.31 PKC seems to regulate the subcellular localization, phosphorylation states, and transcription of several tight junction-associated proteins,32 although the isozyme specificity has not been clearly elucidated. At least 11 different isozymes of PKC are known. These can be subdivided in three classes according to their responsiveness to activators.33 The classic or conventional isozymes (α, βI, βII, and γ) are both Ca2+- and diacylglycerol-dependent. The novel isozymes (δ, ε, θ, η, and μ) are Ca2+-independent but diacylglycerol-dependent. The atypical isozymes (ι/λ and ζ) are neither Ca2+- nor DAG-dependent. In the human intestinal epithelial cell lines HT-29 and Caco-2, stimulation with Toll-like receptor 2 ligands leads to activation of the specific PKC isoforms PKC-α and PKC-δ and enhances barrier function through translocation of ZO-1 on activation.34 Furthermore, activation of PKC by 12-O-tetradecanoylophorbol-13-acetate (TPA) causes increases in transcription of occludin, ZO-1, and claudin-1 in T84 cells and melanoma cells.35,36 Although activation of PKC exerts its effect directly at the transcriptional level, the responsible transcription factors related to PKC activation remain unknown in regulation of tight junctions.
The introduction of the catalytic subunit of human telomerase, human telomerase reverse transcriptase (hTERT), into human somatic cells such as fibroblasts and retinal pigment epithelial cells typically extends their lifespan without altering the growth requirements, disturbance of the cell-cycle checkpoints, tumorigenicity, or chromosomal abnormality.37,38,39,40 Lee et al41 reported that immortalized cells with hTERT of normal human cells derived from the adult pancreas were diploid, displayed normal growth controls, and produced wild-type p16INK4a, K-Ras, and p53. We also established hTERT-transfected human nasal epithelial cells with an extended life span.42
In the present study, we introduced the hTERT gene into HPDE cells in primary culture. The properties of the hTERT-HPDE cells, including pancreatic duct epithelial (PDE) markers CK7, CK19, and carbonic anhydrase isozyme 2 (CA-II) and epithelial tight junction molecules claudin-1, -4, -7 and -18, occludin, JAM-A, ZO-1, ZO-2 and tricellulin, were similar to those of HPDE cells in vivo. In hTERT-HPDE cells, the up-regulation of tight junction molecules and the barrier function were regulated via a PKC signal pathway. The up-regulation of tight junction molecules by TPA was in part controlled via the specific PKC isoforms PKC-α and PKC-δ. By GeneChip analysis and small interfering RNAs (siRNAs), the transcription factor ELF3 was found to be closely associated with the expression of claudin-7 in hTERT-HPDE cells.
Materials and Methods
Inhibitors and PKC Activator
A pan-PKC inhibitor (GF109203X), mitogen-activated protein kinase (MAPK) inhibitor (PD98059), p38 MAPK inhibitor (SB203580), phosphatidylinositol 3-kinase inhibitor (LY294002), PKC-α inhibitor (Gö6976), PKC-δ inhibitor (rottlerin), PKC-θ inhibitor (myristoylated PKC-θ pseudosubstrate peptide inhibitor), and PKC-ε inhibitor (PKC-ε translocation inhibitor peptide) were purchased from Calbiochem-Novabiochem Corporation (San Diego, CA). A nuclear factor-κB inhibitor (IMD-0354) and PKC activator, 12-O-tetradecanoylphorbol 13-acetate, were purchased from Sigma-Aldrich (St. Louis, MO).
Human Pancreatic Tissues
Human pancreatic tissues were obtained from patients with pancreatic or biliary tract diseases who underwent pancreatic resection in the Sapporo Medical University Hospital. Informed consent was obtained from all patients, and the study was approved by the ethics committee of Sapporo Medical University. For the study, approximately 500 mg of tissue was sampled from the macroscopically nonlesion parts of pancreatic specimens, avoiding the main pancreatic ducts, and it was confirmed microscopically by pathologists that there were no malignant or premalignant lesions according to H&E staining. The patients’ information is summarized in Table 1. It was noted that the diameters of the main pancreatic ducts near sample tissues determined by preoperative multidetector-row computed tomography were within the normal range (<3 mm) in almost all patients.
Table 1.
Patients’ Information
| No. | Age (yr) | Sex | Histopathological diagnosis | Operative procedure | MPD (mm)* | Passage |
|---|---|---|---|---|---|---|
| 1 | 78 | F | Cholangiocarcinoma | Pylorus preserving PD | 1.1 | P4 |
| 2 | 67 | F | IPMN | Spleen preserving DP | 2.5 | P3 |
| 3 | 79 | F | Cholangiocarcinoma | Pylorus preserving PD | 1.5 | P3 |
| 4 | 84 | F | Pancreatic adenocarcinoma | DP | 1.9 | P2 |
| 5 | 72 | M | IPMN | Duodenum preserving subTP | 2.2 | P3 |
| 6 | 53 | M | Well differentiated endocrine tumor | Central pancreatectomy | 1.1 | P3 |
| 7 | 79 | M | Pancreatic adenocarcinoma | DP | 1.5 | P2 |
| 8 | 80 | M | Pancreatic adenocarcinoma | DP | 1.5 | P3 |
| 9 | 74 | F | Cholangiocarcinoma | Pylorus preserving PD | 3.6 | P2 |
| 10 | 64 | F | Adenocarcinoma of duodenum | Subtotal stomach preserving PD | 3.4 | P2 |
| 11 | 73 | M | Cholangiocarcinoma | rHPD | 2.3 | P2 |
| 12 | 77 | M | Cholangiocarcinoma | Pylorus preserving PD | 1.2 | P2 |
F, female; M, male; IPMN, intraductal papillary mucinous neoplasm; PD, pancreaticoduodenectomy; DP, distal pancreatectomy; subTP, subtotal pancreatectomy; rHPD, right hepatic lobectomy and pylorus preserving pancreaticoduodenectomy.
Diameters of the main pancreatic ducts (MPD) near sample tissues determined by preoperative multidetector computed tomography.
Isolation and Culture of HPDE Cells
Human pancreatic tissues were minced into pieces 2 to 3 mm3 in volume and washed with PBS containing 100 U/ml penicillin and 100 μg/ml streptomycin (Lonza Walkersville, Walkersville, MD) three times. These minced tissues were suspended in 10 ml of Hanks’ balanced salt solution with 0.5 μg/ml DNase I and 0.04 mg/ml Liberase Blendzyme 3 (Roche, Basel, Switzerland) and then incubated with bubbling of mixed O2 gas containing 5.2% CO2 at 37°C for 10 minutes. The dissociated tissues were subsequently filtered with 300-μm mesh followed by filtration with 70-μm mesh (Cell Strainer, BD Biosciences, San Jose, CA). After centrifugation at 1000 × g for 4 minutes, isolated cells were cultured in bronchial epithelial basal medium (BEBM, Lonza Walkersville) containing 10% fetal bovine serum (FBS) (CCB, Nichirei Bioscience, Tokyo, Japan) and supplemented with BEGM SingleQuots (Lonza Walkersville; including 0.4% bovine pituitary extract, 0.1% insulin, 0.1% hydrocortisone, 0.1% gentamicin, amphotericin-B [GA-1000], 0.1% retinoic acid, 0.1% transferrin, 0.1% triiodothyronine, 0.1% epinephrine, and 0.1% human epidermal growth factor), 100 U/ml penicillin and 100 μg/ml streptomycin on 60-mm culture dishes (Corning Life Sciences, Acton, MA), coated with rat tail collagen (500 μg of dried tendon/ml of 0.1% acetic acid). Following the above protocol, tissue dissociation and cell isolation were repeated for the same sample a maximum of seven times. The cells were placed in a humidified 5% CO2:95% air incubator at 37°C.
The retroviral vector BABE-hygro-hTERT (kindly provided by Dr. Robert Weinberg) was used. The viral supernatant was produced from an ecotropic packaging cell line by transfection of plasmid DNA as reported previously.43 The packaging cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% FBS and supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. At 24 hours after plating on 60-mm dishes, HPDE cells in primary culture were exposed overnight to the viral supernatant containing the retrovirus. After being washed with serum-free BEBM medium, the hTERT-transfected HPDE (hTERT-HPDE) cells were cultured in serum-free BEBM medium supplemented with the above-mentioned factors and 2.5 μg/ml amphotericin-B. The hTERT-HPDE cells became confluent on the 60-mm culture dishes in 2 to 3 weeks, and the first passage was done using 0.05% trypsin-EDTA (Sigma-Aldrich) in 60-mm culture dishes. At day 5 after the first passage, the second passage was done in the same manner in 60- or 35-mm culture dishes and the second-passaged cells were used for the experiments at days 5 to 7 after plating.
The hTERT-HPDE cells were treated with 1, 5, and 10% FBS or 1, 10, and 100 nmol/L TPA for 24 hours. Some cells were pretreated with 10 μmol/L GF109203X, 10 μmol/L PD98059, 10 μmol/L SB203580, 10 μmol/L LY294002, 5 μmol/L IMD-0354, 5 μmol/L Gö6976, 1 μmol/L rottlerin, 5 μmol/L myristoylated PKC-θ pseudosubstrate peptide inhibitor, or 10 μmol/L PKC-ε translocation inhibitor peptide for 1 hour before treatment with 10% FBS or 100 nmol/L TPA.
Pancreatic Cancer Cell Line
The human pancreatic cancer cell line HPAC was purchased from American Type Culture Collection (Manassas, VA) and maintained with Dulbecco’s modified Eagle’s medium containing 10% FBS and supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin on 60-mm culture dishes coated with rat tail collagen.
RNA Isolation, RT-PCR Analysis, and Real-Time PCR Analysis
Total RNA was extracted and purified using TRIzol (Invitrogen, Carlsbad, CA). One microgram of total RNA was reverse-transcribed into cDNA using a mixture of oligo(dT) and SuperScript II reverse transcriptase according to the manufacturer’s recommendations (Invitrogen). Synthesis of each cDNA was performed in a total volume of 20 μl for 50 minutes at 42°C and terminated by incubation for 15 minutes at 70°C. PCR was performed in a 20-μl total mixture containing 100 pmol/L primer pairs, 1.0 μl of the 20-μl total RT product, PCR buffer, dNTPs, and TaqDNA polymerase according to the manufacturer’s recommendations (Takara, Kyoto, Japan). Amplifications were for 25 to 35 cycles, depending on the PCR primer pair with cycle times of 15 seconds at 96°C, 30 seconds at 55°C, and 60 seconds at 72°C. Final elongation time was 7 minutes at 72°C. Seven microliters of the total 20-μl PCR product was analyzed by 1% agarose gel electrophoresis with ethidium bromide staining and standardized using a GeneRuler 100-bp DNA ladder (Fermentas, Burlington, ON, Canada). The PCR primers used to detect CA-II, CK7, CK19, amylase, glucagon, insulin, somatostatin, claudin-1, -2, -3, -4, -5, -6, -7, -8, -9, and -18, occludin, JAM-A, ZO-1, ZO-2, tricellulin, ELF1, ELF2, ELF3, ELF4, ELF5 and glucose-3-phosphate dehydrogenase (G3PDH) are indicated in Table 2.
Table 2.
Primers of RT-PCR
| Gene | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|
| CA-II | 5′-AAGGAACCCATCAGCGTCAG-3′ | 5′-AAAGCACCAACCAGCCACAG-3′ | 357 |
| CK7 | 5′-TGAGATCGACAACATCAAGAAC-3′ | 5′-CGGATGGAATAAGCCTTCAG-3′ | 414 |
| CK19 | 5′-GCCACTACTACACGACCATCC-3′ | 5′-GAATCCACCTCCACACTGACC-3′ | 334 |
| Amylase | 5′-GACTTTCCAGCAGTCCCATA-3′ | 5′-GTTTACTTCCTGCAGGGAAC-3′ | 310 |
| Glucagon | 5′-ATCTGGACTCCAGGCGTGCC-3′ | 5′-AGCAATGAATTCCTTGGCAG-3′ | 179 |
| Insulin | 5′-CAGCCGCAGCCTTTGTGAAC-3′ | 5′-CAGGCTGCCTGCACCAGGG-3′ | 170 |
| Somatostatin | 5′-CGTCAGTTTCTGCAGAAGTCC-3′ | 5′-CCATAGCCGGGTTTGAGTTA-3′ | 196 |
| Claudin-1 | 5′-AACGCGGGGCTGCAGCTGTTG-3′ | 5′-GGATAGGGCCTTGGTGTTGGGT-3′ | 593 |
| Claudin-2 | 5′-GCTTCTACTGAGAGGTCTG-3′ | 5′-TTCTTCACACATACCCTG-3′ | 718 |
| Claudin-3 | 5′-TGCTGTTCCTTCTCGCCGCC-3′ | 5′-CTTAGACGTAGTCCTTGCGG-3′ | 300 |
| Claudin-4 | 5′-AGCCTTCCAGGTCCTCAACT-3′ | 5′-AGCAGCGAGTCGTACACCTT-3′ | 249 |
| Claudin-5 | 5′-GACTCGGTGCTGGCTCTGAG-3′ | 5′-CGTAGTTCTTCTTGTCGTAG-3′ | 451 |
| Claudin-6 | 5′-TGAGGCCCAAAAGCGGGAGC-3′ | 5′-CGTAATTCTTGGTAGGGTAC-3′ | 200 |
| Claudin-7 | 5′-AGGCATAATTTTCATCGTGG-3′ | 5′-GAGTTGGACTTAGGGTAAGAGCG-3′ | 252 |
| Claudin-8 | 5′-TCATCCCTGTGAGCTGGGTT-3′ | 5′-TGGAGTAGACGCTCGGTGAC-3′ | 261 |
| Claudin-9 | 5′-AGGCCCGTATCGTGCTCACC-3′ | 5′-ACGTAGTCCCTCTTGTCCAG-3′ | 310 |
| Claudin-18 | 5′-TTCCATCCCAGTACCAAAGC-3′ | 5′-CCGTTCTTTCCCCAGACATA-3′ | 226 |
| Occludin | 5′-TCAGGGAATATCCACCTATCACTTCAG-3′ | 5′-CATCAGCAGCAGCCATGTACTCTTCAC-3′ | 189 |
| JAM-A | 5′-GGTCAAGGTCAAGCTCAT-3′ | 5′-CTGAGTAAGGCAAATGCAG-3′ | 582 |
| ZO-1 | 5′-CGGTCCTCTGAGCCTGTAAG-3′ | 5′-GGATCTACATGCGACGACAA-3′ | 371 |
| ZO-2 | 5′-GCCAAAACCCAGAACAAAGA-3′ | 5′-ACTGCTCTCTCCCACCTCCT-3′ | 213 |
| Tricellulin | 5′-AGGCAGCTCGGAGACATAGA-3′ | 5′-TCACAGGGTATTTTGCCACA-3′ | 240 |
| ELF1 | 5′-GCTCTTCCGGACTGTTCATG-3′ | 5′-CCACACTGACGGTTCCATTG-3′ | 425 |
| ELF2 | 5′-ACTGCATCTGTGTCAGCAAC-3′ | 5′-CAGGCTGCATGGTGATTTTG-3′ | 244 |
| ELF3 | 5′-CACTGATGGCAAGCTCTTC-3′ | 5′-GGAGCGCAGGAACTTGAAG-3′ | 262 |
| ELF4 | 5′-CGTTCACAATGGCATCATAAC-3′ | 5′-CTGCCTTTGCCATCCTTTG-3′ | 453 |
| ELF5 | 5′-CAAGACTGTCACAGTCATAG-3′ | 5′-GTCAACCCGCTCCAAAATTC-3′ | 261 |
| GATA-1 | 5′-TCAATTCAGCAGCCTATTCC-3′ | 5′-TTCGAGTCTGAATACCATCC-3′ | 377 |
| GATA-2 | 5′-TGTTGTGCAAATTGTCAGACG-3′ | 5′-CATAGGTGCCATGTGTCCAGC-3′ | 279 |
| GATA-3 | 5′-AAGTGCATGACTCACTGGAGG-3′ | 5′-TAGGCTTCATGATACTGCTCC-3′ | 251 |
| GATA-4 | 5′-CTGGCCTGTCATCTCACTACG-3′ | 5′-GGTCCGTGCAGGAATTTGAGG-3′ | 263 |
| GATA-5 | 5′-TCGCCAGCACTGACAGCTCAG-3′ | 5′-TGGTCTGTTCCAGGCTGTTCC-3′ | 290 |
| GATA-6 | 5′-TTCTAACTCAGATGATTGCAGC-3′ | 5′-GCTGCACAAAAGCAGACACG-3′ | 300 |
| G3PDH | 5′-ACCACAGTCCATGCCATCAC-3′ | 5′-TCCACCACCCTGTTGCTGTA-3′ | 452 |
Real-time PCR detection was performed using a TaqMan Gene Expression Assay kit with a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). The amount of 18S ribosomal RNA (rRNA) (Hs99999901) mRNA in each sample was used to standardize the quantity of the following mRNAs: claudin-1 (Hs00221623), claudin-4 (Hs00533616), claudin-7 (Hs00154575), occludin (Hs00170162), JAM-A (F11 receptor, Hs00170991), ZO-1 (Hs00543824), and ZO-2 (Hs00178081). The relative mRNA expression levels between the control and treated samples were calculated by the difference of the threshold cycle (comparative CT [ΔΔCT] method) and presented as the average of triplicate experiments with a 95% confidence interval.
Western Blot Analysis
For Western blotting of total cell lysates, the dishes were washed with PBS and 250 or 120 μl of sample buffer (1 mmol/L NaHCO3 and 2 mmol/L phenylmethylsulfonyl fluoride) was added to 60- or 35-mm culture dishes, respectively. The cells were scraped and collected in microcentrifuge tubes and then sonicated for 10 seconds. The protein concentrations of samples were determined using a BCA Protein Assay Reagent Kit (Pierce Chemical, Rockford, IL). Aliquots of 15 μg of protein/lane for each sample were separated by electrophoresis in 4/20% SDS-polyacrylamide gels (Cosmo Bio Co., Tokyo, Japan). After electrophoretic transfer to nitrocellulose membranes (Immobilon, Millipore, Billerica, MA), the membranes were saturated with blocking buffer (Tris-buffered saline with 0.1% Tween 20 and 4% skim milk) for 30 minutes at room temperature and incubated with anti-claudin-1, anti-claudin-4, anti-claudin-7, anti-occludin, anti-JAM-A, anti-ZO-1, anti-ZO-2, anti-CK19, and anti-actin antibodies diluted 1:1000 in PBS, an anti-phospho-pan-PKC antibody diluted 1:1000 in Tris-buffered saline, and anti-CK7 and anti-CA-II antibodies diluted 1:2000 in PBS (Table 3) for 1 hour at room temperature. The membranes were incubated with horseradish peroxidase-conjugated anti-rabbit or mouse IgG (Dako A/S, Copenhagen, Denmark) for 1 hour at room temperature. The immunoreactive bands were detected using an ECL Western blotting analysis system (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Table 3.
Antibodies
| Antibody | Type | Dilution
|
Company | |
|---|---|---|---|---|
| IC | WB | |||
| CK7 | mAb (OV-TL 12/13) | 1:200 | 1:2000 | Dako A/S (Copenhagen, Denmark) |
| CK19 | mAb (RCK108) | 1:200 | 1:1000 | Dako A/S (Copenhagen, Denmark) |
| CA-II | pAb | 1:2000 | Abcam (Cambridge, UK) | |
| Claudin-1 | pAb | 1:100 | 1:1000 | Zymed Laboratories (San Francisco, CA) |
| Claudin-4 | pAb | 1:1000 | Zymed Laboratories (San Francisco, CA) | |
| Claudin-4 | mAb (3E2C1) | 1:100 | 1:1000 | Zymed Laboratories (San Francisco, CA) |
| Claudin-7 | pAb | 1:100 | 1:1000 | Zymed Laboratories (San Francisco, CA) |
| Occludin | mAb (OC-3F10) | 1:100 | Zymed Laboratories (San Francisco, CA) | |
| Occludin | pAb | 1:100 | 1:1000 | Zymed Laboratories (San Francisco, CA) |
| JAM-A | pAb | 1:1000 | Zymed Laboratories (San Francisco, CA) | |
| ZO-1 | pAb | 1:100 | 1:1000 | Zymed Laboratories (San Francisco, CA) |
| ZO-2 | pAb | 1:1000 | Zymed Laboratories (San Francisco, CA) | |
| E-cadherin | mAb (36/E-cadherin) | 1:200 | BD Biosciences (San Jose, CA) | |
| Phospho-pan-PKC | pAb | 1:1000 | Cell Signaling Technology (Danvers, MA) | |
| Actin | pAb | 1:1000 | Sigma-Aldrich (St. Louis, MO) | |
IC, immunocytochemistry; WB, Western blotting; mAb, mouse monoclonal antibody; pAb, rabbit polyclonal antibody.
Immunocytochemistry
The cells grown on 35-mm glass-based dishes (Asahi Glass Co., Ltd, Tokyo, Japan) were fixed with an ethanol and acetone mixture (1:1) at −20°C for 10 minutes. After the cells were rinsed with PBS, they were incubated overnight at 4°C with anti-CK7, anti-CK19, and anti-E-cadherin antibodies diluted 1:200 in PBS, and anti-claudin-1, anti-claudin-4, anti-claudin-7, anti-occludin, and anti-ZO-1 antibodies diluted 1:100 in PBS (Table 3). Alexa Fluor 488 (green)-conjugated anti-rabbit IgG or Alexia Fluor 592 (red)-conjugated anti-mouse IgG (Invitrogen) diluted 1:200 in PBS was used as a secondary antibody for 1 hour at room temperature. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (Sigma-Aldrich). The specimens were examined using an epifluorescence microscope (Olympus, Tokyo, Japan). Phase-contrast photomicrographs were taken with an Axiovert 200 inverted microscope (Carl Zeiss Co., Ltd., Göttingen, Germany).
Freeze-Fracture Analysis
The cells grown on 60-mm dishes were centrifuged into pellets and then immersed in 40% glycerin solution after fixation in 2.5% glutaraldehyde/0.1 mol/L PBS (pH 7.3). The specimens mounted on a copper stage were frozen in liquid nitrogen, fractured at −150°C to −160°C and replicated by platinum/carbon from an electron beam gun positioned at a 45° angle followed by carbon applied from overhead in a JFD-7000 freeze-fracture device (JEOL Ltd., Tokyo, Japan). After replicas were thawed, they were floated on filtered 10% sodium hypochlorite solution for 30 minutes in Teflon dishes. The replicas were then washed in distilled water for 30 minutes, mounted on copper grids, and examined at an acceleration voltage of 100 kV with a JEOL-1200EX transmission electron microscope (JEOL Ltd.).
Measurement of Transepithelial Electrical Resistance
The cells were cultured to confluence on inner chambers of a 12-mm Transwell with 0.4-μm pore-sized filters (Corning Life Sciences). TER was measured using an EVOM voltmeter with an EndOhm-12 chamber (World Precision Instruments, Sarasota, FL) on a heating plate (Fine, Tokyo, Japan) adjusted to 37°C. Values are expressed in standard units of ohms per square centimeter and presented as the mean ± SD of triplicate experiments. For calculation, the resistance of blank filters was subtracted from that of filters covered with cells.
Measurement of Permeability
To determine the paracellular flux, the cells were cultured on a 12-mm Transwell with 0.4-μm pore-sized filters, and then fluorescein isothiocyanate-labeled dextran (molecular mass 7 kDa)-containing medium was added to the inner chamber. Samples were collected from the outer chamber at 15, 30, 60, and 90 minutes and were measured with a Wallac 1420 multilabeled counter (PerkinElmer Life and Analytical Sciences, Waltham, MA).
GeneChip Analysis
Total RNA was extracted from samples and evaluated for quality and integrity using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Gene transcripts expressed by the cells were analyzed and compared using OpArray Human v4.0 (Operon Biotechnologies Inc., Tokyo, Japan). The chips were scanned in a GenePix 4000B (MDS Analytical Technologies, Toronto, ON, Canada) to detect hybridization signals, and the data were analyzed using Array-Pro Analyzer v4.5 (Media Cybernetics, Inc., Bethesda, MD) and Microarray Data Analysis Tool v3.0 (Filgen, Inc., Nagoya, Japan).
siRNA Experiment
For knockdown of human ELF3 and GATA-3, Stealth Select RNAi against human ELF3 and GATA-3 was synthesized by Invitrogen and the sequences for sense and antisense strands were as follows: ELF3-siRNA1, sense 5′-GCGCCAUUGACUUCUCACGAUGUGA-3′ and antisense 5′-UCACAUCGUGAGAAGUCAAUGGCGC-3′; ELF3-siRNA2, sense 5′-GAGGGUACAGAGAAGGCCAGCUGGU-3′ and antisense 5′-ACCAGCUGGCCUUCUCUGUACCCUC-3′; GATA-3-siRNA1, sense 5′-UUGGCAUUCCUCCUCCAGAGUGUGG-3′ and antisense 5′-CCACACUCUGGAGGAGGAAUGCCAA-3′; GATA-3-siRNA2, sense 5′-AUAUUGUGAAGCUUGUAGUAGAGCC-3′ and antisense 5′-GGCUCUACUACAAGCUUCACAAUAU-3′; and GATA-3-siRNA3, sense 5′-UAGGGAUCCAUGAAGCAGAGGCGGG-3′ and antisense 5′-CCCGCCUCUGCUUCAUGGAUCCCUA-3′.
The cells at 24 hours after plating were transfected with 100 nmol/L siRNA using Lipofectamine RNAiMAX Reagent (Invitrogen). The transfected hTERT-HPDE cells were treated with 10 nmol/L TPA for 12 hours at 48 hours after transfection, and the HPAC cells were harvested at 48 hours after transfection.
Results
Characterization of hTERT-HPDE Cells
To establish a stable, reproducible culture method for HPDE cells, we introduced the hTERT gene into HPDE cells in primary culture. In the present study, hTERT-HPDE cells were maintained in serum-free conditioned medium and could be passaged four times at the maximum (Table 1). During these experiments, we used second passaged cells with hTERT transfection.
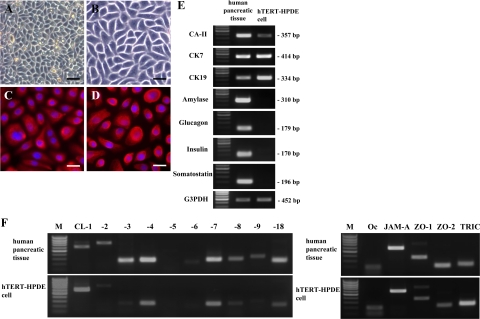
The cells had a small cobblestone appearance in phase-contrast images (Figure 1, A and B) and expressed the PDE markers CK7 and CK19 in immunocytochemical analysis (Figure 1, C and D). In RT-PCR, hTERT-HPDE cells expressed mRNAs of the PDE markers CA-II, CK7, and CK19, whereas mRNAs of the pancreatic exocrine marker amylase and the endocrine markers glucagon, insulin, and somatostatin were not detected (Figure 1E).
Figure 1.
Phase-contrast images (A and B) and immunocytochemical analysis for CK7 (C) and CK19 (D) in hTERT-HPDE cells cultured with serum-free conditioned medium. hTERT-HPDE cells show a small cobblestone appearance and are stained with CK7 and CK19. Scale bars: 80 μm (A); 40 μm (B); 20 μm (C and D). E: RT-PCR for CA-II, CK7, CK19, amylase, glucagon, insulin, and somatostatin in hTERT-HPDE cells. hTERT-HPDE cells express mRNAs of CA-II, CK7, and CK19. Human pancreatic tissues were used as positive controls. F: RT-PCR for tight junction molecules in hTERT-HPDE cells. mRNAs of claudin-1, -4, -7, and -18, occludin, JAM-A, ZO-1, ZO-2, and tricellulin are detected in hTERT-HPDE cells. Human pancreatic tissues were used as positive controls. CL, claudin; Oc, occludin; TRIC, tricellulin; M, 100-bp ladder DNA marker.
Previous reports show that mRNAs of claudin-1, -2, -3, -4, -5, -7, -8, -9, -10, -11, and -12 can be detected by real-time PCR in normal human pancreatic tissues.44 and that claudin-1, -3, -4, and -7 were observed in acini by immunostaining, as are claudin-1, -2, -3, -4, and -7 in ducts, and claudin-3 and -7 in endocrine.45 We investigated the expression pattern of tight junction molecules in hTERT-HPDE cells compared with human pancreatic tissues. In RT-PCR of human pancreatic tissues, mRNAs of claudin-1, -2, -3, -4, -7, -8, -9, and -18, occludin, JAM-A, ZO-1, ZO-2, and tricellulin were detected (Figure 1F). In hTERT-HPDE cells, mRNAs of claudin-1, -4, -7, and -18, occludin, JAM-A, ZO-1, ZO-2, and tricellulin were detected among tight junction molecules expressed in human pancreatic tissues (Figure 1F).
Induction of Tight Junction Proteins at the Transcriptional Level by FBS in hTERT-HPDE Cells
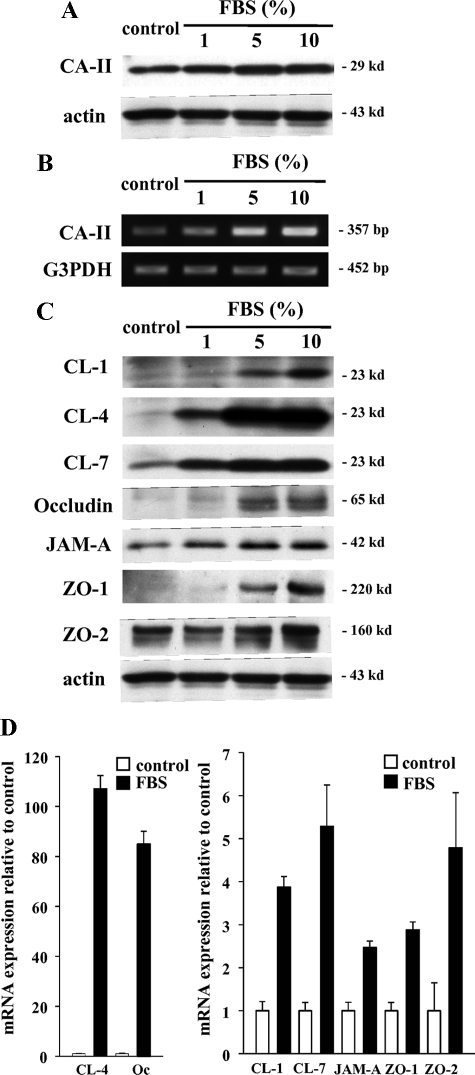
In this culture system, hTERT-HPDE cells in serum-free conditioned medium had growth potential and a long lifespan. Treatment with FBS induced an increase in protein and mRNA of CA-II dependent on the FBS concentration (Figure 2, A and B), whereas proteins of CK7 and CK19 were stably expressed independent of the FBS concentration (data not shown). To investigate changes in tight junction proteins induced by FBS, Western blotting and real-time PCR were performed. In Western blotting, claudin-4 and -7 and JAM-A were increased by 1% FBS, claudin-1, occludin, and ZO-1 were increased by 5% FBS, and ZO-2 was increased by 10% FBS (Figure 2C). In real-time PCR, mRNAs of claudin-4 and occludin were increased in 10% FBS-treated cells 107.1- and 84.9-fold, respectively, compared with the control, whereas mRNAs of claudin-1 and -7, JAM-A, ZO-1, and ZO-2 were increased in 10% FBS-treated cells 3.9-, 5.3-, 2.5-, 2.8-, and 4.8-fold, respectively, compared with the control (Figure 2D).
Figure 2.
Western blotting (A) and RT-PCR (B) for CA-II in hTERT-HPDE cells at 24 hours after treatment with 1, 5, and 10% FBS. Protein and mRNA of CA-II increased depending on the FBS concentration. C: Western blotting for claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 in hTERT-HPDE cells at 24 hours after treatment with 1, 5, and 10% FBS. Proteins of claudin-4 and -7 and JAM-A increased from 1% FBS, claudin-1, occludin, and ZO-1 increased from 5% FBS and ZO-2 increased at 10% FBS. D: Real-time PCR for claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 in hTERT-HPDE cells at 24 hours after treatment with 10% FBS. mRNAs of claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 increased in FBS-treated cells 3.9-, 107.1-, 5.3-, 84.9-, 2.5-, 2.8- and 4.8-fold, respectively, compared with the control. CL, claudin; Oc, occludin.
Localization of Tight Junction Proteins, Formation of Tight Junction Strands, and Up-Regulation of Barrier Function by FBS in hTERT-HPDE Cells
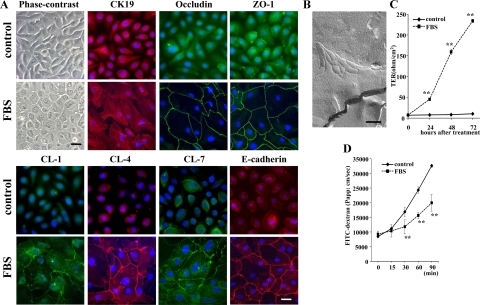
In 10% FBS-treated cells, the cells were changed from a cobblestone appearance to a round shape (Figure 3A). With immunocytochemical analysis, 10% FBS-treated cells expressed CK19 as in the control. In 10% FBS-treated cells, lines of the tight junction proteins occludin, ZO-1, and claudin-1, -4, and -7 were observed at cell borders as well as adherens junction protein E-cadherin, whereas in the control they were not detected at cell borders (Figure 3A). In freeze-fracture replicas of 10% FBS-treated cells, a network composed of several continuous tight junction strands was observed at subapical membranes (Figure 3B). However, in the control, tight junction strands were not detected (data not shown).
Figure 3.
A: Phase-contrast images and immunocytochemical analysis for CK19, occludin, ZO-1, claudin-1, -4, and -7, and E-cadherin in hTERT-HPDE cells at 24 hours after treatment with 10% FBS compared with the control. In FBS-treated cells, the cytoplasm is increased compared with the control and CK19 is expressed as in the control. Lines of occludin, ZO-1, claudin-1, -4, and -7, and E-cadherin are observed at cell borders in FBS-treated cells, whereas they are not detected at cell borders in the control. Black bar = 40 μm; White bar = 20 μm. B: Freeze-fracture replica of hTERT-HPDE cells at 24 hours after treatment with 10% FBS. A network composed of several continuous tight junction strands is observed at subapical membranes. Scale bar = 200 nm. Barrier function measured as TER (C) and paracellular flux using FITC-dextran (D) in hTERT-HPDE cells after treatment with 10% FBS. The value of TER gradually increased until 72 hours after treatment with 10% FBS, whereas in the control, it was still very low at 72 hours. The paracellular flux of FITC-dextran was significantly decreased at 24 hours after treatment with 10% FBS. CL, claudin. **P < 0.01.
To investigate the effects of FBS on the tight junction barrier function of hTERT-HPDE cells, the cells at day 5 after plating were examined for TER and the paracellular flux of FITC-labeled dextran after treatment with 10% FBS. In the control, the value of TER was still very low at 72 hours (Figure 3C). In 10% FBS-treated cells, the value of TER gradually increased and reached 242.3 ± 143.7 Ω/cm2 at 72 hours after treatment (Figure 3C).
When the paracellular flux of FITC-labeled dextran was measured in the cells at 24 hours after treatment with 10% FBS, in which the TER was significantly increased compared with the control, it was significantly decreased compared with the control (Figure 3D).
Pan-PKC Inhibitor Prevents Up-Regulation of Tight Junction Proteins and Barrier Function by FBS in hTERT-HPDE Cells
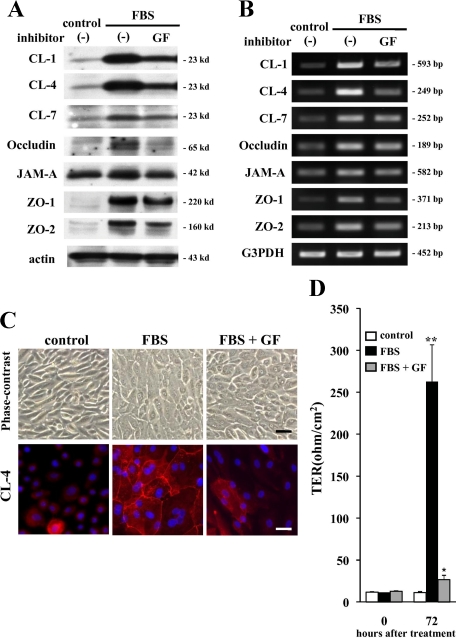
To investigate the mechanisms of induction of tight junctions, we focused on signal transduction pathways and used inhibitors of pan-PKC, MAPK, p38 MAPK, phosphatidylinositol 3-kinase and nuclear factor-κB (GF109203X, PD98059, SB203580, LY294002, and IMD-0354, respectively). The up-regulation of proteins and mRNAs of claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 by 10% FBS was inhibited by GF109203X (Figure 4, A and B), whereas no effect was seen with PD98059, SB203580, LY294002, and IMD-0354 (Supplemental Figure S1, see http://ajp.amjpathol.org). In immunocytochemical analysis, GF109203X inhibited the expression of claudin-4 at cell borders induced by 10% FBS (Figure 4C). Furthermore, GF109203X inhibited up-regulation of barrier function by 10% FBS measured as TER (Figure 4D). These results obtained using inhibitors of signal transduction suggested that induction of tight junctions by FBS in hTERT-HPDE cells was, in part, regulated via a PKC signal pathway.
Figure 4.
Western blotting (A) and RT-PCR (B) for claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 in hTERT-HPDE cells pretreated with the pan-PKC inhibitor GF109203X (GF) before treatment with FBS. Up-regulation of proteins and mRNAs of claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 at 24 hours after treatment with 10% FBS was inhibited by GF109203X. C: Phase-contrast images and immunocytochemical analysis for caludin-4 in hTERT-HPDE cells pretreated with the pan-PKC inhibitor GF109203X before treatment with FBS. GF109203X inhibits the expression of claudin-4 at cell borders at 24 hours after treatment with 10% FBS. Black bar = 80 μm; white bar = 20 μm. D: Barrier function measured as TER in hTERT-HPDE cells pretreated with the pan-PKC inhibitor GF109203X before treatment with FBS. GF109203X inhibited the increase of TER at 72 hours after treatment with 10% FBS. CL, claudin. *P < 0.05, **P < 0.01.
Up-Regulation of Tight Junction Proteins at the Transcriptional Level by PKC Activator TPA in hTERT-HPDE Cells
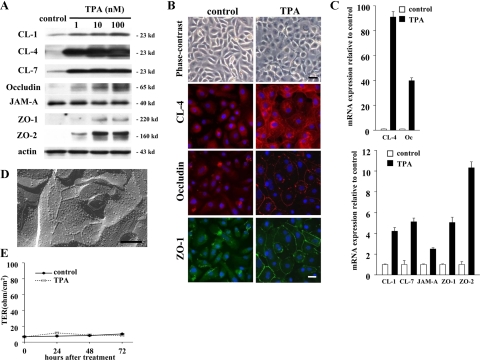
To further evaluate whether a PKC signal pathway was associated with the regulation of tight junctions in hTERT-HPDE cells, we used the PKC activator TPA. In Western blotting, proteins of claudin-1, -4, and -7, occludin, and ZO-2 were increased from 1 nmol/L TPA and ZO-1 protein was increased from 10 nmol/L TPA, whereas no change of JAM-A protein was observed (Figure 5A).
Figure 5.
A: Western blotting for claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 in hTERT-HPDE cells at 24 hours after treatment with 1, 10, and 100 nmol/L TPA. Proteins of claudin-1, -4 and -7, occludin, and ZO-2 increased from 1 nmol/L TPA and ZO-1 protein increased from 10 nmol/L TPA, whereas no change of JAM-A protein was observed. B: Phase-contrast images and immunocytochemical analysis for claudin-4, occludin, and ZO-1 in hTERT-HPDE cells at 24 hours after treatment with 10 nmol/L TPA compared with the control. TPA-treated cells change from a cobblestone appearance to a round shape. In TPA-treated cells, continuous lines of claudin-4 and discontinuous lines of occludin and ZO-1 are observed at cell borders. Black bar = 80 μm; white bar = 20 μm. C: Real-time PCR for claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 in hTERT-HPDE cells at 24 hours after treatment with 100 nmol/L TPA. mRNAs for claudin-1, -4, and -7, occludin, JAM-A, ZO-1, and ZO-2 increased in TPA-treated cells 4.2-, 91.0-, 5.1-, 39.9-, 2.5-, 5.0- and 10.3-fold, respectively, compared with the control. D: Freeze-fracture replica of hTERT-HPDE cells at 24 hours after treatment with 100 nmol/L TPA. Small networks of tight junction strands are observed. Scale bar = 200 nm. E: Barrier function measured as TER in hTERT-HPDE cells after treatment with 100 nmol/L TPA. CL, claudin; Oc, occludin.
In phase-contrast images, the cells were changed from a cobblestone appearance to a round shape by 10 nmol/L TPA (Figure 5B). In immunocytochemical analysis of 10 nmol/L TPA-treated cells, continuous lines of claudin-4 and discontinuous lines of occludin and ZO-1 were observed at cell borders (Figure 5B).
In real-time PCR, mRNAs of claudin-4 and occludin were increased in 100 nmol/L TPA-treated cells 91.0- and 39.9-fold, respectively, compared with the control, and mRNAs of claudin-1 and -7, JAM-A, ZO-1, and ZO-2 were also increased in 100 nmol/L TPA-treated cells 4.2-, 5.1-, 2.5-, 5.0-, and 10.3-fold, respectively, compared with the control (Figure 5C).
In freeze-fracture replicas of 100 nmol/L TPA-treated cells, fragments of tight junction strands and small networks were observed (Figure 5D).
To investigate the effects of TPA on the tight junction barrier function of hTERT-HPDE cells, the cells at day 5 after plating were examined for TER after treatment with 100 nmol/L TPA. In these cells, the value of TER was still very low at 72 hours, similar to the control (Figure 5E).
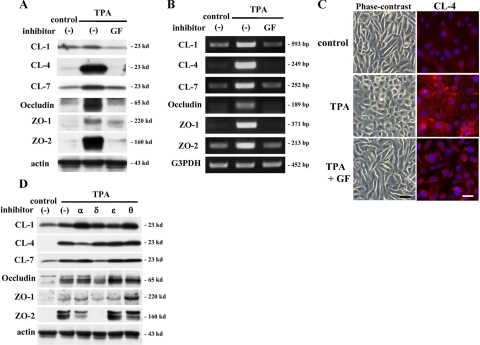
Pan-PKC Inhibitor Prevents Up-Regulation of Tight Junction Proteins by TPA in hTERT-HPDE Cells
To confirm whether the up-regulation of tight junction proteins by TPA was induced via a PKC signal pathway, the cells were pretreated with the pan-PKC inhibitor GF109203X before treatment with 100 nmol/L TPA. In Western blotting and RT-PCR, GF109203X completely inhibited up-regulation of proteins and mRNAs of claudin-1, -4, and -7, occludin, ZO-1, and ZO-2 by 100 nmol/L TPA (Figure 6, A and B). In phase-contrast images, GF109203X prevented TPA-treated cells from becoming round, and they retained a cobblestone appearance similar to the control (Figure 6C). In immunocytochemical analysis, lines of claudin-4 at cell borders in TPA-treated cells disappeared after treatment with GF109203X (Figure 6C).
Figure 6.
Western blotting (A) and RT-PCR (B) for claudin-1, -4, and -7, occludin, ZO-1, and ZO-2 in hTERT-HPDE cells pretreated with the pan-PKC inhibitor GF109203X (GF) before treatment with TPA. Up-regulation of proteins and mRNAs of claudin-1, -4, and -7, occludin, ZO-1, and ZO-2 at 24 hours after treatment with 100 nmol/L TPA was completely inhibited by GF109203X. C: Phase-contrast images and immunocytochemical analysis for claudin-4 in hTERT-HPDE cells pretreated with the pan-PKC inhibitor GF109203X before treatment with TPA. GF109203X prevents the change to round cell shape and the expression of claudin-4 at cell borders at 24 hours after treatment with 100 nmol/L TPA. Black bar = 80 μm; white bar = 20 μm. D: Western blotting for claudin-1, -4, and -7, occludin, ZO-1, and ZO-2 in hTERT-HPDE cells pretreated with specific isoform inhibitors of PKC-α, PKC-δ, PKC-θ, and PKC-ε before treatment with 100 nmol/L TPA. Treatment with the PKC-α inhibitor prevented up-regulation of claudin-4 by TPA. Treatment with the PKC-δ inhibitor prevented up-regulation of claudin-7, occludin, ZO-1, and ZO-2 by TPA. CL, claudin.
Inhibitors of Specific PKC Isoforms Prevent Up-Regulation of Tight Junction Proteins by TPA in hTERT-HPDE Cells
The classic and novel isoforms of PKC respond to TPA. To investigate which PKC isoforms play key roles in the up-regulation of tight junction proteins by TPA, the cells were pretreated with the PKC-α inhibitor Gö6976, PKC-δ inhibitor rottlerin, PKC-θ inhibitor myristoylated PKC-θ pseudosubstrate peptide inhibitor, and PKC-ε inhibitor PKC-ε translocation inhibitor peptide before treatment with 100 nmol/L TPA. Treatment with the PKC-α inhibitor prevented up-regulation of claudin-4 by 100 nmol/L TPA (Figure 6D). Furthermore, treatment with the PKC-δ inhibitor prevented up-regulation of claudin-7, occludin, ZO-1, and ZO-2 by 100 nmol/L TPA (Figure 6D). The up-regulation of claudin-1 by 100 nmol/L TPA was not affected by any inhibitor (Figure 6D).
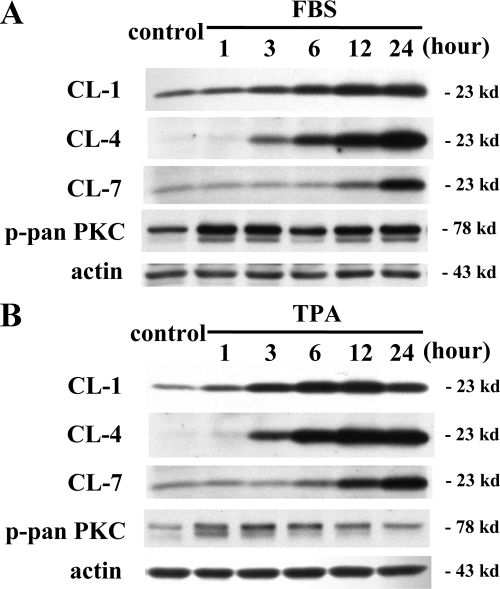
Different Expression Patterns of Claudin-1, -4, and -7 in a Time-Dependent Manner after Treatment with FBS or TPA in hTERT-HPDE Cells
It is thought that claudins are regulated by various factors and that there is differential regulation among claudin family members.9,31,32 Therefore we investigated the time-dependent changes in proteins of claudin-1, -4, and -7 in hTERT-HPDE cells after treatment with 10% FBS or 100 nmol/L TPA. Interestingly, with both treatments, claudin-1 was increased from 1 hour, claudin-4 was increased from 3 hours, and claudin-7 was increased from 12 hours, whereas phospho-pan-PKC was increased from 1 hour (Figure 7, A and B).
Figure 7.
Western blotting for claudin-1, -4, and -7 and phospho-pan-PKC in hTERT-HPDE cells at 1, 3, 6, 12, and 24 hours after treatment with 10% FBS (A) or 100 nmol/L TPA (B). In both FBS- and TPA-treated cells, protein of claudin-1 increased from one hour, claudin-4 increased from three hours, and claudin-7 increased from 12 hours, whereas phospho-pan-PKC increased from one hour. CL, claudin; p-pan PKC, phospho-pan PKC.
Gene Expression Changes Induced by FBS or TPA in hTERT-HPDE Cells
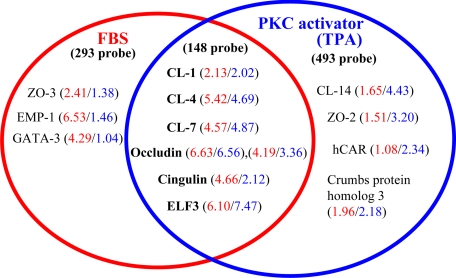
In real-time PCR analysis in the present study, mRNAs of claudin-4 and occludin were markedly increased in hTERT-HPDE cells after treatment with 10% FBS or 100 nmol/L TPA. To investigate the candidate transcription factors associated with the regulation of the expression of tight junction molecules, including claudin-4 and occludin, we performed GeneChip analysis, comparing the cells treated with 10% FBS or 100 nmol/L TPA and the controls.
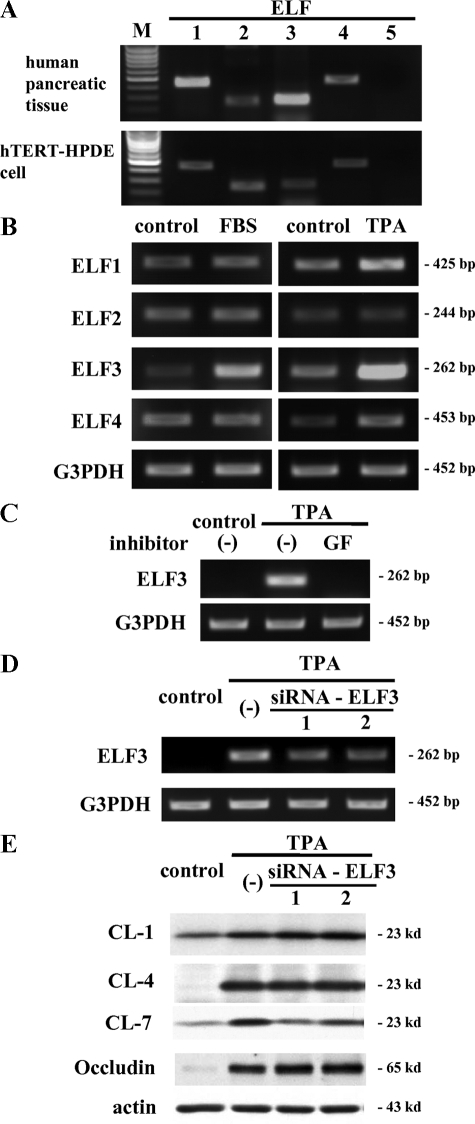
In this GeneChip analysis, we selected gene probes that were up-regulated more than twofold compared with the controls. Altogether 293 probes were up-regulated only in FBS-treated cells, 493 probes were up-regulated only in TPA-treated cells, and 148 probes were up-regulated in both FBS- and TPA-treated cells (Supplemental Table S1, see http://ajp.amjpathol.org). In both FBS- and TPA-treated cells, up-regulation of tight junction molecules claudin-1, -4, and -7, occludin and cingulin was confirmed, whereas ZO-3 and EMP-1 were detected in only FBS-treated cells, and claudin-14, ZO-2, hCAR, and crumbs protein homolog 3 were detected in only TPA-treated cells (Figure 8). Furthermore, in the transcription factors associated with the regulation of the expression of tight junction molecules, up-regulation of ELF3 in both FBS- and TPA-treated cells was detected (Figure 8).
Figure 8.
GeneChip analysis of hTERT-HPDE cells at 24 hours after treatment with 10% FBS or 100 nmol/L TPA. Up-regulated gene probes associated with tight junction molecules are shown in the Venn diagram. The 148 probes that were up-regulated in both FBS- and TPA-treated cells contained claudin-1, -4 and -7, occludin, cingulin, and ELF3. The 293 probes that were up-regulated only in FBS-treated cells contained ZO-3 and EMP-1, and the 493 probes that were up-regulated only in TPA-treated cells contained claudin-14, ZO-2, hCAR, and crumbs protein homolog 3. The fold changes of each probe are depicted in red for FBS-treated cells and in blue for TPA-treated cells. CL, claudin.
Possible Regulation of Claudin-7 via ELF3 in hTERT-HPDE Cells
More recently, it was reported that the expression of claudin-7 at epithelial structures in synovial sarcoma was regulated by transcription factor ELF3.33 In the GeneChip analysis of the present study, up-regulation of ELF3 was observed in both FBS- and TPA-treated hTERT-HPDE cells. We investigated the mRNA expression patterns of ELF family members ELF1, ELF2, ELF3, ELF4, and ELF5 in hTERT-HPDE cells compared with human pancreatic tissues. In RT-PCR, mRNAs of ELF1, ELF2, ELF3, and ELF4, but not ELF5, were detected in both human pancreatic tissues and hTERT-HPDE cells (Figure 9A). In both 10% FBS- and 100 nmol/L TPA-treated cells, ELF3 mRNA was increased (Figure 9B). Furthermore, the pan-PKC inhibitor GF109203X prevented up-regulation of ELF3 mRNA by treatment with 100 nmol/L TPA (Figure 9C).
Figure 9.
A: RT-PCR for ELF1–5 in hTERT-HPDE cells. mRNAs of ELF1, ELF2, ELF3, and ELF4, but not ELF5, are detected in both hTERT-HPDE cells and human pancreatic tissues. B: RT-PCR for ELF1, ELF2, ELF3, and ELF4 in hTERT-HPDE cells at 24 hours after treatment with 10% FBS or 100 nmol/L TPA. mRNA of ELF3 increased in both FBS- and TPA-treated cells compared with the control. C: RT-PCR for ELF3 in hTERT-HPDE cells pretreated with the pan-PKC inhibitor GF109203X (GF) before treatment with TPA. Up-regulation of mRNA of ELF3 at 24 hours after treatment with 100 nmol/L TPA was inhibited by GF109203X. D: RT-PCR for ELF3 in hTERT-HPDE cells pretreated with two siRNAs of ELF3 before treatment with TPA. Up-regulation of mRNA of ELF3 by 10 nmol/L TPA was inhibited by two siRNAs of ELF3. E: Western blotting for claudin-1, -4, and -7 and occludin in hTERT-HPDE cells pretreated with two siRNAs of ELF3 before treatment with TPA. Up-regulation of protein of claudin-7, but not claudin-1 or -4, and occludin by 10 nmol/L TPA was inhibited by two siRNAs of ELF3. CL, claudin; M, 100-bp ladder DNA marker.
To investigate whether ELF3 directly regulated PKC-mediated tight junction molecules in hTERT-HPDE cells, we used two siRNAs of ELF3. In RT-PCR, up-regulation of ELF3 mRNA by treatment with 10 nmol/L TPA was inhibited by the two siRNAs of ELF3 (Figure 9D). In Western blotting, the two siRNAs of ELF3 prevented up-regulation of protein of claudin-7 but not claudin-1 or -4, and occludin by treatment with 10 nmol/L TPA (Figure 9E).
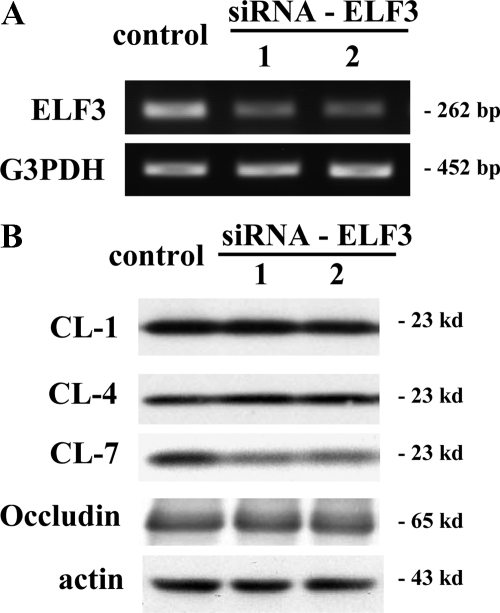
Possible Regulation of Claudin-7 via ELF3 in HPAC Cells
To investigate whether ELF3 also regulated expression of tight junction molecules in the human pancreatic cancer cell line HPAC, we used two siRNAs of ELF3. In RT-PCR, ELF3 mRNA in HPAC cells was decreased by the two siRNAs of ELF3 (Figure 10A). In Western blotting, claudin-1, -4, and -7 and occludin proteins were strongly expressed in HPAC cells, and the two siRNAs of ELF3 prevented expression of protein of claudin-7 but not of claudin-1 or -4 and occludin (Figure 10B).
Figure 10.
A: RT-PCR for ELF3 in pancreatic cancer cell line HPAC at 48 hours after treatment with two siRNAs of ELF3. mRNA of ELF3 was detected in the control and decreased by the two siRNAs of ELF3. B: Western blotting for claudin-1, -4, and -7 and occludin in HPAC cells at 48 hours after treatment with the two siRNAs of ELF3. Proteins of claudin-1, -4, and -7 and occludin were detected in the control and protein of claudin-7, but not claudin-1 or -4, and occludin was decreased by the two siRNAs of ELF3. CL, claudin.
Discussion
In the present study, by using hTERT-HPDE cells that showed properties similar to those of HPDE cells in vivo, we first demonstrated that tight junctions of normal HPDE cells were at least in part regulated via a PKC signal pathway by transcriptional control. Tight junction proteins are elaborately regulated via distinct signal transduction pathways.9 To investigate the mechanisms of induction of tight junctions by FBS in hTERT-HPDE cells, we used various signal transduction inhibitors. The up-regulation of tight junction proteins and barrier function by FBS was inhibited by a pan-PKC inhibitor but not by inhibitors of MAPK, p38 MAPK, phosphatidylinositol 3-kinase, or nuclear factor-κB (Figure 4, Supplemental Figure S1, see http://ajp.amjpathol.org).
PKC is a family of serine-threonine kinases known to regulate epithelial barrier function via tight junctions.46,47 PKC has been shown to induce both assembly and disassembly of tight junctions depending on the cell type and conditions of activation.28,30,47 To confirm whether the PKC signal pathway was closely associated with the regulation of tight junctions in HPDE cells, hTERT-HPDE cells were treated with the PKC activator TPA. Treatment with TPA enhanced expression of most tight junction proteins with a marked increase of claudin-4 and occludin mRNAs, and the changes were similar to those of FBS-treated cells. The up-regulation of tight junction proteins by TPA was inhibited completely by the pan-PKC inhibitor. These findings indicated that expression of most tight junction proteins and the barrier function in HPDE cells were up-regulated via a PKC signal pathway. Furthermore, because claudin-4 expression in normal HPDE cells was enhanced by PKC activators in the present study, these results should help to develop safer and more effective diagnostic and therapeutic methods targeting claudin-4 in pancreatic cancer.
It is known that there is differential regulation of tight junction molecules, including claudins.9,48,49 PKC seems to regulate the subcellular localization, phosphorylation states, and transcription of several tight junction-associated proteins,24,31,32 although the PKC isozyme specificity has not been clearly elucidated.32 PKC-α participates in tight junction disassembly, whereas PKC-ε plays a role in tight junction formation of kidney epithelial cells.47 In the human intestinal epithelial cell lines HT-29 and Caco-2, stimulation with Toll-like receptor 2 ligands leads to activation of PKC-α and PKC-δ and enhances barrier function through translocation of ZO-1 on activation.34 In human nasal primary cells, treatment with inhibitors of PKC-δ and PKC-θ prevents the up-regulation of tight junction proteins by TPA.50
In the present study, mRNAs of claudin-4 and occludin were markedly increased in both FBS- and TPA-treated hTERT-HPDE cells compared with the other tight junction molecules. The proteins of the claudin family were up-regulated in the order claudin-1, -4, and -7 after treatment with FBS or TPA in a time-dependent manner. Treatment with the PKC-α inhibitor prevented up-regulation of claudin-4 by TPA. Furthermore, treatment with the PKC-δ inhibitor prevented up-regulation of claudin-7, occludin, ZO-1, and ZO-2 by TPA, whereas the up-regulation of claudin-1 by TPA was not prevented by inhibitors of PKC-α, PKC-δ, PKC-θ, and PKC-ε. These results suggested that there was differential regulation in tight junction molecules of HPDE cells and that the pathways of PKC-α and PKC-δ were also important in the expression of tight junctions of HPDE cells.
When we selected gene probes that were up-regulated more than twofold compared with the control by GeneChip analysis, the transcription factor ELF3 was detected in both FBS- and TPA-treated cells. ELF3 belongs to the ELF (E74-like factor) subfamily of the ETS transcription factors, but it is distinguished from most ETS family members by its expression pattern, which is specific in epithelial tissues of the lung, liver, kidney, pancreas, prostate, small intestine, and colon mucosa.51 ELF3 controls intestinal epithelial differentiation.52 It is reported that the expression of claudin-7 at epithelial structures in synovial sarcoma is regulated by ELF3.53 Consistently, we demonstrated that ELF3 mRNA was increased in FBS- or TPA-treated cells and a pan-PKC inhibitor prevented up-regulation of ELF3 mRNA by TPA treatment. When knockdown of ELF3 was caused by siRNAs in hTERT-HPDE cells, up-regulation of claudin-7 by TPA was inhibited.
Claudin-7 is expressed in various types of epithelial cells and directly interacts with EpCAM, forming a complex with CD44 variant isoforms and tetraspanins outside of tight junction areas.54,55 Furthermore, EpCAM is one of the surface markers in pancreatic cancer stem cells,56 and claudin-7 regulates the EpCAM-mediated functions in tumor progression, including those in pancreatic cancer, such as proliferation, migration, and anti-apoptosis.57,58 In the present study, in the human pancreatic cancer cell line HPAC, ELF3 mRNA and claudin-1, -4, and -7 and occludin proteins were strongly expressed. When ELF3 mRNA in HPAC cells was decreased by treatment with siRNAs of ELF3, protein expression of claudin-7 but not of claudin-1 or -4 and occludin was decreased. These findings suggested that ELF3 was associated with claudin-7 not only in normal HPDE cells but also in pancreatic cancer cells. Thus, the expression of claudin-7 and its regulation via ELF3 may be important as potential therapeutic targets for pancreatic cancer.
Although PKC exerts its effect directly at the transcriptional level, the transcription factors related to PKC activation in regulation of tight junctions remain unknown. For example, in primary human distal lung epithelial cells, mRNA of claudin-4 is increased by TPA treatment, depending on PKC activation.59 The activation of PKC by TPA causes increases in transcription of occludin, ZO-1, and claudin-1 in T84 cells35 and melanoma cells.36
In the present study, when we selected gene probes that were shown to be up-regulated more than twofold compared with the control by GeneChip analysis, the transcription factor GATA-3 was also detected in FBS-treated hTERT-HPDE cells (Figure 8). The GATA family, included in zinc finger transcription factors, has been recognized in the differentiation of the endoderm in several evolutionarily diverse organisms, and it is in part mediated via a PKC pathway.60,61 In primary human nasal epithelial cells, claudin-1, occludin, ZO-1, and ZO-2 are up-regulated at the transcriptional level by TPA and the transcription factor GATA-3 up-regulates ZO-1 and ZO-2 via a PKC signal pathway.50
In the present study, we investigated the mRNA expression patterns of GATA family members GATA-1-6 in hTERT-HPDE cells. In RT-PCR, mRNAs of GATA-3 and GATA-6 were detected in hTERT-HPDE cells and GATA-3 mRNA was increased after treatment with 10% FBS compared with the control (Supplemental Figure S2, see http://ajp.amjpathol.org). Furthermore, in Western blotting, knockdown of GATA-3 by siRNAs inhibited up-regulation of ZO-2 by treatment with 10% FBS (Supplemental Figure S2, see http://ajp.amjpathol.org). These results suggest that in HPDE cells, GATA-3 may be one of the transcription factors related to PKC activation in regulation of tight junctions, although it regulates only ZO-2 in part.
In conclusion, using hTERT-HPDE cells, we found that the expression of tight junction molecules, including claudins, and barrier function in normal HPDE cells were regulated via a PKC signal pathway. The hTERT-HPDE cells had good growth potential and a longer lifespan. These cells could be passaged four times at the maximum but were not immortalized. Investigation of the regulation of claudins via the PKC signal pathway in normal HPDE cells is essential for their use as biomarkers for diagnosis and molecular therapeutic targets in pancreatic cancer. The hTERT-HPDE cells in this culture system provide us with an indispensable and stable model for studying the regulation of tight junctions in normal HPDE cells.
Acknowledgments
We thank Ms. Emiko Suzuki (Sapporo Medical University) for technical support.
Footnotes
Address reprint requests to Takashi Kojima, Ph.D., Department of Pathology, Sapporo Medical University School of Medicine, S1, W17, Sapporo 060-8556, Japan. E-mail: ktakashi@sapmed.ac.jp.
Supported by the Pancreas Research Foundation of Japan and by grants-in-aid from the National Project “Knowledge Cluster Initiative” (second stage, “Sapporo Biocluster Bio-S”) program for developing a supporting system for upgrading education and research, the Ministry of Education, Culture, Sports, Science, and Technology, and the Ministry of Health, Labour and Welfare of Japan.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–9606. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Yamazaki Y, Katsuno T, Tamura A, Tsukita S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene. 2008;27:6930–6938. doi: 10.1038/onc.2008.344. [DOI] [PubMed] [Google Scholar]
- van Meer G, Simons K. The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J. 1986;5:1455–1464. doi: 10.1002/j.1460-2075.1986.tb04382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido M, Valdés J, Shoshani L, Contreras RG. Role of tight junctions in establishing and maintaining cell polarity. Annu Rev Physiol. 1998;60:161–177. doi: 10.1146/annurev.physiol.60.1.161. [DOI] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. Structure, function, and regulation of cellular tight junctions. Am J Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Breaking through the tight junction barrier. J Cell Biol. 1993;123:1631–1633. doi: 10.1083/jcb.123.6.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–237. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- Kojima T, Murata M, Yamamoto T, Lan M, Imamura M, Son S, Takano K, Yamaguchi H, Ito T, Tanaka S, Chiba H, Hirata K, Sawada N. Tight junction proteins and signal transduction pathways in hepatocytes. Histol Histopathol. 2009;24:1463–1472. doi: 10.14670/HH-24.1463. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol. 2005;171:939–945. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tsukita S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000;476:258–261. doi: 10.1016/s0014-5793(00)01744-0. [DOI] [PubMed] [Google Scholar]
- Michl P, Buchholz M, Rolke M, Kunsch S, Löhr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM. Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology. 2001;121:678–684. doi: 10.1053/gast.2001.27124. [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Illei PB, Ashfaq R, Infante JR, Murphy K, Pandey A, Schulick R, Winter J, Sharma R, Maitra A, Goggins M, Hruban RH. New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am J Surg Pathol. 2008;32:188–196. doi: 10.1097/PAS.0b013e31815701f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Koslowski M, Dhaene K, Usener D, Brandenburg G, Seitz G, Huber C, Türeci Ö. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res. 2008;14:7624–7634. doi: 10.1158/1078-0432.CCR-08-1547. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kato-Nakano M, Kawamoto S, Furuya A, Abe Y, Misaka H, Kimoto N, Nakamura K, Ohta S, Ando H. Therapeutic antitumor efficacy of monoclonal antibody against claudin-4 for pancreatic and ovarian cancers. Cancer Sci. 2009;100:1623–1630. doi: 10.1111/j.1349-7006.2009.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem. 1997;272:26652–26658. doi: 10.1074/jbc.272.42.26652. [DOI] [PubMed] [Google Scholar]
- Yuan X, Lin X, Manorek G, Kanatani I, Cheung LH, Rosenblum MG, Howell SB. Recombinant CPE fused to tumor necrosis factor targets human ovarian cancer cells expressing the claudin-3 and claudin-4 receptors. Mol Cancer Ther. 2009;8:1906–1915. doi: 10.1158/1535-7163.MCT-09-0106. [DOI] [PubMed] [Google Scholar]
- Saeki R, Kondoh M, Kakutani H, Tsunoda S, Mochizuki Y, Hamakubo T, Tsutsumi Y, Horiguchi Y, Yagi K. A novel tumor-targeted therapy using a claudin-4-targeting molecule. Mol Pharmacol. 2009;76:918–926. doi: 10.1124/mol.109.058412. [DOI] [PubMed] [Google Scholar]
- Andreeva AY, Krause E, Müller EC, Blasig IE, Utepbergenov DI. Protein kinase C regulates the phosphorylation and cellular localization of occludin. J Biol Chem. 2001;276:38480–38486. doi: 10.1074/jbc.M104923200. [DOI] [PubMed] [Google Scholar]
- Seth A, Sheth P, Elias BC, Rao R. Protein phosphatases 2A and 1 interact with occludin and negatively regulate the assembly of tight junctions in the CACO-2 cell monolayer. J Biol Chem. 2007;282:11487–11498. doi: 10.1074/jbc.M610597200. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Nigam SK. Role of tyrosine phosphorylation in the reassembly of occludin and other tight junction proteins. Am J Physiol. 1999;276:F737–F750. doi: 10.1152/ajprenal.1999.276.5.F737. [DOI] [PubMed] [Google Scholar]
- Stuart RO, Nigam SK. Regulated assembly of tight junctions by protein kinase C. Proc Natl Acad Sci USA. 1995;92:6072–6076. doi: 10.1073/pnas.92.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis B, Schneeberger EE, Rabito CA. Cellular variability in the development of tight junctions after activation of protein kinase C. Am J Physiol. 1992;263:F293–F300. doi: 10.1152/ajprenal.1992.263.2.F293. [DOI] [PubMed] [Google Scholar]
- Clarke H, Soler AP, Mullin JM. Protein kinase C activation leads to dephosphorylation of occludin and tight junction permeability increase in LLC-PK1 epithelial cell sheets. J Cell Sci. 2000;113:3187–3196. doi: 10.1242/jcs.113.18.3187. [DOI] [PubMed] [Google Scholar]
- Sjö A, Magnusson KE, Peterson KH. Distinct effects of protein kinase C on the barrier function at different developmental stages. Biosci Rep. 2003;23:87–102. doi: 10.1023/a:1025524323842. [DOI] [PubMed] [Google Scholar]
- Yoo J, Nichols A, Mammem J, Calvo I, Song JC, Worrell RT, Matlin K, Matthews JB. Bryostatin-1 enhances barrier function in T84 epithelia through PKC-dependent regulation of tight junction proteins. Am J Physiol Cell Physiol. 2003;285:C300–C309. doi: 10.1152/ajpcell.00267.2002. [DOI] [PubMed] [Google Scholar]
- Banan A, Zhang LJ, Shaikh M, Fields JZ, Choudhary S, Forsyth CB, Farhadi A, Keshavarzian A. θ isoform of protein kinase C alters barrier function in intestinal epithelium through modulation of distinct claudin isotypes: a novel mechanism for regulation of permeability. J Pharmacol Exp Ther. 2005;313:962–982. doi: 10.1124/jpet.104.083428. [DOI] [PubMed] [Google Scholar]
- Newton AC. Regulation of protein kinase C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology. 2004;127:224–238. doi: 10.1053/j.gastro.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Weiler F, Marbe T, Scheppach W, Schauber J. Influence of protein kinase C on transcription of the tight junction elements ZO-1 and occludin. J Cell Physiol. 2005;204:83–86. doi: 10.1002/jcp.20268. [DOI] [PubMed] [Google Scholar]
- Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, Dissanayake SK, Earley R, Indig FE, Nickoloff BJ, Taub DD, Kallioniemi OP, Meltzer P, Morin PJ, Weeraratna AT. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26:3846–3856. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- Jiang XR, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, Chiu CP. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun. 2003;301:1038–1044. doi: 10.1016/s0006-291x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- Kurose M, Kojima T, Koizumi J, Kamekura R, Ninomiya T, Murata M, Ichimiya S, Osanai M, Chiba H, Himi T, Sawada N. Induction of claudins in passaged hTERT-transfected human nasal epithelial cells with an extended life span. Cell Tissue Res. 2007;330:63–74. doi: 10.1007/s00441-007-0453-z. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kobune M, Yamaguchi M, Nakamura K, Ito Y, Sasaki K, Takahashi S, Nakamura T, Chiba H, Sato T, Matsunaga T, Azuma H, Ikebuchi K, Ikeda H, Kato J, Niitsu Y, Hamada H. Ex vivo expansion of human umbilical cord hematopoietic progenitor cells using a coculture system with human telomerase catalytic subunit (hTERT)-transfected human stromal cells. Blood. 2003;101:532–540. doi: 10.1182/blood-2002-04-1268. [DOI] [PubMed] [Google Scholar]
- Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186–193. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borka K, Kaliszky P, Szabó, Lotz G, Kupesulik P, Schaff Z, Kiss A. Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas. Virchows Arch. 2007;450:549–557. doi: 10.1007/s00428-007-0406-7. [DOI] [PubMed] [Google Scholar]
- Balda MS, Gonzalez-Mariscal L, Matter K, Cereijido M, Anderson JM. Assembly of the tight junction: the role of diacylglycerol. J Cell Biol. 1993;123:293–302. doi: 10.1083/jcb.123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AY, Piontek J, Blasig IE, Utepbergenov DI. Assembly of tight junction is regulated by the antagonism of conventional and novel protein kinase C isoforms. Int J Biochem Cell Biol. 2006;38:222–233. doi: 10.1016/j.biocel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life. 2009;61:431–437. doi: 10.1002/iub.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi J, Kojima T, Ogasawara N, Kamekura R, Kurose M, Go M, Harimaya A, Murata M, Osanai M, Chiba H, Himi T, Sawada N. Protein kinase C enhances tight junction barrier function of human nasal epithelial cells in primary culture by transcriptional regulation. Mol Pharmacol. 2008;74:432–442. doi: 10.1124/mol.107.043711. [DOI] [PubMed] [Google Scholar]
- Tymms MJ, Ng AY, Thomas RS, Schutte BC, Zhou J, Eyre HJ, Sutherland GR, Seth A, Rosenberg M, Papas T, Debouck C, Kola I. A novel epithelial-expressed ETS gene. ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q322 and expression in tissues and cancer. Oncogene. 1997;15:2449–2462. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Gutierrez-Hartmann A. Ets transcription factors in intestinal morphogenesis, homeostasis and disease. Histol Histopathol. 2008;23:1417–1424. doi: 10.14670/hh-23.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno Y, Okamoto T, Ishibe T, Nagayama S, Shima Y, Nishijo K, Shibata KR, Fukiage K, Otsuka S, Uejima D, Araki N, Naka N, Nakashima Y, Aoyama T, Nakayama T, Nakamura T, Toguchida J. Expression of claudin7 is tightly associated with epithelial structures in synovial sarcomas and regulated by an Ets family transcription factor. ELF3 J Biol Chem. 2006;281:38941–38950. doi: 10.1074/jbc.M608389200. [DOI] [PubMed] [Google Scholar]
- Ladwein M, Pape UF, Schmidt DS, Schnölzer M, Fiedler S, Langbein L, Franke WW, Moldenhauer G, Zöller M. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp Cell Res. 2005;309:345–357. doi: 10.1016/j.yexcr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Koch M, Nübel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, Weitz J, Zöller M. A complex of EpCAM, claudin-7. CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res. 2007;5:553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26:2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- Nübel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, Langbein L, Zöller M. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res. 2009;7:285–299. doi: 10.1158/1541-7786.MCR-08-0200. [DOI] [PubMed] [Google Scholar]
- Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- Wray C, Mao Y, Pan J, Chandrasena A, Piasta F, Frank JA. Claudin-4 augments alveolar epithelial barrier function and is induced in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2009;297:L219–L227. doi: 10.1152/ajplung.00043.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- Sugden PH. An overview of endothelin signaling in the cardiac myocyte. J Mol Cell Cardiol. 2003;35:871–886. doi: 10.1016/s0022-2828(03)00153-6. [DOI] [PubMed] [Google Scholar]